© Daniela Furrer Soliz Urrutia, 2017
The human epidermal growth factor receptor 2 (HER2) in
the breast cancer: from measurement to targeted
treatment
Thèse
Daniela Furrer Soliz Urrutia
Doctorat en médecine expérimentale
Philosophiæ doctor (Ph. D.)
The human epidermal growth factor receptor 2 (HER2) in
breast cancer: from measurement to targeted treatment
Thèse
Daniela Furrer Soliz Urrutia
Sous la direction de :
iii
Résumé
La surexpression du récepteur 2 du facteur de croissance épidermique humain (HER2) et/ou l’amplification du gène HER2 sont des facteurs prédictifs du cancer du sein. Avec l’introduction du traitement ciblé au trastuzumab, l’évaluation fiable d’HER2 est devenue essentielle. Malheureusement, jusqu’à 50% des patientes HER2-positives développent une résistance envers ce médicament.
Les objectifs étaient : 1) déterminer la façon la plus fiable et économique pour évaluer le statut HER2 (cohorte de 521 cas consécutifs de cancer du sein); 2) examiner l’association entre deux polymorphismes d’HER2 (Ile655Val et Ala1170Pro), la consommation de tabac et d’alcool et la réponse au trastuzumab (cohorte de 236 patientes HER2-positives traitées au trastuzumab). De plus, dans une étude pilote, nous avons examiné l’association entre les patrons de méthylation d’ADN dans la tumeur et la réponse au trastuzumab (cohorte de 12 patientes HER2-positives traitées au trastuzumab).
Le statut HER2 a été évalué par immunohistochimie (IHC), hybridation fluorescente in situ (FISH) et essai TaqMan. Nous avons comparé le statut HER2 déterminé par FISH sur lame complète (LC, un tissu par lame) et par matrice tissulaire (TMA, 60 tissus par lame), ainsi que le statut HER2 évalué par IHC et FISH sur le bloc ayant servi pour le diagnostic (bloc diagnostique) et sur un bloc choisi aléatoirement (bloc aléatoire). Les informations cliniques ont été obtenues dans les dossiers médicaux, celles sur la consommation de tabac et d’alcool par des questionnaires validés. Le patron de méthylation d’ADN a été évalué en utilisant la micropuce Illumina Infinium HumanMethylation450 BeadChip.
La concordance générale entre le statut HER2 déterminé par FISH sur LC et TMA était de 98,2%, et celle entre les blocs diagnostiques et aléatoires était de 98,0% au FISH et de 93,6% à l’IHC. La consommation de tabac et l’allèle Val étaient associés à une moins bonne réponse, tandis que la consommation d’alcool était associée à une meilleure réponse. Le patron de méthylation dans les tumeurs de patientes atteintes d’un cancer du sein HER2-positif qui ont développé une résistance au trastuzumab diffère de celui des patientes qui répondent au traitement. Cependant, ces résultats semblent dépendre de la méthode bioinformatique d’analyse utilisée.
Nous concluons que l’évaluation d’HER2 par FISH sur TMA représente une méthode fiable et économique. Les taux de concordances obtenus par FISH, mais pas ceux observés à
iv
l’IHC, satisfont l’exigence du Collège des pathologistes américains d’au moins 95% de concordance entre les résultats obtenus avec la méthode de référence et la nouvelle méthode. Le tabagisme, la consommation d’alcool et le polymorphisme HER2 Ile655Val pourraient influencer la réponse au traitement au trastuzumab.
v
Abstract
The overexpression of the human epidermal growth factor receptor 2 (HER2) and/or HER2 gene amplification are predictive factors in breast cancer. Following the HER2-targeted treatment with trastuzumab, the reliable evaluation of HER2 has become essential. Unfortunately, up to 50% of HER2-positive breast cancer patients develop resistance towards this drug.
The objectives were: 1). To determine the most reliable and economical method to evaluate HER2 status (cohort of 521 consecutive breast cancer cases); 2). To examine the association between tobacco and alcohol consumption, and two HER2 polymorphisms (Ile655Val and Ala1170Pro), and the response to trastuzumab (cohort of 236 HER2-positive breast cancer patients treated with trastuzumab). Moreover, in a pilot study, we explored the association between genome-wide DNA methylation patterns in breast cancer tissues and the response to trastuzumab (cohort of 12 breast cancer patients treated with trastuzumab). HER2 status was evaluated by immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), and TaqMan assay. We compared HER2 status determined by FISH on whole tissue (WT, one tissue per slide) section and tissue microarray (TMA, 60 tissues per slide) section, and HER2 status evaluated by IHC and FISH on the block used for diagnostic (diagnostic block) and on a randomly chosen additional block (random block). Clinicopathological information were assessed by review of medical records, tobacco and alcohol consumption by an administered validated questionnaire. DNA methylation patterns were evaluated using the Illumina Infinium HumanMethylation450 BeadChip.
Overall concordance between HER2 status determined by FISH on WT and TMA sections was 98.2% and that between diagnostic and random blocks was 98.0% for FISH and 93.6% for IHC. Tobacco consumption and the Val allele were associated with a worse response, whereas alcohol consumption was associated with a better response. Methylation pattern in tumor tissues of HER2-positive breast cancer patients who acquired resistance to trastuzumab treatment differed from that of HER2-positive breast cancer patients who responded to trastuzumab treatment. However, this observation seemed to depend upon the method of bioinformatics analysis used.
We conclude that FISH performed on TMA section represents a reliable and economical method for the evaluation of HER2. Results obtained by FISH, but not those obtained by
vi
IHC, fulfill the recommendations of the College of American Pathologists of concordance greater than 95% between the reference method and the new method. Tobacco use, alcohol consumption and Ile655Val HER2 polymorphism might influence the response to trastuzumab treatment.
vii
Table of contents
Résumé ... iii
Abstract ... v
Table of contents ... vii
Tables index ... xiii
Figures index ... xv
List of abbreviations ... xvi
Acknowledgments ... xxii
Foreword ... xxiv
Chapter 1: Introduction ... - 1 -
Literature overview ... - 2 -
1.1. Breast cancer intrinsic subtypes ... - 2 -
1.1.1. Molecular classification of breast cancer ... - 2 -
1.1.2. Molecular classification of breast cancer subtypes using immunohistochemical surrogates ... - 5 -
1.1.3. HER2-enriched subtype vs. breast cancer clinically evaluated as HER2-positive ... - 7 -
1.1.4. Breast cancer clinically evaluated as HER2-positive ... - 8 -
1.2. Methods for the evaluation of HER2 status in breast cancer specimens... - 9 -
1.2.1. Immunohistochemistry (IHC) and in situ hybridization (ISH) methods ... - 9 -
1.2.2. Methodological worries related to HER2 testing ... - 11 -
1.2.3. Concordance of HER2 status determined by IHC, FISH, CISH and SISH in the literature ... - 13 -
1.3. HER2 biology... - 14 -
1.3.1. HER2 structure and function ... - 14 -
1.3.2. Consequences of constitutive HER2 receptor activation... - 18 -
1.4. Treatment of HER2-positive breast cancer patients ... - 19 -
1.4.1. Anti-HER2 agents ... - 19 -
1.4.2. Endocrine therapy ... - 24 -
1.4.3. Chemotherapeutic drugs ... - 26 -
1.5. Resistance to trastuzumab ... - 27 -
1.5.1. Molecular mechanisms of trastuzumab resistance ... - 28 -
1.5.2. Genetic and epigenetic factors and survival of HER2-positive breast cancer patients ... - 34 -
1.5.2.1. Single nucleotide polymorphisms (SNPs) ... - 34 -
1.5.2.2. HER2 SNPs ... - 34 -
viii
1.5.3. Epigenetics ... - 35 -
1.5.3.1. DNA methylation ... - 37 -
1.5.3.2. Distribution of CpG dinucleotides and CpG methylation in the human genome ... - 39 -
1.5.3.3. Mechanisms of control of gene expression through DNA methylation ... - 40 -
1.5.3.4. Aberrant DNA methylation in cancer ... - 42 -
1.5.3.5. Link between DNA methylation and histone modifications ... - 43 -
1.5.3.6. Link between DNA methylation and miRNAs ... - 44 -
1.5.3.7. Methods for DNA methylation analysis ... - 46 -
1.5.3.8. Aberrant methylation in breast cancer ... - 50 -
1.5.3.9. DNA methylation signature in HER2-positive breast cancer ... - 51 -
1.5.3.10. Genome-wide DNA methylation analysis and association with survival in breast cancer patients ... - 52 -
1.5.3.11. Genome-wide DNA methylation analysis in breast cancer and response to chemotherapy- 53 - 1.5.3.12. DNA methylation in breast cancer tissues and trastuzumab response in HER2-positive breast cancer patients ... - 53 -
1.5.3.13. Epigenetic therapies for treatment of breast cancer patients ... - 54 -
1.5.3. Lifestyle factors and survival of HER2-positive breast cancer patients ... - 55 -
1.5.3.1. Tobacco and alcohol consumption ... - 55 -
1.5.3.2. Molecular and epidemiological ink between tobacco and alcohol exposure and HER2 ... - 56 -
Chapter 2: Advantages and disadvantages of technologies for HER2 testing in breast cancer specimens . - 58 - Résumé ... - 59 -
Abstract ... - 60 -
Introduction ... - 61 -
Principles of the methods of analysis, advantages and disadvantages of each described technique ... - 62 -
Southern blot ... - 62 -
Northern blot ... - 63 -
Enzyme-linked immunosorbent assay (ELISA) ... - 63 -
Western blot ... - 65 -
Polymerase chain reaction (PCR)-based assays ... - 65 -
Multiplex ligation-dependent probe amplification (MLPA) ... - 67 -
Immunohistochemistry (IHC) ... - 68 -
Fluorescence in situ hybridization (FISH) ... - 70 -
Bright-field in situ hybridization (BRISH) methods ... - 73 -
ix
Silver-enhanced in situ hybridization (SISH) ... - 76 -
Gold-facilitated autometallographic in situ hybridization (GOLDFISH) ... - 77 -
Bright-field double in situ hybridization (BDISH) ... - 77 -
HER2 gene-protein assay ... - 79 -
mRNA in situ hybridization ... - 80 -
Instant-quality fluorescence in situ hybridization (IQFISH) ... - 81 -
Automated HER2 FISH assay ... - 82 -
Performance of assays used for the HER2 status determination in breast cancer in predicting response to anti-HER2 therapies ... - 82 -
Conclusion ... - 83 -
Acknowledgements ... - 85 -
Conflict of interest ... - 85 -
References ... - 86 -
Chapter 3: Contextualisation, hypothesis and objectives ... - 106 -
3.1. Contextualisation ... - 106 -
3.2. Hypothesis ... - 106 -
3.3. Objectives ... - 108 -
Chapter 4: Tissue microarray is a reliable tool for the evaluation of HER2 amplification in breast cancer . - 110 - Résumé ... - 111 -
Abstract ... - 112 -
Introduction ... - 113 -
Materials and Methods ... - 114 -
Specimen collection and patient population ... - 114 -
Tissue microarray construction and processing ... - 114 -
Fluorescence in situ hybridization ... - 115 -
HER2 evaluation on TMA ... - 116 -
Statistical analysis ... - 116 -
Results ... - 116 -
Discussion ... - 117 -
Acknowledgements ... - 120 -
References ... - 121 -
Chapter 5: Concordance between immunohistochemistry and fluorescence in situ hybridization in the determination of human epidermal growth factor receptor 2 (HER2) status using tissue microarray in breast cancer specimens ... - 129 -
x
Résumé ... - 130 -
Abstract ... - 131 -
1. Introduction ... - 132 -
2. Materials and methods ... - 133 -
2.1. Specimen collection and patient population ... - 133 -
2.2. Tissue microarray (TMA) construction ... - 133 -
2.3. Immunohistochemistry ... - 134 -
2.4. Fluorescence in situ hybridization ... - 134 -
2.5. HER2 evaluation on TMA ... - 134 -
2.6. Statistical analysis ... - 135 -
3. Results ... - 135 -
4. Discussion ... - 137 -
5. Conclusions ... - 140 -
6. Conflict of interest and sources of funding... - 140 -
7. Acknowledgments ... - 141 -
References ... - 142 -
Chapter 6: Validation of a new classifier for the automated analysis of the human epidermal growth factor receptor 2 (HER2) gene amplification in breast cancer specimens ... - 152 -
Résumé ... - 153 -
Abstract ... - 154 -
Introduction ... - 155 -
Material and Methods ... - 156 -
Case selection ... - 156 -
Fluorescence In Situ Hybridization ... - 156 -
Manual scoring (reference method) ... - 156 -
Tile-sampling classifier ... - 157 -
Nuclei-sampling classifier ... - 158 -
Results ... - 159 -
Determination of the accuracy of the nuclei-sampling classifier on special specimens ... - 160 -
Determination of the accuracy of the nuclei-sampling classifier on equivocal specimens... - 161 -
Reproducibility of results ... - 161 -
Discussion ... - 161 -
xi List of abbreviations... - 167 - Competing interests... - 167 - Authors’ contributions ... - 167 - Acknowledgements ... - 167 - References ... - 168 -
Chapter 7: Evaluation of human epidermal growth factor receptor 2 (HER2) single nucleotide polymorphisms (SNPs) in normal and breast tumor tissues and their link with breast cancer prognostic factors ... - 174 -
Résumé ... - 175 -
Abstract ... - 176 -
Introduction ... - 177 -
Materials and Methods ... - 178 -
Study population and data collection... - 178 -
Ile655Val and Ala1170Pro polymorphisms ... - 178 -
Statistical analysis ... - 179 -
Results ... - 179 -
Discussion ... - 180 -
Conclusions ... - 182 -
Acknowledgments ... - 183 -
Conflict of interest statement ... - 183 -
Funding ... - 183 -
References ... - 183 -
Chapter 8: Association between tobacco and alcohol consumption, HER2 polymorphisms and response to trastuzumab in HER2-positive breast cancer patients ... - 193 -
Résumé ... - 194 -
Abstract ... - 195 -
Introduction ... - 196 -
Material and Methods ... - 197 -
Study population and data collection... - 197 -
Polymorphism substudy ... - 198 -
Study end points ... - 198 -
Statistical analysis ... - 198 -
Results ... - 199 -
HER2 polymorphism substudy ... - 200 -
xii
Conclusion ... - 204 -
Acknowledgments ... - 205 -
References ... - 206 -
Chapter 9: Association between genome-wide DNA methylation pattern and response to trastuzumab in HER2-positive breast cancer patients ... - 216 -
Résumé ... - 217 -
Abstract ... - 218 -
Introduction ... - 219 -
Material and Methods ... - 220 -
Study population ... - 220 - DNA extraction ... - 220 - Bioinformatic analysis ... - 221 - Statistical analysis ... - 223 - Results ... - 223 - Discussion ... - 225 - Acknowledgments ... - 226 - References ... - 227 -
Chapter 10 : Discussion and conclusion ... - 232 -
10.1. General discussion ... - 232 -
10.2. Perspectives ... - 243 -
10.3. Conclusion ... - 245 -
References ... - 247 -
xiii
Tables index
Table 1.1. Surrogate definition of intrinsic subtypes of breast cancer as defined by the 2011 St Gallen
International Expert Consensus Panel ... - 6 - Table 2.1. Main characteristics of the described techniques ... - 105 - Table 4.1. Interpretation criteria for fluorescence in situ hybridization according to the 2007 and the 2013 American Society of Clinical Oncology/College of American Pathologists scoring systems ... - 124 - Table 4.2. Concordance of human epidermal growth factor receptor 2 (HER2) gene amplification status between fluorescence in situ hybridization performed on whole tissue sections (reference method) and on diagnostic tissue microarray (TMA) sections according to the 2013 American Society of Clinical
Oncology/College of American Pathologists scoring system ... - 125 - Table 4.3. Concordance of human epidermal growth factor receptor 2 (HER2) gene amplification status between fluorescence in situ hybridization performed on whole tissue sections (reference method) and on diagnostic tissue microarray (TMA) sections according to the 2013 American Society of Clinical
Oncology/College of American Pathologists scoring system ... - 126 - Table 4.4. Concordance of human epidermal growth factor receptor 2 (HER2) gene amplification status between fluorescence in situ hybridization performed on whole tissue sections (reference method) and on diagnostic tissue microarray (TMA) sections according to the number of informative cores using the 2007 American Society of Clinical Oncology/College of American Pathologists scoring system ... - 127 - Table 4.5. Concordance of human epidermal growth factor receptor 2 (HER2) status determined by
fluorescence in situ hybridization on whole tissue section (reference method) and on random TMA section according to the 2013 ASCO/CAP scoring system ... - 128 - Table 5.1. Concordance of HER2 status determined by IHC and FISH on diagnostic TMA section according to the 2013 ASCO/CAP scoring system ... - 146 - Table 5.2. Concordance of HER2 status determined by IHC and FISH on random TMA section according to the 2013 ASCO/CAP scoring system ... - 147 - Table 5.3. Concordance of HER2 status determined by FISH on diagnostic TMA section and random TMA section according to the 2013 ASCO/CAP scoring system ... - 148 - Table 5.4. Concordance of HER2 status determined by IHC on diagnostic TMA section and random TMA section according to the 2013 ASCO/CAP scoring system ... - 149 - Table 5.A.1. Comparison of HER2 status determined by IHC and FISH on diagnostic TMA section according to the 2007 ASCO/CAP scoring system ... - 150 - Table 5.A.2. Concordance of HER2 status according to the combined IHC and FISH between diagnostic TMA section and random TMA section according to the 2013 ASCO/CAP scoring system ... - 151 - Table 6.1. Comparison of results obtained by different methods for non-amplified and amplified cases .... - 171 - Table 6.2. Comparison of results obtained by different methods for amplified cases without HSR ... - 171 - Table 6.3. Comparison of results obtained by different methods for equivocal cases ... - 171 - Table 7.1. Characteristics of the study population ... - 189 - Table 7.2. Association of HER2 Ile655Val and Ala1170Pro polymorphisms in normal and tumor breast tissues with prognostic factors ... - 190 - Table 7.3. Distribution of HER2 Ile655Val and Ala1170Pro polymorphisms in normal breast and tumor breast tissues ... - 192 - Table 8.1. Baseline characteristics of the study population ... - 211 -
xiv
Table 8.2. Unadjusted and adjusted hazard ratios for disease-free survival according to the tobacco
consumption before breast cancer diagnosis and during trastuzumab treatment ... - 212 - Table 8.3. Unadjusted and adjusted hazard ratios for disease-free survival according to the tobacco
consumption before breast cancer diagnosis and during trastuzumab treatment, stratified by ER status .. - 213 - Table 8.4. Unadjusted and adjusted hazard ratios for disease-free survival according to the alcohol
consumption before breast cancer diagnosis and during trastuzumab treatment ... - 214 - Table 8.5. Unadjusted and adjusted hazard ratios for disease-free survival according to the HER2
polymorphisms ... - 215 - Table 9.1. Baseline characteristics of cases and controls ... - 231 - Table 10.1. Impact of the 2013 ASCO/CAP scoring criteria on the classification of cases used for the validation of the software programming algorithm (results obtained at manual counting) ... - 236 - Table 10.2. Comparison of results obtained by different methods for non-amplified and amplified cases (HER2 gene amplification status according to the 2013 ASCO/CAP scoring criteria) ... - 237 - Table S1. Concordance of HER2 status : IHC vs. SISH, FISH vs. SISH, CISH vs. SISH ... - 299 -
xv
Figures index
Figure 1.1. Immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) results of HER2. ... - 10 -
Figure 1.2. Construction of Tissue microarray ... - 13 -
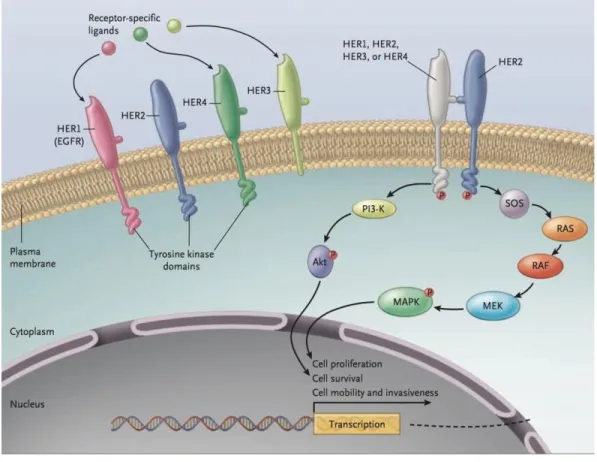
Figure 1.3. Signal transduction by the HER family members. ... - 17 -
Figure 1.4. Proposed mechanisms of action of trastuzumab ... - 21 -
Figure 1.5. Agents that target HER2 receptor. ... - 22 -
Figure 1.6. Proposed mechanisms of trastuzumab resistance ... - 33 -
Figure 1.7. Waddington’s classical epigenetic landscape. ... - 36 -
Figure 1.8. Conversion of cytosine to 5-methylcytosine by DNA methyltransferase (DNMT). ... - 38 -
Figure 1.9. Proposed mechanisms of silencing of CpG islands (CGI) promoters by DNA methylation. ... - 41 -
Figure 1.10. Overview of the Infinium I and Infinium II assays ... - 49 -
Figure 6.1. Image analysis of fluorescence signals ... - 172 -
xvi
List of abbreviations
ADCC antibody-dependent cellular cytotoxicity AI aromatase inhibitor
Ala Alanine
ASCO/CAP American Society of Clinical Oncology/College of American Pathologists ATP adenosine triphosphate
BDISH bright-filed double in situ hybridization
BMI body mass index
BER base excision repair
Cas9 CRISPR-associated protein 9 Cdk cyclin-dependent kinase
CEP17 chromosome enumeration probe 17
CGI CpG island
CI confidence interval
CIMP CpG island methylator phenotype CISH chromogenic in situ hybridization
CRISPR clustered regularly interspaced short palindromic repeat ddNTP dideoxynucleotide triphosphates
DFS disease-free survival DNMT DNA methyltransferase ECD extracellular domain
xvii EGF epidermal growth factor
EGFR epidermal growth factor receptor
EPG epigen
ER estrogen receptors
ERBB2 erb-b2 receptor tyrosine kinase 2 ERK extracellular signal-regulated kinase FcgR Fc gamma receptor
FDA Food and Drug Administration FDR false dicovery rate
FFPE formalin-fixed, paraffin-embedded FGFR fibroblast growth factor receptor FISH fluorescence in situ hybridization GEP gene expression profiling
HAT histone acetyltransferase HDAC histone deacetylase H&E hematoxylin & eosin
HER1 human epidermal growth factor receptor 1 HER2 human epidermal growth factor receptor 2 HER3 human epidermal growth factor receptor 3 HER4 human epidermal growth factor receptor 4 HMT histone methyltransferase
xviii
HPLC high performance liquid chromatography
HR hazard ratio
IAP intracisternal A-particles
IGF-1R insulin-like growth factor I receptor
IgG immunoglobulin G
Ile Isoleucine
IHC immunohistochemistry IR insulin receptor
LHRH luteinizing hormone-releasing hormone LINE long interspersed nuclear elements LVEF left ventricular ejection fraction
M methylated
MAPK mitogen-activated protein kinase MBD methyl-CpG binding domain MBP methyl-CpG-binding proteins
MCM2 mini-chromosome maintenance complex component 2 MGMT O6-methylguanine-DNA methyltransferase
MSP methylation-specific PCR MUCA Mucin 4
NGS next-generation sequencing NK Natural killer
xix
NNK 4-(methylnitrosamino)-1-3-(3-pyridyl)-1-butanon OS overall survival
PAM50 Prediction Analysis of Microarray 50 PCR polymerase chain reaction
PCR-RFLP polymerase chain reaction- restriction fragment length polymorphism PFS progression-free survival
PI3K phosphatidylinositol 3’kinase PR progesterone receptor
Pro Proline
PTEN phosphatase and tensin homolog deleted on chromosome 10 qRT-PCR quantitative reverse transcription polymerase chain reaction
RR response rate
RTK receptor tyrosine kinase SAM S-adenosyl-methionine
SERD selective estrogen receptor down-regulator SERM selective estrogen receptor modulator SINE short interspersed nuclear elements SISH silver-enhanced in situ hybridization SNP single nucleotide polymorphism TET ten-eleven translocation
xx TKI tyrosine kinase inhibitor
TMA tissue microarray TME tumor microenvironment TSG tumor suppressor gene TSS transcription start site
U unmethylated
Val Valine
VEGF vascular endothelial growth factor
VEGFR vascular endothelial growth factor receptor WGA whole genome amplification
WHO World Health Organization
xxi
To Jorge
To Gabriel and Léa
“Life is not measured by the number of breaths we take, but by the moments that take our breath away.”
xxii
Acknowledgments
Foremost, I would like to express my gratitude to my thesis supervisor, Dr. Caroline Diorio, for her continuous guidance and support throughout my Ph.D. studies. I am grateful for her availability, her wise advice, her sensibility and friendship. I would also thank her for encouraging me to pursue my ideas and suggested directions for research.
I would like to thank the pathologists of the Service de Pathologie of the St-Sacrement Hospital, including Drs. Chantal Caron, Anne Choquette, Michel Beauchemin, Mohamed Amin Hashem, Simon Jacob, Sophie Laberge, Mohib Morcos, Nathalie Mourad, Alexandre Odashiro, and Ion Popa. In particular, I would like to thank Dr. Simon Jacob for his support, his kindness, for enthusiastically sharing his broad knowledge of breast pathology and for always making time for me despite his very busy schedule.
I am grateful to the personnel of the Service de Pathologie of the St-Sacrement Hospital for their technical support. Special thanks to François Sanschagrin and Claudie Paquet for introducing me to the fascinating world of molecular diagnostics and for checking the FISH results.
I am grateful to Annick Michaud and Geneviève Ouellette for their great technical support during laboratory work. I would like to thank Sue-Ling Chang for her support with statistical analysis and for the English revision of the manuscripts. Many thanks to Isabelle Dumas for her help in the comprehension of epidemiological concepts and for her wise advice. I am also grateful to the clinicians and to the personnel of the Clinique des Maladies du Sein Deschênes-Fabia, in particular to Drs. Louise Provencher and Julie Lemieux. I would also like to thank Christian Laflamme for his support during manuscript submission. I am also grateful to our great research nurses Danièle Audet and Lucie Tellier for their availability and kindness.
Many thanks also to Drs. Frédéric Barabé and Marc-André Côté for the critical review of one manuscript.
I would like to thank Caty Blanchette, Myrto Mondor and Eric Demers for their support in statistical analysis.
xxiii
Many thanks to Frédéric Fournier and Arnaud Droit for performing the bioinformatics analysis using the R statistical programming environment. I am also grateful to Yvan Labrie and Marie-Christine Pouliot for introducing me to the GenomeStudio software and software for the analysis of pathways and networks.
I am grateful to the Fondation du cancer du sein du Québec and the Banque de tissus et de données of the Réseau de recherche sur le cancer of the FRQS, which is affiliated with the Canadian Tumor Repository Network, for providing clinical specimens.
I am grateful to all my friends at the Research Unit at St-Sacrement Hospital, including Carlotta Lunghi, Kaoutar Ennour-Idrissi, Danielle Larouche, Ludivine Soguel Alexander, Thyphavone Oudanonh, Sofia Laforest, Geneviève Larouche, and Elissar Issa for the many pleasant hours spent together and for creating a productive and enjoyable work environment.
I am grateful to Denis Guillette for his prompt informatics support. I would also like to thank the unit secretary, Ginette Desbiens, for her efficient work and her contagious good mood. I am grateful to all women who participated in our studies.
I am indebted to the Fonds de recherche du Québec –Santé (FRQS) (doctoral fellowship), the Laval University Cancer Research Center (Bourse de distinction Luc Bélanger), the Fondation des Hôpitaux Enfant-Jésus – St-Sacrement and Hoffmann-La Roche Limited for financial support.
Special thanks to our friends, including Rossana, Vincent, Caroline, Gaëlle, Samuel, Aida, Richard, Chantal and Rumiana, who represent our extended family here in Quebec.
I am grateful to my extended family and my family-in-law for their constant love and support, and for their patience and comprehension. Special thanks to my mother Adriana for being a great example of strength and determination.
Special thanks go to my husband Jorge for being such an amazing companion and for sharing with me the joys and difficulties of this great Ph.D. adventure. A very special thanks to little Gabriel and Léa who allow us to re-discover and appreciate the very essence of this miraculous thing called life.
xxiv
Foreword
The research work presented in this Ph.D. thesis was carried out at the St-Sacrement Hospital in Quebec City. The aim of my research work was multifaceted. In the first part of my research, we aimed to identify the most reliable and economical method to assess HER2 status in breast cancer specimens. The results of this research question are presented in Chapters 4 and 5. With the exception of the writing of the grant research proposal, I participated to all aspects for the accomplishment of this project. I was responsible for the literature review, the laboratory work, the analysis of the results and the writing of the manuscripts. Drs. Simon Jacob and Chantal Caron, François Sanschagrin and Caroline Diorio participated in the conception of the study, and in the analysis and interpretation of results. Caroline Diorio supervised each of these stages. All co-authors provided constructive feedback and made specific suggestions to improve the final version of the manuscripts. Both articles have been published in the journal Anticancer Research. This part of the research was funded by the Fondation des Hôpitaux Enfant-Jésus – St-Sacrement and Hoffmann La Roche Limited. In addition, I also reviewed all existing methods for the evaluation of HER2 status in breast cancer specimens and I wrote a review article that has been published in the American Journal of Clinical Pathology (Chapter 2). All co-authors provided constructive feedback and made specific suggestions to improve the final version of the manuscript. In an attempt to further identify the best method to reliably determine HER2 status in breast cancer specimens, we validated a software programming algorithm that analyses fluorescent signals in single tumor cell nuclei within breast cancer tissue sections (Chapter 6). For this project, I participated in the conception of the study and I was responsible for the literature review, the laboratory work, the analysis of the results and the writing of the manuscript that has been published in the journal Diagnostic
Pathology. Drs. Simon Jacob and Chantal Caron, François Sanschagrin and Caroline Diorio
participated in the conception of the study, and in the analysis and interpretation of results. Caroline Diorio supervised each of these stages. All co-authors provided constructive feedback and made specific suggestions to improve the final version of the manuscript. This project was partly funded by the MetaSystems company.
In the second part of this thesis we aimed to investigate the factors that could affect the response to trastuzumab in HER2-positive breast cancer patients. We concentrated on several aspects, including HER2 polymorphisms, alcohol and tobacco consumption and genome-wide DNA methylation pattern in breast cancer tissues.
xxv
As a first step, we analyzed the association between HER2 polymorphisms and breast cancer prognostic factors. For this part of the project, I wrote the manuscript that has been published in the review The Breast (Chapter 7). Annick Michaud performed the laboratory work. Christian Laflamme collected clinico-pathological information. Éric Demers performed statistical analysis. Caroline Diorio and Julie Lemieux supervised each of these stages. All co-authors provided constructive feedback and made specific suggestions to improve the final version of the manuscript. This project was funded by the Fondation des Hôpitaux Enfant-Jésus – St-Sacrement and Hoffmann La Roche Limited.
In a second step, I proposed to analyze the association between HER2 polymorphisms, alcohol and tobacco consumption and genome-wide DNA methylation pattern in breast cancer tissues and the response to trastuzumab in HER2-positive breast cancer patients (Chapters 8 and 9). For this part of the thesis, I was responsible for the literature review, the collection of clinicopathological information (histopathological data, information about treatment received, follow-up data), the laboratory work (only Chapter 9), the analysis of methylation data using GenomeStudio software and the analysis of pathways, conducting statistical analysis, and the writing of the manuscripts. Annick Michaud assessed HER2 genotyping in breast cancer tissues and normal breast tissues (Chapter 8). Frédéric Fournier and Arnaud Droit analysed methylation data using the statistical environment R and performed paired statistical analysis (Chapter 9). Caroline Diorio supervised each of these stages. All co-authors provided constructive feedback and made specific suggestions to improve the final version of the manuscript. The article on the association between HER2 polymorphisms, alcohol and tobacco intake and response to trastuzumab has been submitted to the journal Clinical Breast Cancer (24th May, 2017, under revision), while the
manuscript on the association between DNA methylation pattern in breast cancer tissues and response to trastuzumab is finalized but is not yet submitted for publication. These projects were funded by the Fondation des Hôpitaux Enfant-Jésus – St-Sacrement and Hoffmann La Roche Limited.
Although it does not appear in this document, I also participated in another project, whose goal was to evaluate the association between the phosphorylated form of HER2 and the response to trastuzumab in HER2-negative breast cancer patients. For this project, I performed the literature review and I generated preliminary results. Moreover, I also contributed to the writing of research grant proposals based on these preliminary results.
xxvi
The articles listed below represent my contribution to the field of molecular epidemiology. All these manuscripts were written during my doctoral studies. Articles 1, 2, 4, 5, 7, 8 and 9 are included in this Ph.D. thesis.
1. Furrer D., Jacob S., Caron S., Sanschagrin F., Provencher L., Diorio C. 2013. Validation of a new classifier for the automated analysis of the human epidermal growth factor receptor
2 (HER2) gene amplification in breast cancer specimens. Diagnostic pathology 8: 17.
2. Furrer D., Jacob S., Sanschagrin F., Diorio C. 2015. Advantages and disadvantages of technologies for HER2 testing in breast cancer specimens. American Journal of Clinical
Pathology 144(5): 686-703.
3. Diorio C., Furrer D., Michaud A., Laberge S., Popa I., Jacob S., Provencher L., Hogue JC. 2016. Validation of EP1 antibody clone for estrogen receptor immunohistochemistry for breast cancer. Anticancer Research 36(1): 435-7.
4. Furrer D., Jacob S., Caron C., Sanschagrin F., Provencher L., Diorio C. 2016. Tissue microarray is a reliable tool for the evaluation of HER2 status in breast cancer specimens.
Anticancer Research 36(9): 4661-6.
5. Furrer D., Côté M., Provencher L., Laflamme C., Barabé F., Jacob S., Michaud A., Lemieux J., Diorio C. 2016. Evaluation of human epidermal growth factor receptor 2 (HER2) single nucleotide polymorphisms (SNPs) in normal and breast tumor tissues and their link with breast cancer prognostic factors. The Breast 30: 191-196.
6. Yousef E.M., Furrer D., Laperrière D., Tahir M.R., Mader S., Diorio C., Gaboury L.A. 2017. MCM2: an alternative to Ki-67 for measuring breast cancer cell proliferation. Modern
pathology 30(5): 682-697.
7. Furrer D., Jacob S., Caron C., Sanschagrin F., Provencher L., Diorio C. 2017. Concordance between immunohistochemistry and fluorescence in situ hybridization in the determination of human epidermal growth factor receptor 2 (HER2) status using tissue microarray in breast cancer specimens. Anticancer Research 37(6): 3323-3329.
xxvii
8. Furrer D., Jacob S., Michaud A., Provencher L., Lemieux J., Diorio C. 2017. Association between tobacco and alcohol consumption and HER2 polymorphisms and response to trastuzumab in HER2-positive breast cancer patients. Clinical Breast Cancer, submitted. 9. Furrer D., .Fournier F., Jacob S., Droit A., Diorio C. Association between genome-wide DNA methylation pattern and response to trastuzumab in HER2-positive breast cancer patients. In preparation.
10. Odashiro P.P., Orain M., Furrer D., Diorio C., Simonyan D., Moreira A., Joubert P. PD-L1 in thymomas and thymic carcinomas. In preparation.
- 1 -
Chapter 1: Introduction
Over the past decades, microarray-based gene expression studies have highlighted the fact that breast cancer comprises a heterogeneous group of diseases in term of differentiation and proliferation, prognosis and treatment. These studies allowed the identification of breast cancer intrinsic subtypes (1-3). One of these subtypes is the so-called human epidermal growth factor receptor 2 (HER2)-enriched subtype. Clinically, HER2-positive breast cancer is characterized by the overexpression of the HER2 receptor and/or HER2 gene amplification (4). HER2-positive breast cancer patients have a particular worse prognosis. In addition, HER2-positive breast cancer patients are eligible to receive targeted treatment with trastuzumab, a monoclonal antibody specifically directed against the HER2 receptor (5).
Evaluation of the HER2 status in breast cancer specimens is crucial for defining patient management. Considering the clinical and economic implications of targeted anti-HER2 treatments, reliable HER2 test results are essential. False negative results would deny the patients access to the potential benefits of trastuzumab, whereas false positive results would expose patients to the potential cardiotoxic side effects of this expensive agent without experiencing any therapeutic advantages (6).
Immunohistochemistry (IHC) and in situ hybridization (ISH) techniques are the most commonly used techniques for the determination of HER2 status in breast cancer specimens (7, 8). As each technique has its own advantages and disadvantages, currently there is still no consensus on which method is the best for evaluating the HER2 status in breast cancer specimens (6). The first aim of this project, therefore, was to identify the most reliable and economical method to evaluate HER2 status in breast cancer specimens. Trastuzumab treatment significantly improved survival in HER2-positive breast cancer patients. Two Cochrane systematic reviews reported that, when combined with standard chemotherapy, trastuzumab significantly improved overall survival (OS) and disease-free survival (DFS) in HER2-positive women with early and locally advanced breast cancer (9) and OS and progression-free survival (PFS) in metastatic HER2-positive breast cancer patients (10). .
Despite this noteworthy achievement, a subset of HER2-positive breast cancer patients does not respond to trastuzumab-based treatment (11). Although several molecular
- 2 -
mechanisms of trastuzumab resistance have been proposed in the literature (12), no clinically effective strategies to overcome trastuzumab resistance have been identified yet (11).
It has been reported that genetic and epigenetic factors can influence the efficacy of antineoplastic drugs in breast cancer patients (13-15). It has been shown that HER2 polymorphisms have an impact on HER2 function (16, 17). It has also been reported that methylation pattern in breast cancer tissues of HER2-positive breast cancer patients is heterogeneous (18). Given that trastuzumab response in HER2-positive breast cancer patients is also heterogeneous, we hypothesize that methylation pattern in tumor tissues of patients that respond to trastuzumab treatment is different to that of patients that develop resistance to this agent.
Evidence suggests that also lifestyle factors including tobacco and alcohol use might influence the survival of breast cancer patients (19-21). Interestingly, it has been reported the existence of molecular and epidemiological links between tobacco and ethanol exposures and HER2 (22-26). We hypothesize that HER2 polymorphisms, tobacco and alcohol consumption may represent factors influencing the efficacy of targeted therapies in HER2-positive breast cancer patients.
The second aim of our study was therefore to analyze the association of HER2 polymorphisms and methylation patterns in breast cancer tissues, and tobacco and alcohol consumption with the response to trastuzumab in HER2-positive breast cancer patients.
Literature overview
1.1. Breast cancer intrinsic subtypes
1.1.1. Molecular classification of breast cancer
Breast cancer is the most frequently diagnosed cancer and the leading cause of cancer-related death amongst women worldwide (27). In 2015, it is estimated that in Canada 25,000 new cases of breast cancer have been diagnosed and 5,000 women have died from this malignancy (28).
It is now well recognized that breast cancer is a heterogeneous disease characterized by various pathological features, different treatment response, and substantial differences in survival (29). Based on the World Health Organization (WHO) classification, there are at
- 3 -
least 20 different histological types of breast cancer (30). A major drawback of this classification is that around 90% of breast cancers will eventually belong to the most common morphological subtypes, namely invasive ductal carcinomas or invasive lobular carcinomas, which represent approximatively 80% and 10% of invasive breast cancers, respectively. As patients with tumors showing similar histological appearance may have very different clinical outcomes, the histopathological classification presents limited prognostic and predictive values, and its clinical utility is relatively modest (31).
Given that histological diversity cannot entirely explain the difference in breast cancer behavior, at the beginning of the new century novel approaches, particularly gene expression profiling, have been used to refine breast cancer classification independently of histomorphological criteria. Perou and collaborators performed cDNA microarray analysis of RNA extracted from frozen tissue to characterize the gene expression of 65 breast cancer specimens obtained from 42 patients (1). The central hypothesis of this study was that tumors could be characterized on the up- or down-regulation of sets of genes, the so-called “intrinsic genes”. Intrinsic genes were defined as genes with significantly greater variability in expression between different tumors than between duplicate samples from the same tumor. This resulted in the selection of 496 genes representing an “intrinsic gene list” chosen from an initial list encompassing 8,102 genes that allowed classifying tumor specimens into subtypes. This seminal work showed for the first time that breast tumors could be classified into molecular subtypes distinguished by differences in their gene expression profiles. A subsequent study that similarly analyzed the expression profiles of 115 independent breast tumor samples using RNA extracted from frozen tissue and an intrinsic set of 534 genes allowed a refined molecular classification of breast tumors as reported by Perou et al. (3). Using hierarchical clustering analyses tumors showing similar gene expression patterns were grouped together. This analysis allowed identifying two distinctive groups of breast cancers based on the expression of the estrogen receptors (ER) and ER-related genes (GATA3, X-box binding protein 1, HNF3A) (3): whereas one cluster expressed ER and ER-associated genes, the other one did not. The first group of tumors was defined as ER-positive, the second one as ER-negative. This finding was consistent with breast cancer cell biology, since the binding of estrogen to nuclear ER and the subsequent binding of the dimerized ER to estrogen responsive elements (EREs) located in the promoter of target genes regulate cellular growth and proliferation in several target tissues of the human body, including breast tissue (32).
- 4 -
Tumors grouped in the ER-positive cluster showed a gene expression profile similar to that of luminal cells found in the normal breast (3). For this reason, they were named as luminal tumors. Luminal tumors were subdivided into two categories: luminal A tumors, characterized by highest ER expression, and luminal B tumors that expressed ER-related genes at a lower level. Moreover, compared to luminal A tumors, luminal B tumors showed higher expression of proliferation/cell-cycle related genes or proteins, including MKI67 and AURKA, and lower expression of several luminal-related genes or proteins (e.g. progesterone receptor (PR) and FOXA) (33). In the ER-negative cluster, three subtypes were observed: the HER2-enriched tumors, the basal-like tumors, and the normal breast-like tumors. The HER2-enriched tumors were characterized by high expression of the
human epidermal growth factor receptor 2 (HER2) gene and other genes associated with
the HER2 pathway and/or HER2 amplicon located in the 17q12 chromosome. The basal-like tumors were named as such as they express cytokeratins (CK5/6, CK14, and CK17) that are usually expressed in normal breast myoepithelial cells, cells that underlie the breast luminal cells. The normal breast-like tumor group has been described to express genes that are usually expressed in adipose tissue and other non-epithelial cell types. These tumors were also shown to express basal epithelial genes. However, it has been postulated that this was likely an artificial category caused by poorly sampled tumor tissue (2).
As illustrated above, the intrinsic breast cancer subtypes were initially identified using unsupervised clustering analysis of gene expression using RNA isolated from frozen tissues. In order to improve applicability of gene expression profiling to routine pathology specimens, Parker and collaborators developed a quantitative reverse transcription polymerase chain reaction (qRT-PCR) assay to identify the intrinsic molecular subtypes using RNA extracted from more easily available formalin-fixed, paraffin-embedded (FFPE) tissues (34). They proposed an efficient 50-gene classifier, referred to as the Prediction Analysis of Microarray 50 (PAM50), that reanalyzed the previous five intrinsic subtypes defining the four major subtypes currently known: luminal A, luminal B, HER2-enriched and basal-like. The PAM50 classifier is an often used method to classify intrinsic subtypes (35). The prognostic value of the PAM50 classifier was validated in a test set of 761 node-negative breast cancer patients who did not receive any adjuvant therapy. Using statistical models, the risk of relapse (ROR) score was estimated for each test case. From this analysis it emerged that intrinsic subtypes, as defined with the PAM50 classifier, showed prognostic significance. Moreover, the results remained significant in multivariate analyses that incorporated standard parameters,
- 5 -
including ER status, histological grade, tumor size, and node status. In addition, in a set of 113 patients treated with neoadjuvant chemotherapy, the intrinsic subtype model predicted neoadjuvant chemotherapy (taxane and anthracycline regimen) efficacy with a negative predictive value for pathologic complete response of 97% (34).
Molecular understanding of breast cancer paved the way to personalized medicine. Since the initial description of the intrinsic subtypes, several studies have validated their prognostic and predictive significance (5, 36-43). It has emerged that patients with basal-like or HER2-enriched breast cancer showed the worst outcome, patients with luminal A tumors were associated with a favourable prognosis and patients with luminal B tumors showed intermediate outcome (3, 44). In addition, patients with luminal B tumors were less sensitive to endocrine therapy (the therapy for hormone receptor-positive breast cancer) than patients with luminal A tumors. However, patients with luminal B tumors showed a better response to neoadjuvant chemotherapy compared to patients with luminal A tumors, achieving higher pathologic complete response rate (5). Breast cancer patients with HER2-enriched tumors benefited from the treatment with an anti-HER2 agent, trastuzumab (33).
1.1.2. Molecular classification of breast cancer subtypes using
immunohistochemical surrogates
Given that methods for gene expression profiling present some limitations in the context of routine clinical pathology (unavailability of these techniques in many laboratories, specialized knowledge required to interpret results, requirement of fresh or frozen tissue), an alternative approach to identify breast cancer subtypes has been developed (33). Intrinsic breast cancer subtypes as primarily identified by gene expression profiling can, to a certain extent, be approximated using evaluation of molecular markers that can be assessed using routine clinical pathology methods, including immunohistochemistry (IHC) and in situ hybridization (ISH) techniques (37, 45-48). IHC allows the evaluation of protein expression in tissue sections by the utilization of antibodies directed against the specific protein (49). ISH methods allow the determination of the gene copy number within cell nuclei using a DNA probe specifically directed against the gene of interest (50).
This simplified clinicopathological classification was included in the 2011 St Gallen International Expert Consensus Panel in order to propose treatment strategies (51). In this classification breast cancer subtypes are defined on the basis of immunohistochemical analysis of ER, PR, the evaluation of overexpression and/or amplification of HER2, and the
- 6 -
assessment of the Ki-67 index, defined as the percentage of cancer cells positive for Ki-67 staining (Table 1.1.). Ki-67 is a cell proliferation marker used for the evaluation of cell proliferation in clinical specimens and it is considered one of the most important cell proliferation-related genes (52).
Table 1.1. Surrogate definition of intrinsic subtypes of breast cancer as defined by the 2011 St Gallen International Expert Consensus Panel
Intrinsic subtype (GEP) IHC and FISH classification (St. Gallen)
Luminal A “Luminal A”
ER-positive and/or PR-negative, HER2-negative, Ki-67 low (< 14%)a
Luminal B “Luminal B (HER2-negative)”
ER and/or PR-positive, HER2-negative, Ki-67 ≥ 14% “Luminal B (HER2-positive)”
ER and/or PR-positive, Any Ki-67, HER2 overexpressed or Amplified
HER2-enriched “HER2-positive”
HER2 overexpressed or amplified, ER and PR absent Basal-like “Triple negative”
ER and PR absent, HER2-negative
Abbreviations: GEP: gene expression profiling; IHC: immunohistochemistry; FISH: fluorescence in situ hybridization; ER: Estrogen Receptor; PR: Progesterone receptor; HER2: human epidermal growth factor receptor 2.
a If a reliable Ki-67 measurement is not available, some alternative assessment of tumor proliferation such as tumor grade may be used to distinguish between “Luminal A” and “Luminal B” subtypes.
Adapted from Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011 (51).
- 7 -
A Ki-67 cut-off value of 14% was recommended by the Panel with the aim of distinguishing “luminal A” from “luminal B (HER2-negative)” breast carcinomas (51). The differentiation between luminal A and luminal B tumors carries important therapeutic implications. Luminal A and luminal B tumors represent in fact clinically distinct subtypes in term of differentiation and proliferation, prognosis and treatment. Luminal A tumors tend to be more differentiated and show a lower proliferation rate compared to luminal B tumors. Moreover, luminal B patients show a worse prognosis compared to luminal A patients and are treated with chemotherapy in addition to hormone blockage (5).
Major drawbacks of Ki-67 are its lack of reproducibility, in addition to the difficulty to find an appropriate cut-off (10%, 14%, and 20%) (37, 53-55). Therefore, other markers for the evaluation of breast cancer proliferation have been proposed in the literature, including cyclins, topoisomerase II and the mini-chromosome maintenance complex component 2 (MCM2) (56, 57). A recent study has highlighted the clinical relevance of the evaluation of MCM2 in breast cancer specimens (58). MCM2 is a protein that belongs to the MCM complex. This complex functions as a replicative helicase (59, 60), an enzyme that unwinds DNA during chromosomal replication, a crucial step in the initiation of DNA replication (61). MCMs proteins are expressed throughout the G1 phase and have emerged as biomarkers of cell cycle state, as they allow distinguishing cycling cells from quiescent cells (60). Yousef and collaborators demonstrated that MCM2 assessment (using a cut-off of 40%) allowed a clear distinction between luminal A and luminal B/HER2-negative breast cancer specimens (58).
In these few pages, I have briefly highlighted the characteristics of the different breast cancer subtypes. However, since the molecule of interest of the present Ph.D. thesis is the transmembrane receptor HER2, in the following subsection I will focus on the HER2-enriched breast cancer subtype.
1.1.3. enriched subtype vs. breast cancer clinically evaluated as
HER2-positive
The HER2-enriched subtype is characterized by high expression of HER2-related and proliferation-related genes (e.g. ERBB/HER2 and GRB7) and shows low expression of luminal clusters (e.g. ESR1 and PGR) compared to luminal A and B tumors (62). In the absence of treatment, patients harboring tumors of the HER2-enriched subtype have a worse prognosis than those with luminal A tumors (5).
- 8 -
It is important to note that the simplified clinicopathological classification of breast cancer subtypes based on IHC and ISH results does not completely overlap with the HER2-enriched subtype based on gene expression profiling since only 50% of tumors belonging to the HER2-enriched subtype are HER2-positive and ER-negative (34, 63-65). It has been reported that 15% of HER2-enriched subtype are both HER2 and ER-positive, 16% are HER2-negative and ER-positive and 18% are negative for both receptors (64). It has been postulated that the subgroup of HER2-enriched tumors that are clinically evaluated as HER2-negative (around 30%) might present mutations of some downstream pathway component that mimic HER2 amplification (64).
1.1.4. Breast cancer clinically evaluated as HER2-positive
The HER2 receptor is a 185 kDa transmembrane protein that is encoded by the HER2 (also known as erb-b2 receptor tyrosine kinase 2 [ERBB2]) gene, which is located on the long arm of chromosome 17 (17q12-21.32) (66). HER2 is normally expressed on cell membranes of epithelial cells of several organs like the breast and the skin, as well as gastrointestinal, respiratory, reproductive, and urinary tract (67). Whereas in normal breast epithelial cells HER2 is expressed at low levels (two copies of the HER2 gene and up to 20,000 HER2 receptors) (68), in HER2-positive breast cancer cells there is an increase in the number of
HER2 gene copies (up to 25-50, termed gene amplification) and HER2 receptors (up to 40
to 100 fold increase, termed protein overexpression), resulting in up to 2 million receptors expressed at the tumor cell surface (69). Besides breast cancer, HER2 overexpression has also been reported in other types of tumors, including stomach, ovary, colon, bladder, lung, uterine cervix, head and neck, and oesophageal cancer as well as uterine serous endometrial carcinoma (70).
In human breast cancer, HER2 gene amplification and/or receptor overexpression, which occur in 15% to 20% of patients, are important markers for poor prognosis, including a more aggressive disease and shorter survival (4). Moreover, HER2-positive status is considered a predictive marker of response to HER2-targeted drugs, including trastuzumab and lapatinib (71). Patients are eligible for targeted anti-HER2 therapies when their breast cancer specimens overexpress the protein at the IHC and/or are HER2 gene amplified at ISH. Consequently, accurate testing of HER2 status is essential for the clinical management of breast cancer patients.
- 9 -
1.2. Methods for the evaluation of HER2 status in breast cancer
specimens
1.2.1. Immunohistochemistry (IHC) and in situ hybridization (ISH) methods
Currently, several diagnostic methods are approved for the evaluation of HER2 status in breast cancer specimens in routine clinical practice: immunohistochemistry (IHC) and in situ hybridization (ISH) techniques, most commonly fluorescent ISH (FISH) (7, 8).
IHC allows the evaluation of the HER2 protein expression in formalin-fixed, paraffin-embedded (FFPE) tissues using specific antibodies (8). By this method, it is then possible to estimate the number of cells showing membranous staining in the tissue section as well as the intensity of the staining (72). Membranous staining is scored on a semi-quantitative scale.
ISH techniques, instead, allow the quantification of HER2 gene copy number within tumor cell nuclei using a DNA probe coupled to a fluorescent, chromogenic, or silver detection system (i.e., FISH, chromogenic ISH [CISH], or silver-enhanced ISH [SISH]), or a combination of CISH and SISH systems (bright-field double ISH [BDISH]) (6). ISH is effectuated either as a single-color assay (HER2 probe only) to evaluate HER2 gene copies per nucleus or as a dual-color assay using differentially labeled HER2 and chromosome 17 centromere (chromosome enumeration probe 17, CEP17) probes simultaneously. The dual-color assay allows the determination of the HER2/CEP17 ratio (73). The HER2/CEP17 ratio is often regarded as a better reflection of the HER2 amplification status, as the latter may be influenced by abnormal chromosome 17 copy number (mainly polysomy) (74).
In 2007, the American Society of Clinical Oncology (ASCO)/ the College of American Pathologists (CAP) published guidelines for HER2 testing in breast cancer to improve reliability of HER2 test results (8). These guidelines included recommendations regarding specimen handling and testing requirements. The guidelines also included scoring criteria of immunohistochemical and ISH staining to classify breast cancer specimens into three categories: positive, equivocal and negative (Figure 1.1.). A case is considered positive at IHC when the HER2 expression is scored as 3+ (strong, complete, homogeneous membrane staining in >30% of tumor cells). A case is evaluated as positive (amplified) at ISH when the mean HER2 gene copy number is > 6 signals/nucleus or HER2 gene/chromosome 17 copy number ratio (HER2/CEP17) is > 2.2. Equivocal cases at IHC show a strong, complete membrane staining in ≤ 30% of tumor cells or weak/moderate
- 10 -
heterogeneous complete membrane staining in ≥ 10% of tumor cells (IHC score 2+). Equivocal cases at IHS show a mean HER2 gene copy number of ≥ 4 and ≤ 6 signals/nucleus or a HER2/CEP17 ratio between 1.8 and 2.2. A case is evaluated as negative at IHC when the HER2 expression is scored as 0/1+ (no staining/ weak or incomplete membrane staining in any percentage of tumor cells). A sample is evaluated as negative (non-amplified) at ISH when the mean HER2 gene copy number is < 4 signals/nucleus or the HER2/CEP17 ratio is < 1.8. The equivocal category requires additional testing with the alternative assay (IHC or ISH) for final determination. In 2013, the ASCO/CAP revised the guidelines published in 2007 to clarify the recommendations for HER2 testing in breast cancer specimens (7). The updated guidelines included new scoring criteria for IHC and ISH.
Figure 1.1. Immunohistochemistry (IHC) and fluorescence in situ
hybridization (FISH) results of HER2.
Representative IHC HER2 staining are presented in the first row : A. Negative staining (IHC score 0/1+); B. Equivocal staining (IHC score 2+); C. Positive staining (IHC score 3+); Representative HER2 FISH results are presented in second row : D. Negative FISH result (non-amplified); E. Equivocal FISH result (equivocal); F. positive FISH result (amplified).
A detailed description of the IHC and the ISH methods for the evaluation of HER2 status in breast cancer specimens, including the commercially available diagnostic tests and antibody clones as well as scoring criteria for reporting IHC and ISH results according to the ASCO/CAP recommendations (2007 and 2013 ASCO/CAP scoring criteria) are presented in Chapter 2.
A
B
C
- 11 -
The updated ASCO/CAP guidelines also presented a new approach for the determination of HER2 status in breast cancer specimens. While the 2007 ASCO/CAP guidelines recommended performing HER2 testing on resection specimens and to retest when results were equivocal (8), the updated guidelines recommend effectuating an initial test (IHC or ISH) on core biopsy (7). If test results are equivocal or if there is an apparent histopathological discordance with the test result, reflex testing on tumor specimen section with an alternative assay (IHC or ISH) should be performed.
Although in clinical practice IHC and ISH are the most frequently used techniques for the evaluation of HER2, other methods have been developed over the years for the determination of HER2 status in breast cancer specimens. An exhaustive review of all existing methods to assess HER2 status in breast cancer, including a discussion on the advantages and disadvantages of each described technique, is presented in Chapter 2.
1.2.2. Methodological worries related to HER2 testing
Although the FISH assay is considered a more objective and quantitative method compared to IHC, this technique presents several disadvantages (6). For example, manual counting of fluorescent signals is time-consuming. To overcome this limitation, several automated image analysis software have been developed for the enumeration of fluorescent signals (75, 76). Some of these software employ a programming algorithm through which the software quantifies fluorescent signals in images on the basis of square tiles of fixed dimensions (77). The major drawback for this method of analysis is that the size of the tile does not always correspond to the size of a single tumor cell nucleus. This method might therefore not completely reflect the biology of cells (78). In the literature, one study has analyzed the utility of an image analysis software (EIKONA3D, Alpha Tec Ltd) for the evaluation of HER2 amplification in single tumor cell nuclei in a cohort of 100 breast cancer cases (79). The authors found a very good concordance (100.0%) between the results obtained by manual scoring and those obtained with the image analysis software for non-amplified cases. However, the concordance between the two methods for amplified cases was moderate (74.1%). Therefore, better performing software programming algorithms that analyse fluorescent signals in single tumor cell nuclei within breast cancer tissue section are warranted.
The high cost of the FISH technique represents a further limitation of this method. Since tissue microarray (TMA) technology facilitates the simultaneous molecular characterization
- 12 -
of a large amount of specimens, this technique might represent an economical replacement for whole tissue section analysis for breast cancer specimens. This technique has already been extensively described elsewhere (80-82), therefore it is only briefly described here (Figure 1.2.). Tissue core cylinders obtained from several formalin-fixed, paraffin-embedded (FFPE) specimens are placed into a single, empty paraffin block using manual, automated or semiautomatic tissue microarrayers (82). Before taking tissue cores from the donor blocks, regions of interest are marked on hematoxylin & eosin (H&E) slides. The corresponding regions on the matching paraffin block (donor block) is then identified and marked. Tissue cores are removed from these areas of the donor block using hollow needles of the microarray instrument and placed at defined array coordinates in the paraffin recipient block. The diameter of tissue cores ranges from 0.6 to 2.0 mm. Location and details of each single core are then recorded to generate a clinical database. To date, only a few studies have analyzed the utility of TMA in the evaluation of HER2 gene amplification by FISH in breast cancer specimens (83-86). In these studies, the authors reported excellent agreement rates (ranging from 91% to 97%) between HER2 gene amplification status determined on whole tissue section and on TMA section. To date, no study evaluating this concordance using the 2013 ASCO/CAP scoring criteria has been conducted.
- 13 -
Figure 1.2. Construction of Tissue microarray
Cylindric tissue cores are removed from donor paraffin block and transferred into premade holes of an empty recipient paraffin block using a tissue microarrayer. For details, see text. Source:http://apps.pathology.jhu.edu/blogs/pathology/tissue-microarrays.
1.2.3. Concordance of HER2 status determined by IHC, FISH, CISH and SISH in
the literature
Although several methods have been approved by the FDA for the evaluation of HER2 status in breast cancer specimens, measure variation between these techniques has been reported. Evaluation of the concordance of HER2 status determined by IHC and FISH has already been studied in numerous studies (for a systematic review and a meta-analysis see (87) and (88), respectively). The concordance between IHC and FISH was 96% for negative cases (IHC 0/1+) and 91% for positive cases (IHC 3+) cases. Among the IHC-equivocal cases (IHC 2+), 36% showed HER2 gene amplification and 64% were considered non-amplified (88). The concordance rate was influenced by several factors, including the antibody clone and the scoring criteria used (7, 8, 89). The majority of the studies that analyzed the concordance between IHC and FISH have been conducted on cohort of selected breast cancer specimens, where cases with equivocal immunostaining or with borderline HER2 gene amplification were overrepresented. Only a few studies have analyzed the concordance between IHC and FISH for all IHC categories in a cohort of consecutive breast cancer cases (72, 90-93).