HAL Id: inserm-00854329
https://www.hal.inserm.fr/inserm-00854329
Submitted on 26 Aug 2013
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
decisions.
Marco-Antonio Mendoza-Parra, Hinrich Gronemeyer
To cite this version:
Marco-Antonio Mendoza-Parra, Hinrich Gronemeyer. Genome-wide studies of nuclear receptors in cell
fate decisions.. Seminars in Cell and Developmental Biology, Elsevier, 2013, 24 (10-12), pp.706-15.
�10.1016/j.semcdb.2013.07.001�. �inserm-00854329�
Pleasecitethisarticleinpressas:Mendoza-ParraM-A,GronemeyerH.Genome-widestudiesofnuclearreceptorsincellfatedecisions.Semin CellDevBiol(2013),http://dx.doi.org/10.1016/j.semcdb.2013.07.001
ARTICLE IN PRESS
GModelYSCDB-1461; No.ofPages10
SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx
ContentslistsavailableatScienceDirect
Seminars
in
Cell
&
Developmental
Biology
j ou rn a l h o m e pa g e :w w w . e l s e v i e r . c o m / l o c a t e / s e m c d b
Review
Genome-wide
studies
of
nuclear
receptors
in
cell
fate
decisions
夽
Marco-Antonio
Mendoza-Parra
∗,
Hinrich
Gronemeyer
∗DepartmentofCancerBiology,InstitutdeGénétiqueetdeBiologieMoléculaireetCellulaire(IGBMC)/CNRS/INSERM/UniversitédeStrasbourg,BP10142, 67404IllkirchCedex,France
a
r
t
i
c
l
e
i
n
f
o
Articlehistory: Available online xxx Keywords: Nuclearreceptors Retinoicacidreceptor Functionalgenomics Systemsbiology
a
b
s
t
r
a
c
t
Nuclearreceptors(NRs)areimportantmediatorsoftheinformationencodedinthechemicalstructure ofitscorrespondingligand,astheyinterpretsuchinformationinthecontextofthecellidentityand physiologicalstatusandconvertitintosequentialtranscriptionregulatoryevents.Atthecelllevelthis canresultintemporallycoordinatedprocessessuchascellfatetransitions,whichcomprisetheregulation ofaplethoraofgeneprogramsincludingamongothersregulationofcellproliferation,metabolismand specificfunctionalitiesthatareacquiredbythedifferentiatedcell.Whileboththeearlystepsofnuclear receptorfunctionandtheirimpactonanimal/organphysiologyisratherwellunderstood,littleisknown aboutthedynamicgenenetworksthatultimatelycauseaparticular(cell)physiologicalphenomenon inducedbythecognateNRligand/hormone.
Thankstoadvancesinmassiveparallelsequencingandbioinformaticsanalysesofgenome-widedata sets,timehascomeforthedevelopmentofNRsystemsbiology.Indeeditisnowpossibletointegrate globaltranscriptionfactorbinding,epigeneticchromatinhistoneandDNAmodificationpatternswith transcriptomesand3-dimensionalchromatinstructures,extractdecisionpointsintemporalstudiesand decipherthetemporalcontrolofgenenetworksthataretheultimategeneticreadoutsofNR ligand-inducedphysiologicalphenomena.Inthisreviewwewillsummarizethechronologyofthedevelopment ofincreasinglylargerdatasetsforNRaction,withaparticularfocusonstudiesperformedwiththe RAR/RXRnuclearreceptorfamily,anddiscussthepresentattemptstointegrateamultitudeof genome-widedatasetsintheultimatecontextofthetemporal3-dimensionalchromatinstructure.
© 2013 The Authors. Published by Elsevier Ltd. All rights reserved.
Contents
1. Introduction... 00
2. Nuclearreceptorsinapost-genomicera... 00
2.1. AssessingtheglobalgeneexpressionsignaturesdrivenbyNRs... 00
2.2. Mappingthechromatinbindingsitesofnuclearreceptors... 00
3. NRgeneexpressionprogramsandtheirassociatedkeyfactorsinvolvedinsignalingdiversification... 00
3.1. StudyingNR-drivencellfatetransitionsasdynamicgeneexpressionprograms... 00
3.2. Additionalmechanismsinvolvedincontrollingtheretinoids-drivendynamicdiversifiedgeneprograms... 00
3.2.1. MultipleRXR–RARheterodimersmediateRA-signaling... 00
3.2.2. Epigeneticmodificationsandco-regulatorsestablishregulatoryprinciplesaffectingRA-regulatedgeneprograms upstreamanddownstreamofRXR–RARheterodimeraction ... 00
3.2.3. Thethree-dimensionalchromatinorganizationanditsdynamicchangesdrivenbyNR-signaling... 00
4. FuturedirectionsinthestudyofNR-drivencellfatetransition ... 00
References... 00
夽 Thisisanopen-accessarticledistributedunderthetermsoftheCreativeCommonsAttribution-NonCommercial-NoDerivativeWorksLicense,whichpermits non-commercialuse,distribution,andreproductioninanymedium,providedtheoriginalauthorandsourcearecredited.
∗ Correspondingauthors.Tel.:+33388653473;fax:+33388653437.
E-mailaddresses:marco@igbmc.fr(M.-A.Mendoza-Parra),hg@igbmc.u-strasbg.fr(H.Gronemeyer). 1084-9521/$–seefrontmatter © 2013 The Authors. Published by Elsevier Ltd. All rights reserved.
2 M.-A.Mendoza-Parra,H.Gronemeyer/SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx
1. Introduction
Nuclear receptors(NRs; 48 NRs exist in humans)constitute amajorclassoftranscriptionalregulatorsinmetazoansthatare believedtohaveevolvedpriortothedivergenceofvertebratesand invertebrates.Thefirstreceptor(estrogenreceptor,ER)was iden-tified1958byElwoodJensenbutonlyaftercloningseveralNRs inthe1980sitbecameapparentthatthesereceptorsforsteroids, thyroids,retinoicacidsandothersmallmoleculeligands,several ofwhichactinanintracrinefashion,constituteasuperfamilyof transcription factors(TFs) that includesreceptorsfor which no naturalligandisknownormaynotexist.NRsbindina ligand-dependent(e.g.,estrogen receptor)or ligand-independent (e.g., retinoicacidreceptor)mannertocis-actingDNAregulatory ele-ments,whichmaybepositionedproximaltopromoterregionsof targetgenesorregulategenesduetostructuralproximityinthe contextofchromatinarchitecture,actasactivatorsand/or repres-sorsoftranscription,andmayexertnon-genomicactivities.NRsare ofmajorsocial(the“pill”,NRantagonistsforabortion), pharmaceu-ticaland clinicalimportance(e.g.,endocrine-dependentcancers ormetabolicdiseases),asalargemajorityof physiological pro-cessesandpathologiesinvolve(aberrant)NRaction[forreviews see1,2–13].
Conceptually, ligand binding modulates the communication of the nuclear receptor with the intracellular environment, which entails essentiallyreceptor-protein and receptor-DNA or receptor-chromatininteractions.Duringthisprocess,receptorsare importantmediators ofthe information encoded inthe chemi-calstructureofagivenligand,astheyinterpretthisinformation inthe context ofcellular identityand cell-physiologicalstatus; thus transforming it into a dynamic chain of receptor-protein andreceptor-DNAinteractions.NRspresentamodularstructure mainlycharacterizedbyaDNA-binding(DBD)andligand-binding (LBD)domains,whose3Dstructuresinpresenceandabsenceof cognateDNAresponseelementsandvariousagonistsor antago-nists,respectively,havebeendetermined[14–19].TheLBDserves as dual input–output information processor, as ligand binding (otherinputsare,forexample,receptorphosphorylations)induces allostericchangesofreceptorsurfacesthatrepresentdockingsites for subunits of transcription and/or epigenetic machineries, or enzymecomplexes(output).FurthermoreNRsareactively regu-latedbypost-translationalmodifications(e.g.,phosphorylations; ubiquitinylations)whichmayhaveadirectorindirectroleintheir transcriptionalregulationfunction(reviewedin[20]).
Boththeearly stepsof nuclearreceptor function as wellas theirphysiologicalimpactareingeneralratherwellunderstood. Infact,duetoaplethoraofmolecularandstructuralbiology stud-ies,thesequenceofeventsthatfollowsthebindingofaligandis largelyknown,andweunderstandhowtheseeventscanbe mod-ulatedby liganddesign [3,21,22].Briefly, bindingof anagonist totheNRligandbindingdomain(LBD)inducesanallosteric con-formationalreorganizationwhichaltersurfacesintheligand-free receptor(apoNR)towhichco-repressors(CoRs)bindresultingin dissociationofCoRcomplexes,whichcontainepigeneticenzymes (HDACs,histone deacetylases).Importantly,only some apoNRs, suchasRARsandTRs,recruitCoRcomplexesandthus,canactas transcriptionalrepressorswhenbindingtochromatininabsenceof ligand.OtherNRs,likeERandGRarebelievedtobindtochromatin onlyafterinteractionwiththeircorrespondingligands.
In addition to the molecular/structural insights, extensive mousegenetics providedimportant informationconcerningthe physiologicalrolesofseveralnuclearreceptors[23,24],andofsome of theirco-regulators [25,26].However, how a singlemolecule thatbinds toitscorrespondingreceptorregulatesa plethora of cell-specificdynamicnetworksofgenesandhowtheepigenome contributestotranscriptionalregulationthatultimatelyreadsout
asa(cell)physiologicalphenomenon,isstillaunknown(Fig.1).In thisreview,wesummarizetheeffortsperformedtopavetheway intothedevelopmentofsystemsbiologyofnuclearreceptoraction. Wewilladdressthechronologyofthedevelopmentofincreasingly largeromicsdatasetsforNRs action,withaparticularfocuson theRAR/RXRnuclearreceptors,anddiscussthepresentattempts tointegrateamultitudeofgenomewidedatasetsintheultimate contextofthe4-dimensionalstructureofchromatin.
2. Nuclearreceptorsinapost-genomicera
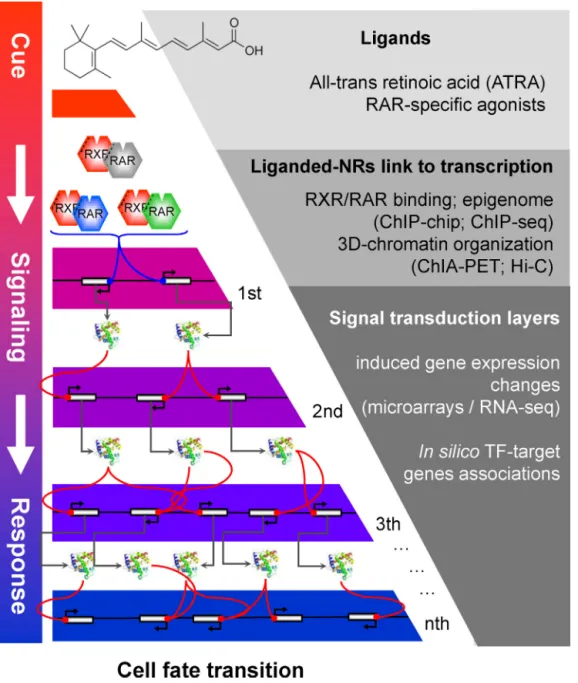
Thepublicationofthefirstdraftofthehumangenomesequence in2001,followedbythoseofvariousothermodelorganisms,gave risetoanewwaytoaddressthemoleculargeneticsofthe homeo-stasisoflivingorganisms.Sincethen,anybiologicalphenomena andits(de)regulationcanbeexplored,inprinciple,ina “genome-wide”context.Indeed, thankstotheadvancesin genome-wide or “omics” approaches, it is now possible to assess the global transcriptionalactivitybyavarietyofapproaches(i.e.,by microar-rays;RNAsequencing,etc.),characterizethegenomiclocalization oftranscriptionfactorsorevaluateepigeneticchromatin modifi-cationinahighresolutionmanner(ChIP-chip;ChIP-seqassays). Furthermore,newmethodologiesusingaproximity-basedligation approacharestartingtogiveinsightsintothe3-dimensional chro-matinstructuresprovidinganewwaytointerrogatethemolecular principlesregulatinglivingsystems, suchas thegenenetworks involvedincellfatedecisionsthataretriggeredbyinternalor exter-nalfactors(Fig.1).
2.1. AssessingtheglobalgeneexpressionsignaturesdrivenbyNRs ThedissectionofNRligand-inducedsignalinginvolvedin var-iousphysiologicalprocesseshasbeenearlyonevaluatedbyhigh throughputgenomicmethods.Importantly,thisnewwayto inter-rogatethemolecularhomeostasisofbiologicalsystemsgenerates highernumberofsignificanttargetsthanthoseidentifiedin pre-viousyears bystandard genetics/molecularbiology approaches, thus providing a more comprehensive view of the regulatory eventsduringNR-signaling.Indeed,in2002BalmerandBlomhoff summarizedmore than1191publishedarticlesonretinoic acid receptorsandclassified532genesasRAregulatedtargets[27]. Inthesameyear,twootherstudiesfocusedonRA-inducedcell differentiationintwo wellknownembryocarcinomacell(ECC) models(F9differentiatesintoparietalendoderm[28],whileP19 differentiatesintoneuronalcells[29])identifiedasimilarnumber ofdifferentiallyregulatedgenesbyincorporatingintheirassays one of the early versions of the microarray technology(cDNA PCR-spottedmicroarrays;reviewed in[30]).Furthermore,these twostudiesaswellasothersfocusedintheglobaltranscriptional regulation response driven by various other NRs (anextensive reviewconcerningtheuseofmicroarraysforthegenomicprofiling inaNR-mediated contexthasbeenpresentedin[31]),assessed thechangesintranscriptionalactivityoverdifferenttime-points asawaytoidentifygene-specificsignaturesaswellastemporal associations,pavingthewaytowardsaspatio-temporalviewof cell-fatetransitions(furtherdiscussedinSection3).
DespitetheimportantnumberofNR-regulatedgenesidentified inthesestudies,themajorlimitationofthis approachwasthat directNRtargetscouldnotbedistinguishedfromgenesthatwere indirectlyregulated.Topartiallycircumventthisproblem,theuse ofproteinsynthesisinhibitors,likecycloheximide,wasintroduced during the assays to avoid transcriptional regulation cascades progression.Inthiscontext,Harrisetal.inducedF9differentiation during6hinpresenceof ATRAandtheproteininhibitor cyclo-heximidewhichgaverisetotheidentificationof109significantly
Pleasecitethisarticleinpressas:Mendoza-ParraM-A,GronemeyerH.Genome-widestudiesofnuclearreceptorsincellfatedecisions.Semin CellDevBiol(2013),http://dx.doi.org/10.1016/j.semcdb.2013.07.001
ARTICLE IN PRESS
GModelYSCDB-1461; No.ofPages10
M.-A.Mendoza-Parra,H.Gronemeyer/SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx 3
Fig.1. Schematicrepresentationoftheretinoicacidsignalingtransductionprocessassessedbyglobalapproaches.Fromtoptobottom:cellfatetransitionisaconsequence ofaninitialcue[all-transretinoicacid(ATRA)andrelatedretinoidligands]thatinitiatessignaltransductionthroughtranscriptionregulationprimarilymediatedthrough thecorrespondingRAR/RXRreceptors.Theinitialsignaltransductioncascadeisdiversifiedoverseveralsignaltransductionlayers,whichtogetherspecifytheassociated cellfatetransition.Boththeprimarysignalingresponse,aswellasthefurthersignalingtransductionlayerscanbeevaluatedthroughglobalapproaches(rightsideofthe panel).Methodslistedforassessingthe3D-chromatinorganizationcorrespondstoHi-C(Highresolutionchromatinconformationcapture[74])andChIA-PET(Chromatin InteractionAnalysisbyPaired-endtagssequencing[72]).
differentiallyregulatedgenes[28].Whiletheuseofsuchinhibitor appears as an elegant way to identify in a selective manner primary/directtargets,thestudyperformedintheRA-inducedF9 differentiationsystemdemonstrated thatonly22 ofthemwere presentintheATRAcontrolassay,suggestingthattheothergenes areartifactuallyinducedbycycloheximidetreatment.
Further global transcriptome studies performed in different modelsystemsincorporatedsystematicallyearlyandlate treat-menttimepoints,undertheassumptionofadirectcorrelationwith putative primary/direct and secondary/indirect NR-responsive genes[32].Otherstookadvantageoftheavailabilityofspecific lig-andsasawaytorestricttheanalysistoagivensetofdifferentially regulatedgenes.Forinstance,thesyntheticpan-RARagonistTTNPB hasbeenusedtodecreasethe‘contamination’withgenes respon-dingtopermissiveRXRheterodimers,asall-transRAisomerizesto theRXRligand9-cisRA[33,34].
Considering that NRs can be expressed as different iso-types/isoforms,evaluatingtheirspecifictranscriptionalregulation cascades through global approaches became a crucial task for understandingthebiologicalroleofNRdiversity.Intheparticular caseoftheRAnuclearreceptors,RARsandRXRsareeachexpressed fromthethreeisotypicgenes(␣,and␥),whichexpressisoforms bydifferentialpromoterusageandsplicing[1].WhileallthreeRAR isotypeswereshowntobepresentinmodelsystemsliketheF9 ECCs,previousstudieshadalreadyprovidedevidenceforspecific rolesofsuchisotypes;forinstanceRXR␣/RAR␥heterodimer iso-typeisessentialforRA-inducedF9differentiation[35–37].
Inthiscontext,SuandGudasaimedatidentifyingthespecific role of RAR␥ by performing global gene expression profiling with wild-type and RAR␥−/− F9 cells in presence or absence of ATRA[38]. While theydemonstratedthat wildtype and the RAR␥−/−cellspresentedsimilarmorphologicalandproliferation
4 M.-A.Mendoza-Parra,H.Gronemeyer/SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx characteristicsin the absenceof RA treatment, important gene
expression differences were observed; this reveals limitations for theuse of RAR knockoutcells to decipher specific roles of RARisotypes.Similarobservationsmadeforglobaltranscription regulationstudiesperformedinRAR␣−/−F9cells[39]andin a gene-centricmannerforseveralRXRandRARisotypeknockouts [35,36]revealed“artifactual”ligandresponsesofspecificRXR–RAR heterodimersthussuggestingthatglobalstudieswithRXR–RAR knockoutcellsneedtobeinterpretedcarefully.
In summary,global approaches for assessing thechanges in geneexpressionregulationinthecontextofaliganded-NRfunction werewidelyusedinthepastprovidingaratherfastandaccurate waytoidentifygeneexpressionsignaturesinthestudiedsystems. Nevertheless,whilethesestudiesidentifieddifferentiallyregulated genes,theydidnotprovideinsightsintothegeneprogramming functionsofligandedNR(isotype)s.
2.2. Mappingthechromatinbindingsitesofnuclearreceptors NRsignalingisbasedontheircapacityofregulating transcrip-tion; thus thedissectionof the effects of NRs onphysiological processesrequiresthecomprehensivemappingoftheirdynamic interactionswiththechromatintoidentifyregulatedtargetgenes. As the DBD is the primary determinant of DNA interaction specificityalargenumberofstudieshasbeendevotedtothe under-standingofthesequence-specificandstructure determinantsof NRDBD–DNAinteraction([40–44];foralistofNRbindingsites see[45]).InthecaseofRAreceptors,invitrobindingand trans-activationstudiesdemonstratedthatRXR/RARheterodimersbind preferentiallytoinverted(IR)ordirectrepeat(DR)sequencesof thehexamericmotif (A/G)G(G/T)TCA,oftenspaced by5, 2or 1 nucleotide(DR5, DR2, DR1) due tothe dimerization character-isticsoftheDNAbindingdomain[41–43,46].Thischaracteristic RA-ResponsiveElement(RARE),hasbeenshowntopresentmajor divergencewhencomparedwiththeRXR/RARbindingsites asso-ciatedtowell-knownRA-inducedgenes[47],indicatingthatthe consensusRAREs maycorrespondto highaffinitybinding sites but occurrarelyin natural RAtarget genes.Indeed, high affin-itybindingsitescanbeisolatedbyco-immunoprecipitationwith DNAbutthosesitesarenotusedforgeneregulation,mostlikely becausetheyarenotaccessibleinthecorrespondingchromatin [48].Furthermore,mappingthepotentialRXR/RARbindingsites bycomparingtheconsensusRAREsdoesnottakeinconsideration additionalmechanisms,liketheepigeneticmechanismsthat regu-lateaccessofRXR/RARheterodimers[49,50]andsteroidreceptor homodimers[51],and/or thesynergisticinteraction withother NR/TFs[52].
Forthesereasons,thecurrentmethodofchoicefor comprehen-siveandunbiasedmappingoftheprotein-chromatininteractions istheuseofchromatinimmunoprecipitationcombinedwithhigh throughputsequencing.Notethatthehybridizationof immunopre-cipitatedchromatintomicroarraychips(knownalsoasChIP-chip) represented the first approaches for global mapping of NR-chromatininteraction.Infact,Delacroixetal.aimedatidentifying theRARbindingsitestodiscriminatebetweendirectandindirect RA-regulatedtargetsusing Taf4lox/− MEFs,which undergo mor-phologicalchangesuponRAtreatmentaccompaniedbyregulation of>1000genes[53].Afterintegrationof3xFlag-HAtaggedRAR␣ orRAR␥isotypestheyperformedChIPassaysandhybridizedthe IPedDNAtoAgilentpromotermicroarrays(ChIP-chip)[54]. Sur-prisingly,theyidentified∼300RAR-occupiedsitesofwhich<25% correspondedtodifferentiallyRA-regulatedgenes.Laterstudies performedin variousothersystems demonstrated thatonly an smallfractionof theRXR/RARbindingsites arelocated in pro-moterregions,thusexplaining,atleastpartially,thelowcorrelation
betweenRARoccupancyandgeneexpressionregulationobserved inthisstudy.
Infact,usingaconceptuallysimilarapproach,Huaetal.have integratedeGFP-taggedRAR␣orRAR␥intohumanMCF-7breast cancercellstocharacterizetheRARisotype-selectivelyregulated pathwaysimplicatedintheanti-proliferativeandapoptoticeffects of RA [55]. Importantly, the RAR-specific IPed chromatin was hybridizedtotilingarrayscontainingmorethan40million oligonu-cleotideprobesvirtuallyinterrogatingtheentirehumangenome. Undertheseconditions,theyfound>3000RAR␥and>7000RAR␣ bindingsitesrespectively,fromwhichmorethan85%ofthe iden-tifiedsiteswerelocatedinintronicorpromoter-distalintergenic regions.
In a similar manner a recent study withmouse embryonic stemcellsaimedatidentifyingtheRA-dependentgeneprograms involvedinneuronaldifferentiation[56],butincontrasttothe pre-viouslymentioned studies,thebindingofendogenous RARwas mappedbyusingapan-RARantibodyandcombiningChIPassays withmassiveparallelsequencing(ChIP-seq); thustheyavoided over-expressionand/or“artefactual”bindingoftaggedconstructs. This assay, performedbefore and after8hof ATRA treatment, revealedbothconstitutiveanddenovobindingsitesuponRA expo-sure,which werethen correlatedwithglobal microarray-based gene expression and RNA polymeraseII initiation and elonga-tion(assessedbyChIP-seq).Thefractionofdifferentiallyregulated genesassociatedwithRARbindingwasestimatedbyusinga5kb proximity criterion; this way only 15% of the identified bind-ing sites could be linked to a (transcriptionally active) coding region.Indeed,itisnowgenerallyacceptedthatdistalenhancers, which cannot be identified by simple binding site proximity, can regulateNR-responsive genes (furtherdiscussed in Section 3.2.3).
Ourownrecentstudyusedthewell-establishedF9modelto dissectthegeneregulatorypathwaysthatareresponsibleforthe RA-inducedendodermaldifferentiationbyintegratingtheglobal RAR bindingand gene regulation information from five differ-enttime-points duringthefirst48hafterRAexposure[57].For eachtime-pointthedifferentialtranscriptionalregulationhasbeen assessedaftertreatmentwithATRAor RAR␣,,␥-specific ago-nists.
GiventheessentialroleofRAR␥inF9celldifferentiation[37],we inferredtheRXR␣–RAR␥heterodimer-genomiclocationby map-pingbothheterodimercomponentsseparatelyatall5time-points (Fig.2A).Overall,RXR␣displayedmorebindingsitesthanRAR␥,as wasexpectedfromthepromiscuousheterodimerizationofRXR␣ withmultiplepartners.WhenevaluatingRXR␣and RAR␥ bind-ingsitesasheterodimercomponents,weidentifiedaconstitutive RXR␣–RAR␥bindingpopulationplusanotherpresentingahighly dynamicbehaviorduringATRAtreatment(Fig.2B).Infact,while theoverallnumbersofRXR␣–RAR␥bindingsitesdecreased dur-ingF9differentiation (∼2000 sites inthe absenceof treatment andlessthan1000sitesafter48hinpresenceofATRA)(Fig.2B andC),wedetectedsignificantamountsofdenovorecruited het-erodimersevenafter24or48hoftreatment,indicatingsustained andhighlydynamic interactionof theRXR␣–RAR␥heterodimer withchromatintargetsduringthiscellphysiologicalprocess. Unex-pectedly,theoveralldecreaseofRXR␣–RAR␥heterodimersbinding sitesdidnotcorrelatewiththeobservedamountsofRXR␣ bind-ingsites,thussuggestingthattheobserveddecreaseofthebinding sitesofRXR␣–RAR␥heterodimersmayresultfromanexchange withotherRXR␣heterodimersduringtheprocessofdifferentiation (Fig.2C).
ComparingRA-inducedgeneexpressionwiththereceptor bind-ing in a 10kb distance interval (Fig. 3A) we found that more than 50%of genesinduced duringthe first24hof ATRA treat-mentshowedaRXR␣oranRXR␣–RAR␥bindingsitewithin10kb
Please cite this article in press as: Mendoza-Parra M-A, Gronemeyer H. Genome-wide studies of nuclear receptors in cell fate decisions. Semin Cell Dev Biol (2013), http://dx.doi.org/10.1016/j.semcdb.2013.07.001
ARTICLE IN PRESS
G Model YSCDB-1461; No. of Pages 10 M.-A. Mendoza-Parra, H. Gronemeyer / Seminars in Cell & Developmental Biology xxx (2013) xxx – xxx 5Fig.2.RXR␣andRAR␥nuclearreceptorspresentahighlydynamicbindingtochromatinduringATRA-inducedF9differentiation.(A)TemporalrecruitmentofRAR␥andRXR␣inproximityoftheCyp26a1locus,asrevealed
byChIP-seq.Notethatbothreceptorsbindtotheidenticalchromatinlocusbutwithdifferentdynamics.(B)ThenumberglobalRXR␣–RAR␥bindingsites(definedbytheco-occurrenceofbothreceptors)areillustratedinthe
contextoftheirtemporalrecruitment,durationofoccupancyanddissociation.RXR␣–RAR␥co-occupiedsitespertimepointaresub-classifiedbasedontheirrecruitmentintervalsanddepictedbycolorcoding.(C)Schematic
modelillustratingthe(i)globalprogressivelossofRXR␣–RAR␥heterodimerasobservedinfigure(B);aswellas(ii)thatofRAR␥butnotofRXR␣fromchromatinbindingsitesobservedduringATRA-inducedF9differentiation;
6 M.-A.Mendoza-Parra,H.Gronemeyer/SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx
Fig.3.DifferentialgeneexpressioninducedbyretinoidsinF9embryonalcarcinomacells.(A)Schematicrepresentationillustratingthetranscriptionregulationactivity associatedtotheproximallocalizationofRXR␣–RARnuclearreceptors.NotethatgeneswereclassifiedasputativetargetgenesifatleastoneRXR␣orRXR␣–RAR␥binding sitewaslocatedinupto10kbdistance.(B)GenesexhibitingATRA-inducedorrepressedmRNAlevelsattheindicatedtimepointsduringF9celldifferentiation(induced genes≥1.8-fold;repressedgenes≤0.5-foldrelativetovehicle)wereclassifiedasputativetargetgenesfollowingthecriterionillustratedin(A).(C)RXR␣bindingsites classifiedintheirgenomiccontextdemonstratesthatmorethan70%ofthemarelocatedfarawayfromcodingregions(>10kbdistance;leftpanel),inadditiontotheirstrong preferenceforintergenicregions(rightpanel).(D)RXR␣–RAR␥ATRA-putativetargetgenesdescribedin(B)werefurtherclassifiedbasedontheirresponsetoRAR-specific agonists.TakeninconsiderationthatonlytheRAR␥agonist(BMS961)canreproducetheF9differentiationphenotypeobservedduringATRAtreatment,thecharacterized RXR␣–RAR␥ATRA-putativetargetgeneswerefurtherclassifiedas“dispensable”and“required”forinducingthedifferentiationphenotype.
proximity(Fig.3B).Incontrast,mostofthedown-regulatedgenes lacked such sites. Importantly, more than 70% of the mapped RXR␣ sites could not be associated to an annotated coding region(Fig.3C), suggesting that theymight regulate transcrip-tionthrough3-dimensionalchromatinstructuresormayregulate asyet non-annotated transcripts. Tofurther confirmthe direct transcriptionalregulationbythecharacterizedRXR␣–RAR␥ bind-ingsites,wecomparedthetranscriptionalresponsesinpresence of ATRA or RAR-specific agonists [57]. Importantly, 67% of the ATRA-inducedputativeRXR␣–RAR␥targetsdidrespondsimilarly tothedifferentiationcompetentRAR␥agonistBMS961. Surpris-ingly, also thetreatment with BMS753 (RAR␣-specific agonist) orBMS641(RAR-specificagonist) induceda responseofsome ATRA-RXR␣–RAR␥ targets,albeit only in a minor fraction(42% and6% of theATRA-inducedgenes, respectively).Thissuggests thatamongthecharacterizedATRA-dependentRXR␣–RAR␥ tar-gets(i)∼30%ofthemaredispensableforinducingtheobserved celldifferentiationphenotype;and (ii)fromthe remaining70% onlyathirdofthemareindeedessentialfordrivingthe differen-tiationprocess(Fig.3D). Importantly,insidethislastpopulation comprisesseveralTFs,likeFoxa1,Foxp1,Hoxa5,Hoxb5,Rarbor RXR␥,indicatingthat RA-signaltransductioninvokesthe induc-tionof“downstream”transcriptionfactors,whichinturnregulate signalingbifurcationeventstoyieldthefinaldifferentiated pheno-type.
3. NRgeneexpressionprogramsandtheirassociatedkey
factorsinvolvedinsignalingdiversification
3.1. StudyingNR-drivencellfatetransitionsasdynamicgene expressionprograms
Asmentioned above,the integrative analysisof global gene expression response to a given ligand and the corresponding NR-chromatin association can, in principle, identify an impor-tant proportion of the NR-mediated gene-regulatory events. Furthermore,theuseofdynamicbindingandtranscription infor-mation provided additional insight in molecular mechanisms occurringduringcell-fatetransition.Infact,thesestudiesrevealed (1) a highlydynamic target gene expression[28,29,57] and (2) similarly dynamic chromatin occupancyof pre-existing and de novo recruited RXR–RAR heterodimers, including heterodimer replacementorevenheterodimerpartnerswaps[57].Similar stud-iesperformedinothermodelsystemslike3T3-L1cellsintegrated theglobalchromatinlocalizationofRXRandPPAR␥NRswiththat assessedforRNApolymeraseIIduringtheinductionofadipocyte differentiation[58].LikeinthepreviouslydiscussedRA-induced F9differentiationstudy[57],thisintegrativeanalysisperformed in a temporal manner revealed a differential recruitment of PPARsandRXRduringadipogenesisandallowedtheclassification genes by their relative transcriptional activity assessed from
Pleasecitethisarticleinpressas:Mendoza-ParraM-A,GronemeyerH.Genome-widestudiesofnuclearreceptorsincellfatedecisions.Semin CellDevBiol(2013),http://dx.doi.org/10.1016/j.semcdb.2013.07.001
ARTICLE IN PRESS
GModelYSCDB-1461; No.ofPages10
M.-A.Mendoza-Parra,H.Gronemeyer/SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx 7 RNA polymerase IIbinding and thepresence of RXR/PPAR␥ in
proximity.
Despitethesefindings,thegainofinformationbythistypeof integrativeomicsanalysesremainsrestrictedtodirectly charac-terizethecorrespondingNR-regulatedtargetsandtheirpotential temporal changes,which representsonlya smallfractionofall differentiallyregulatedgenes.NR-drivencell-fatetransitionsare expectedtotakeplacethroughasignaltransductionmechanism, inwhichthedirecttargetsareinthefrontlineofthesignaling process (‘initiator program’) and the downstream layers com-prisetemporallyspecified(‘executor’)geneprogramsthatresult inamplification,diversificationandspecificationofthesignaling atdifferentlevels[57,59],whichultimatelyleadstotheemergence ofaspecificcellphenotype/functionality.
Thefirstssignaltransductionlayersaremediatedmainly,albeit notexclusivelybytranscriptionfactors(TFs).Whilethe reconstruc-tionoftheexecutorprogramsmayprofitfromthecharacterization ofthecascadeofTFsthatpropagatethesignaltransductionand diversificationprocess,itisvirtuallyimpossibletodirectly charac-terizetheseeventsinagivensystembyapplyingseriesofChIP-seq assaystargetingallpotentiallyrelatedTFs.Nevertheless,the avail-abilityinpublicrepositoriesofTFsinteractomesformultiplecell modelsrepresentsanimportantresourceforinsilicodataset inte-gration;notein this respect theimportant contributionbythe ENCODEconsortium[60].
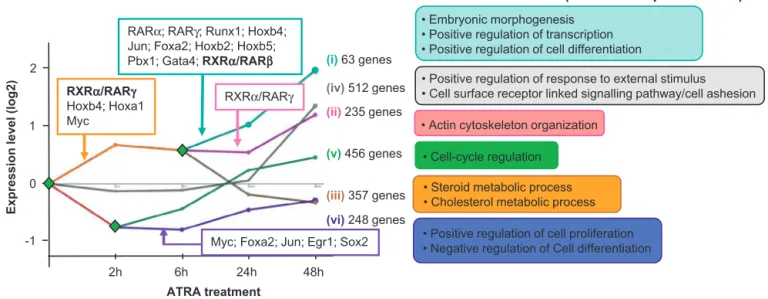
Infact,thedeconvolutionoftheRAsignalingpathwaysduring theF9induceddifferentiationhasbeenperformedbyintegrating TFtargetgeneannotations,includingtheidentifieddirectputative RXR␣–RAR␥targets,withtheATRA-inducedgeneprogramming [57].Thisanalysis,performedwiththeDynamicRegulatoryEvents Miner (DREM; [61]), predicted six distinct gene co-expression paths, which recapitulate the different subprograms generated duringtheRA-inducedsignaltransduction.Inadditiontoclassify thetemporalgeneexpressioninformationinco-expressionpaths, DREMevaluateswhetheragivenco-expressionpathisenriched for genesthat are annotated as targetsof a specific TF, whose action contributes tothe predictedbifurcation.In this manner, DREMpredicted3bifurcationpointsleadingtosignal diversifica-tionandassociatedtocandidateTFs.Asproof-of-principle,DREM associatedRXR␣–RAR␥withupregulatedsubprogramsvalidated bydifferentialgeneexpressionandthechromatin-bindingpattern ofRXR␣–RAR␥(Fig.4).Notably,DREMpredictedtranscription fac-torsoftheHomeoboxfamily(e.g.,Hoxa1,Hoxb2,Hoxb4,Hoxb5) andotherslikeRAR␣orFoxa2,tobeenrichedintheupregulated subprograms,whiletherepressedpathwasassociatewithTFslike Egr1[62]andSox2[63],previouslydescribedaspositively regulat-ingcellproliferationandstemcellpluripotency.
The predictedRA-induced co-expression paths were further evaluatedinthecontextofgeneco-citationinteractionsto con-struct the RA-driven RXR␣–RAR␥-mediated signaling network. This type of analysis provides a global view of the relevant genesinvolvedinsignaltransductionbyintegratinginformation extractedfromtheexistingliteratureonpreviouslyreported inter-actionsandprovidesacomprehensivewaytoassociatefunctional features tothepredictedsubprograms. Importantly,this analy-sisillustratesthecomplex temporalcoordination ofthevariety ofmolecularprocessesinvolvedinRA-induceddifferentiationand predictscriticalnodesassociatedwiththecellfatetransition initi-atedbyRA[57].
3.2. Additionalmechanismsinvolvedincontrollingthe retinoids-drivendynamicdiversifiedgeneprograms 3.2.1. MultipleRXR–RARheterodimersmediateRA-signaling
As mentioned above, the first level of signal diversification resultsfromthemultiplicityofRXR–RARcomplexesthatcanbe
formeddependingontheactualexpressionlevelsof6receptor isotypes(RXR␣;RXR;RXR␥RAR␣;RAR;RAR␥).Anenigmatic aspectcharacterizedin our studyconcerns thehighly dynamic bindingand the potential“heterodimercomponents swapping” ofRXR␣–RAR␥heterodimersduringtheRA-induced differentia-tionprocess.Whilethemethodologiesallowedhighlightingsuch phenomenon,itsbiologicalsignificanceremains elusive.Clearly, exploringtheroleofotherRXR/RARheterodimersduringthis pro-cesswillprovideinsightsintotheheterodimercross-functionalities asbasisfortheobserveddynamics.
SuchstudiesnecessitatereChIPassaystoprovidereliable infor-mationaboutco-occupancyoftheevaluatedheterodimerpartners at agiven chromatinsite. WhilereChIPassays werepreviously shown as a powerful method for evaluating simultaneous co-occupancy events in a locus-centric manner [64,65], their low yieldsarenotcompatiblewiththerequirementsforglobal ChIP-seqassays.Toovercomethisproblemwehaverecentlycombined reChIPswithlinearDNAamplification(LinDA-reChIP-seq)inorder todefinetheglobalbindingpatternofco-occupiedRXR␣andRAR␥ chromatinsitestopredictheterodimerbindingpatterns[66–68]. Usingsuchstrategies,thecomplexityofRXR–RARheterodimers canbedecorticatedtowardthecontributionsofthedifferent com-binationsofreceptors.
3.2.2. Epigeneticmodificationsandco-regulatorsestablish regulatoryprinciplesaffectingRA-regulatedgeneprograms upstreamanddownstreamofRXR–RARheterodimeraction
In addition to theTF-driven decisions for signal diversifica-tionprocessduringRA-induceddifferentiation,severaladditional factorsandregulatoryparadigmsmayimpactonprogram execu-tion.Infact,epigeneticmodificationofchromatinanditsinterplay withRAregulationhasalreadybeendemonstratedingene-centric studieswithPolycombproteinsandH3K27me3[49,69,70].Other epigeneticmodificationsmayalsoregulateNR’srecruitmentand theepigenetic actionof co-activator/co-integrators recruitedby ligandedRXR/RARheterodimers mayexertpioneeringactivities specifyingdownstreamprograms.
A novelmechanism of signalingpathwaydiversification has beenreportedrecentlybyCeschinetal.[59].Studyingestrogen receptor(ER␣)signalinginbreastcancercellstheyanalyzedthe roleoftwoepigeneticfactors,thehistoneacetyltransferase(HAT) CBP and the methyltransferase CARM1/PRMT4. Both CBP and CARM1bindtotheSRC/p160co-activators andareco-recruited by agonist-bound ER␣ tochromatintargets. Based onprevious knowledge that CARM1 methylates CBP at specific arginine residues [71] and the observation that CBP methylation was exclusivelyCARM1-dependent,theymappednotonlythebinding siterepertoiresofER␣,SRC3,CBP,CARM1andacetylatedhistones (H3K18ac)butalsothemethylatedCBPspeciesusingantibodies thatrecognizeselectivelythemethylatedCBPresidues[59]. Inter-estingly,thefirstobservationwasthatmethylationatR2151was requiredforestrogen-dependentrecruitmentofCBPtochromatin. Themolecularbasis forthisrequirementisnotknownanditis unclearifthisisageneralphenomenonorrestrictedtocertaincell (types). Moreover, the subsequent multi-dimensional analysis, whichincludedatime-seriesoftranscriptomics,identifieddistinct “hubs”ofER␣targetgenes.Thesehubsdifferedbytherecruitment oftheparticularmethyl-CBPspecies(whichvaryinHATactivities), thusindicatingthatthecrosstalkbetweenco-recruitedepigenetic factorscanleadtopathwaydiversification.Itwillbeinteresting toassesswhetherthesedifferenthubs,aswellasthose formed bytheotherHATp300correspondtofunctionallyrelatedtarget genesandwhethera“methyl-HATcode”mayindeedexist.
Theabovestudiesshowthatcomprehensivemulti-dimensional omics-derivedinformationtogetherwiththebioinformaticstools todefine dynamic gene regulatorynetworks by integrating NR
8 M.-A.Mendoza-Parra,H.Gronemeyer/SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx 0 2 -1 1 48h 24h 6h 2h Ex p re ss ion le v el ( log 2) ATRA treatment (i)63 genes (iv)512 genes (ii)235 genes (v)456 genes (iii)357 genes (vi)248 genes • Embryonic morphogenesis •Positive regulation of transcription • Positive regulation of cell differentiation
•Steroid metabolic process • Cholesterol metabolic process
•Positive regulation of response to external stimulus • Cell surface receptor linked signalling pathway/cell ashesion
• Cell-cycle regulation
•Positive regulation of cell proliferation • Negative regulation of Cell differentiation • Actin cytoskeleton organization
Asso
ciated
GO ter
ms (enr
ichment
p-value<10
-2)
RXRα/RARγ
Hoxb4; Hoxa1 Myc
RARα; RARγ; Runx1; Hoxb4; Jun; Foxa2; Hoxb2; Hoxb5; Pbx1; Gata4; RXRα/RARβ
RXRα/RARγ
Myc; Foxa2; Jun; Egr1; Sox2
Fig.4. DynamicregulatorymapofATRA-inducedtranscriptome.DREMco-expressionanalysis;color-codedpathssummarizecommoncharacteristics.Diamondsindicate predictedbifurcationpointswhichgiverisetothedifferentco-expressionpaths;transcriptionfactorswhosetargetgenesareover-enrichedinagivenpathareillustrated. Thenumberofgenesperco-expressionpath,aswellastheirrelevantGeneOntologytermsisdisplayedontheright.
chromatinbindingpatterns,epigenomesandtranscriptomeswill shedsignificantlightonthemolecularmechanisms,keyfactors anddecisionpointsthatdefinedecisionsspecifyingcellfateand cellfunction.
3.2.3. Thethree-dimensionalchromatinorganizationandits dynamicchangesdrivenbyNR-signaling
Thedesignationof NRtargetgenes fromChIP-seqstudies is generallybased onlinearproximitycriteria. Howeverthelarge majorityofbindingsitesarelocatedinintergenicregionsandthus, onlya smallfractionofall identifiedbindingeventsare gener-allyconsideredinsuchanalyses.Thefunctionoftheseintergenic bindingsiteshasbecomemuchclearerfromrecentstudies interro-gatingthe3-dimensionalorganizationofchromatininthenucleus; forinstanceintheparticularcaseofER␣[72].Itisnowgenerally acceptedthatthechromatinarchitecture,i.e.,theorganizationof chromatinin“loops’,‘domains’andpossibly‘factories’with dedi-catedfunctionalities,correspondstoastructuralorganizationthat specifiesthephysicalinteractionbetweenpromotersanddistant regulatoryelements, sometimes with the involvement of non-codingRNAs.Indeed,theentirenucleushastobeconsideredasa regulatorynetworkofitsown[73].Importantly,thecombinationof proximityligation-mediatedassayswithmassiveparallel sequenc-ingprovidedthetechnologytoanalyzethisarchitectureglobally (Hi-C[74]; TCC[75]).Furthermore,theincorporationof a prior immunoprecipitationstepduringtheseassaysallowstostudysuch 3D-chromatinorganizationinassociationwithagivensignaling orregulatory/processing component(ER␣[72]; CTCF [76]; RNA polymeraseII[77]).Yet,thedynamicaspectofnuclear architec-tureinprocesses likeRA-induceddifferentiationorthechanges ofnucleararchitectureinrelatedpathologies,withitssubsequent consequencesonsignalingofthediseasedcell/organ,hasnotyet beenaddressed.Itisinterestingtonoteinthisrespectthatlinks betweenchromatinarchitectureandfeaturesofcancercellsare emerging[78,79].
4. FuturedirectionsinthestudyofNR-drivencellfate
transition
Howcanthestructuralinformationthatispresentinasimple chemical molecule, likeall-trans retinoic acid(ATRA), be ‘read’ toset-up the sequence of temporally controlled events, which
finally lead to the cell-physiological changes that characterize a differentiated cell? Our previous study performed with the embryocarcinomaF9cellmodelsystemprovidedforthefirsttime asystems biologyviewoftheATRA-inducedsignalingpathway diversificationthrough differentregulatory decisions character-izedatdifferenttime-pointsduringdifferentiation[57].Yetthe viewoftheretinoicacid(RA)-inducedsignaltransductionevents inferredfromthisstudyisfarfrombeingcomprehensive.Thisisin partduetothereducednumberofmoleculareventsthatcouldbe importedinthespatio-temporalomicsdataanalyses(discussed above), but also a consequence of technicalconstraints related tothecomplexityof thesystemwhichoperates withuptosix receptorsandmultipleheterodimers.
Therapiddevelopmentofnext-generationsequencing(NGS) technologiesposesmultiplechallengesforthebioinformatics anal-ysesoftheenormousamountsofdatathataregatheredbymassive parallelsequencing.Whileinthepastyearsseveralcomputational effortsaimingtoassessthelocalenrichmentconfidenceinsingle NGS-generatedprofileshavebeenreported,anumberofkeyissues concerningmethodologiesformulti-profilecomparisonsare lack-ingorareonlyincompletelyaddressed.Asdiscussedabove,the useofintegrativegenomicsapproachesmaybecomethe method-ologyofchoicefordecorticatingtheNR-drivensignaltransduction events;thustheimplementationofsuitedcomputational meth-odsfocusedonenhancingtheconfidenceinomicsdataassessment atthetimeoftheirintegrationrepresentsanessentialaspectto considerforthiskindofstudies.Importantly,futuredataset analy-sesofNR-drivendifferentiationstudieswillneedtointegratetwo majoradditionalelements:(i)thethree-dimensional chromatin structure revealed bymethodologies like Hi-C(High resolution chromatin conformation capture [74]) or ChIA-PET (Chromatin InteractionAnalysisbyPaired-endtagssequencing[72])and(ii)the temporalnatureoftheevaluatedeventsthroughouttheinduced (cell physiological) process. Importantly, such spatio-temporal analysis willprovide information aboutNR binding withinthe chromatin architecture, the chromatin modification status and nucleosomeoccupancy,and theobserved differential transcrip-tional/translational activity in a given physiological context. In addition,computationalmethodsforreconstructingthedynamic regulatorygenenetworksmaybeappliedwiththeaimof infer-ringtheATRA-inducedsignalingpathwaydiversificationthrough temporallydefinedregulatorydecisionsasillustratedinprevious
Pleasecitethisarticleinpressas:Mendoza-ParraM-A,GronemeyerH.Genome-widestudiesofnuclearreceptorsincellfatedecisions.Semin CellDevBiol(2013),http://dx.doi.org/10.1016/j.semcdb.2013.07.001
ARTICLE IN PRESS
GModelYSCDB-1461; No.ofPages10
M.-A.Mendoza-Parra,H.Gronemeyer/SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx 9 studies[57,61,80].Thesestudieshaveprovidedaninitialinsight
intotheenormouscomplexitythatwearefacingalreadyinmodel systems,suchasstemcells,whentryingtounderstandata molec-ularandmechanisticlevelthe4-dimensionalhierarchiesthatare initiatedbyasingleinducerandgoverncellfatetransition.
References
[1]LaudetV,GronemeyerH.Thenuclearreceptorfactsbook.SanDiego:Academic Press;2002.
[2]deLeraAR,BourguetW,AltucciL,GronemeyerH.Designofselectivenuclear receptormodulators:RARandRXRasacasestudy.NatureReviewsDrug Dis-covery2007;6:811–20.
[3]GronemeyerH,GustafssonJA,LaudetV.Principlesformodulationofthenuclear receptorsuperfamily.NatureReviewsDrugDiscovery2004;3:950–64.
[4]GermainP,ChambonP,EicheleG,EvansRM,LazarMA,LeidM,etal. Inter-nationalunionofpharmacology.LXIII.RetinoidXreceptors.Pharmacological Reviews2006;58:760–72.
[5]GermainP,ChambonP,EicheleG,EvansRM,LazarMA,LeidM,etal. Inter-nationalunionofpharmacology.LX.Retinoicacidreceptors.Pharmacological Reviews2006;58:712–25.
[6]SopranoDR,QinP,SopranoKJ.Retinoicacidreceptorsandcancers.Annual ReviewofNutrition2004;24:201–21.
[7]AltucciL,GronemeyerH.Thepromiseofretinoidstofightagainstcancer. NatureReviewsCancer2001;1:181–93.
[8]AltucciL,LeibowitzMD,OgilvieKM,deLeraAR,GronemeyerHRAR.RXR modulationincancerandmetabolicdisease.NatureReviewsDrugDiscovery 2007;6:793–810.
[9]BookoutAL,JeongY,DownesM,YuRT,EvansRM,MangelsdorfDJ.Anatomical profilingofnuclearreceptorexpressionrevealsahierarchicaltranscriptional network.Cell2006;126:789–99.
[10]EvansRM,BarishGD,WangYX.PPARsandthecomplexjourneytoobesity. NatureMedicine2004;10:355–61.
[11]MakishimaM,LuTT,XieW,WhitfieldGK,DomotoH,EvansRM,etal.Vitamin Dreceptorasanintestinalbileacidsensor.Science2002;296:1313–6.
[12]ShulmanAI,MangelsdorfDJ.Retinoidxreceptorheterodimersinthemetabolic syndrome.NewEnglandJournalofMedicine2005;353:604–15.
[13]RepaJJ,MangelsdorfDJ.TheliverXreceptorgeneteam:potentialnewplayers inatherosclerosis.NatureMedicine2002;8:1243–8.
[14]Bourguet W,GermainP, GronemeyerH.Nuclearreceptor ligand-binding domains:three-dimensionalstructures,molecularinteractionsand pharma-cologicalimplications.TrendsinPharmacologicalSciences2000;21:381–8.
[15]BourguetW,RuffM,ChambonP,GronemeyerH,MorasD.Crystalstructure oftheligand-bindingdomainofthehumannuclearreceptorRXR-␣.Nature 1995;375:377–82.
[16]BourguetW,VivatV,WurtzJM,ChambonP,GronemeyerH,MorasD.Crystal structureofaheterodimericcomplexofRARandRXRligand-bindingdomains. MolecularCell2000;5:289–98.
[17]leMaireA,TeyssierC,ErbC,GrimaldiM,AlvarezS,deLeraAR,etal.Aunique secondary-structureswitchcontrolsconstitutivegenerepressionbyretinoic acidreceptor.NatureStructuralandMolecularBiology2010;17:801–7.
[18]WurtzJM,BourguetW,RenaudJP,VivatV,ChambonP,MorasD,etal.A canonicalstructurefortheligand-bindingdomainofnuclearreceptors.Natural StructuralBiology1996;3:206.
[19]RochelN,CiesielskiF, GodetJ,MomanE,RoessleM,Peluso-IltisC,etal. CommonarchitectureofnuclearreceptorheterodimersonDNAdirectrepeat elementswithdifferentspacings.NatureStructuralandMolecularBiology 2011;18:564–70.
[20]Berrabah W,AumercierP,LefebvreP,StaelsB.Control ofnuclear recep-toractivitiesinmetabolismbypost-translationalmodifications.FEBSLetters 2011;585:1640–50.
[21]ChenW,RoederRG.Mediator-dependentnuclearreceptorfunction.Seminars inCellandDevelopmentalBiology2011;22:749–58.
[22]PerissiV,JepsenK,GlassCK,RosenfeldMG.Deconstructingrepression:evolving modelsofco-repressoraction.NatureReviewsGenetics2010;11:109–23.
[23]MarkM,GhyselinckNB,ChambonP.Functionofretinoidnuclearreceptors: lessons fromgeneticandpharmacologicaldissectionsoftheretinoicacid signalingpathwayduringmouseembryogenesis.AnnualReviewof Pharma-cologyandToxicology2006;46:451–80.
[24]HeldringN,PikeA,AnderssonS,MatthewsJ,ChengG,HartmanJ,etal.Estrogen receptors:howdotheysignalandwhataretheirtargets.PhysiologicalReviews 2007;87:905–31.
[25]YorkB,ReinekeEL,SagenJV,NikolaiBC,ZhouS,LouetJF,etal.Ablationofsteroid receptorcoactivator-3resemblesthehumanCACTmetabolicmyopathy.Cell Metabolism2012;15:752–63.
[26]YorkB,O’MalleyBW.Steroidreceptorcoactivator(SRC)family:mastersof systemsbiology.JournalofBiologicalChemistry2010;285:38743–50.
[27]BalmerJE,BlomhoffR.Geneexpressionregulationbyretinoicacid.Journalof LipidResearch2002;43:1773–808.
[28]HarrisTM,ChildsG.GlobalgeneexpressionpatternsduringdifferentiationofF9 embryonalcarcinomacellsintoparietalendoderm.FunctionalandIntegrative Genomics2002;2:105–19.
[29]WeiY,HarrisT,ChildsG.Globalgeneexpressionpatternsduringneural differ-entiationofP19embryoniccarcinomacells.Differentiation2002;70:204–19.
[30]LyonsP.Advancesinspottedmicroarrayresourcesforexpressionprofiling. BriefingsinFunctionalGenomicsandProteomics2003;2:21–30.
[31]WoodsCG,HeuvelJP,RusynI.Genomicprofilinginnuclearreceptor-mediated toxicity.ToxicologicPathology2007;35:474–94.
[32]EifertC,Sangster-GuityN,YuLM,ChitturSV,PerezAV,TineJA,etal.Global geneexpressionprofilesassociatedwithretinoicacid-induced differentia-tionofembryonalcarcinomacells.MolecularReproductionandDevelopment 2006;73:796–824.
[33]ArimaK,ShiotsuguJ,NiuR,KhandpurR,MartinezM,ShinY,etal.Global anal-ysisofRAR-responsivegenesintheXenopusneurulausingcDNAmicroarrays. DevelopmentalDynamics2005;232:414–31.
[34]MamoonA,Ventura-HolmanT,MaherJF,SubausteJS.Retinoicacidresponsive genesinthemurinehepatocytecelllineAML12.Gene2008;408:95–103.
[35]ChibaH,CliffordJ,MetzgerD,ChambonP.Specificandredundantfunctions ofretinoidXReceptor/Retinoicacidreceptorheterodimersindifferentiation, proliferation,andapoptosisofF9embryonalcarcinomacells.JournalofCell Biology1997;139:735–47.
[36]ChibaH,CliffordJ,MetzgerD,ChambonP.DistinctretinoidXreceptor-retinoic acidreceptorheterodimersaredifferentiallyinvolvedinthecontrolof expres-sionofretinoidtargetgenesinF9embryonalcarcinomacells.Molecularand CellularBiology1997;17:3013–20.
[37]TanejaR,RoyB,PlassatJL,ZusiCF,OstrowskiJ,ReczekPR,etal.Cell-type andpromoter-contextdependentretinoicacidreceptor(RAR)redundancies forRARbeta2andHoxa-1activationinF9andP19cellscanbeartefactually generatedbygeneknockouts.ProceedingsoftheNationalAcademyofSciences oftheUnitedStatesofAmerica1996;93:6197–202.
[38]Su D, Gudas LJ. Gene expressionprofiling elucidates a specific role for RARgammaintheretinoicacid-induceddifferentiationofF9teratocarcinoma stemcells.BiochemicalPharmacology2008;75:1129–60.
[39]LaursenKB,WongPM,GudasLJ.EpigeneticregulationbyRARalpha main-tains ligand-independent transcriptional activity. Nucleic Acids Research 2012;40:102–15.
[40]MaderS,ChenJY,ChenZ,WhiteJ,ChambonP,GronemeyerH.Thepatterns ofbindingofRAR,RXRandTRhomo-andheterodimerstodirectrepeatsare dictatedbythebindingspecificitesoftheDNAbindingdomains.EMBOJournal 1993;12:5029–41.
[41]Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature 1995;375: 203–11.
[42]ZechelC,ShenXQ,ChambonP,GronemeyerH.Dimerizationinterfacesformed betweentheDNAbindingdomainsdeterminethecooperativebindingof RXR/RARandRXR/TRheterodimerstoDR5andDR4elements.EMBOJournal 1994;13:1414–24.
[43]ZechelC,ShenXQ,ChenJY,ChenZP,ChambonP,GronemeyerH.The dimer-izationinterfacesformedbetweentheDNAbindingdomainsofRXR,RARand TRdeterminethebindingspecificityandpolarityofthefull-lengthreceptors todirectrepeats.EMBOJournal1994;13:1425–33.
[44]HuangP,ChandraV,RastinejadF.Structuraloverviewofthenuclear recep-torsuperfamily:insightsintophysiologyandtherapeutics.AnnualReviewof Physiology2010;72:247–72.
[45]Cotnoir-WhiteD,LaperriereD,MaderS.Evolutionoftherepertoireofnuclear receptorbindingsitesin genomes.Molecularand CellularEndocrinology 2011;334:76–82.
[46]UmesonoK,MurakamiKK,ThompsonCC,EvansRM.Directrepeatsas selec-tiveresponseelementsforthethyroidhormone,retinoicacid,andvitaminD3 receptors.Cell1991;65:1255–66.
[47]LaleveeS,AnnoYN,ChatagnonA,SamarutE,PochO,LaudetV,etal. Genome-wideinsilicoidentificationofnewconservedandfunctionalretinoicacid receptorresponseelements(directrepeatsseparatedby5bp).Journalof Bio-logicalChemistry2011;286:33322–34.
[48]RudertF,GronemeyerH.Retinoicacid-responseelementswithahighly repet-itivestructureisolatedbyimmuno-selectionfromgenomicDNA.Journalof SteroidBiochemistryandMolecularBiology1993;46:121–33.
[49]KashyapV,GudasLJ,BrenetF,FunkP,VialeA,ScanduraJM.Epigenomic reorga-nizationoftheclusteredHoxgenesinembryonicstemcellsinducedbyretinoic acid.JournalofBiologicalChemistry2011;286:3250–60.
[50]KashyapV,GudasLJ.Epigeneticregulatorymechanismsdistinguishretinoic acid-mediatedtranscriptionalresponsesinstemcellsandfibroblasts.Journal ofBiologicalChemistry2010;285:14534–48.
[51]Garcia-BassetsI,KwonYS,TeleseF,PrefontaineGG,HuttKR,ChengCS,etal. Histonemethylation-dependentmechanismsimposeliganddependencyfor geneactivationbynuclearreceptors.Cell2007;128:505–18.
[52]Ross-InnesCS,StarkR,HolmesKA,SchmidtD,SpyrouC,RussellR,etal. Coop-erativeinteractionbetweenretinoicacidreceptor-alphaandestrogenreceptor inbreastcancer.GenesandDevelopment2010;24:171–82.
[53]FadlounA,KobiD, DelacroixL, DembeleD, MichelI, Lardenois A,etal. RetinoicacidinducesTGFbeta-dependentautocrinefibroblastgrowth. Onco-gene2008;27:477–89.
[54]DelacroixL,MoutierE,AltobelliG,LegrasS,PochO,ChoukrallahMA,etal. Cell-specificinteractionofretinoicacidreceptorswithtargetgenesinmouse embryonicfibroblastsandembryonicstemcells.MolecularandCellularBiology 2010;30:231–44.
[55]HuaS,KittlerR,WhiteKP.Genomicantagonismbetweenretinoicacidand estrogensignalinginbreastcancer.Cell2009;137:1259–71.
10 M.-A.Mendoza-Parra,H.Gronemeyer/SeminarsinCell&DevelopmentalBiologyxxx (2013) xxx–xxx [56]MahonyS,Mazzoni EO,McCuine S,Young RA,Wichterle H,Gifford DK.
Ligand-dependentdynamicsofretinoicacidreceptorbindingduringearly neu-rogenesis.GenomeBiology2011;12:R2.
[57]Mendoza-ParraMA,WaliaM,SankarM,GronemeyerH.Dissectingthe retinoid-induceddifferentiationofF9embryonalstemcellsbyintegrativegenomics. MolecularSystemsBiology2011:7.
[58]Nielsen R,PedersenTA, HagenbeekD, MoulosP, SiersbaekR, MegensE, etal. Genome-wideprofilingofPPARgamma:RXRandRNApolymerase II occupancyrevealstemporalactivationofdistinctmetabolicpathwaysand changesinRXRdimercompositionduringadipogenesis.Genesand Develop-ment2008;22:2953–67.
[59]CeschinDG,WaliaM,WenkSS,DuboeC,GaudonC,XiaoY,etal. Methyl-ationspecifiesdistinctestrogen-inducedbindingsiterepertoiresofCBPto chromatin.GenesandDevelopment2011;25:1132–46.
[60]DunhamI,KundajeA,AldredSF,CollinsPJ,DavisCA,DoyleF,etal.Anintegrated encyclopediaofDNAelementsinthehumangenome.Nature2012;489:57–74.
[61]ErnstJ,VainasO,HarbisonCT,SimonI,Bar-JosephZ.Reconstructingdynamic regulatorymaps.MolecularSystemsBiology2007;3:74.
[62]MinIM,PietramaggioriG,KimFS,PassegueE,StevensonKE,WagersAJ.The transcriptionfactorEGR1controlsboththeproliferationandlocalizationof hematopoieticstemcells.CellStemCell2008;2:380–91.
[63]OrkinSH,WangJ,KimJ,ChuJ,RaoS,TheunissenTW,etal.Thetranscriptional networkcontrollingpluripotencyinEScells.ColdSpringHarborSymposiaon QuantitativeBiology2008;73:195–202.
[64]ChayaD,HayamizuT,BustinM,ZaretKS.TranscriptionfactorFoxA(HNF3)on anucleosomeatanenhancercomplexinliverchromatin.JournalofBiological Chemistry2001;276:44385–9.
[65]MetivierR,PenotG,HubnerMR,ReidG,BrandH,KosM,etal.Estrogen receptor-alphadirectsordered,cyclical,andcombinatorialrecruitmentofcofactorson anaturaltargetpromoter.Cell2003;115:751–63.
[66]Mendoza-Parra MA, Shankaranarayanan P, Gronemeyer H. Sequential chromatin immunoprecipitation protocol for global analysis through massive parallel sequencing (reChIP-seq). Protocol Exchange 2011,
http://dx.doi.org/10.1038/protex.2011.1256.
[67]ShankaranarayananP,Mendoza-ParraMA,vanGoolW,TrindadeLM, Grone-meyerH.Single-tubelinearDNAamplificationforgenome-widestudiesusing afewthousandcells.NatureProtocols2012;7:328–38.
[68]ShankaranarayananP,Mendoza-ParraMA,WaliaM,WangL,LiN,TrindadeLM, etal.Single-tubelinearDNAamplification(LinDA)forrobustChIP-seq.Nature Methods2011;8:565–7.
[69]AmatR,GudasLJ.RARgammaisrequiredforcorrectdepositionandremovalof Suz12andH2A.Zinembryonicstemcells.JournalofCellularPhysiology2010.
[70]GillespieRF,GudasLJ.Retinoidregulatedassociationoftranscriptional co-regulatorsandthepolycomb groupproteinSUZ12 withtheretinoicacid responseelementsofHoxa1,RARbeta(2),andCyp26A1inF9embryonal carci-nomacells.JournalofMolecularBiology2007;372:298–316.
[71]Chevillard-BrietM,TroucheD,VandelL.ControlofCBPco-activatingactivity byargininemethylation.EMBOJournal2002;21:5457–66.
[72]Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. Anoestrogen-receptor-alpha-boundhumanchromatininteractome.Nature 2009;462:58–64.
[73]FelsenfeldG,DekkerJ.Genomearchitectureandexpression.CurrentOpinion inGeneticsandDevelopment2012;22:59–61.
[74]vanBerkumNL,Lieberman-AidenE,WilliamsL,ImakaevM,GnirkeA,MirnyLA, etal.Hi-C:amethodtostudythethree-dimensionalarchitectureofgenomes. JournalofVisualizedExperiments2010.
[75]KalhorR,TjongH,JayathilakaN,AlberF,ChenL.Genomearchitecturesrevealed bytetheredchromosomeconformationcaptureandpopulation-based model-ing.NatureBiotechnology2012;30:90–8.
[76]HandokoL,XuH,LiG,NganCY,ChewE,SchnappM,etal.CTCF-mediated functional chromatin interactome in pluripotent cells. Nature Genetics 2011;43:630–8.
[77]LiG,FullwoodMJ,XuH,MulawadiFH,VelkovS,VegaV,etal.ChIA-PETtoolfor comprehensivechromatininteractionanalysiswithpaired-endtag sequenc-ing.GenomeBiology2010;11:R22.
[78]RickmanDS,SoongTD,MossB,MosqueraJM,DlabalJ,TerryS,etal. Oncogene-mediatedalterationsinchromatinconformation.ProceedingsoftheNational AcademyofSciencesoftheUnitedStatesofAmerica2012;109:9083–8.
[79]Schuster-BocklerB,LehnerB.Chromatinorganizationisamajorinfluenceon regionalmutationratesinhumancancercells.Nature2012;488:504–7.
[80]DimitrakopoulouK,TsimpourisC,PapadopoulosG,PommerenkeC,WilkE, SgarbasKN,etal.Dynamicgenenetworkreconstructionfromgeneexpression datainmiceafterinfluenzaA(H1N1)infection.JournalofClinical Bioinformat-ics2011;1:27.