HAL Id: inserm-00667508
https://www.hal.inserm.fr/inserm-00667508
Submitted on 7 Feb 2012HAL is a multi-disciplinary open access archive for the deposit and dissemination of sci-entific research documents, whether they are pub-lished or not. The documents may come from teaching and research institutions in France or abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est destinée au dépôt et à la diffusion de documents scientifiques de niveau recherche, publiés ou non, émanant des établissements d’enseignement et de recherche français ou étrangers, des laboratoires publics ou privés.
Heymann
To cite this version:
Kanji Mori, Martine Berreur, Fréderic Blanchard, Catherine Chevalier, Isabelle Guisle-Marsollier, et al.. Receptor activator of nuclear factor-kappaB ligand (RANKL) directly modulates the gene expression profile of RANK-positive Saos-2 human osteosarcoma cells.. Oncology Reports, Spandidos Publications, 2007, 18 (6), pp.1365-71. �inserm-00667508�
Abstract. Receptor activator of nuclear factor κB (RANK)/ RANK ligand (RANKL)/osteoprotegerin (OPG) are the key regulators of bone metabolism. Recent findings demonstrated a crucial role of RANK in several bone-associated tumors. Indeed, we have recently demonstrated functional RANK expression both in a mouse and several human osteosarcoma cell lines. However, RANKL effects on osteosarcoma cells remain to be determined. In this study, we determined RANKL effects on RANK-positive Saos-2 human osteosarcoma cells. cDNA microarray and quantitative RT-PCR analyses clearly demonstrated that RANK-positive osteosarcoma cells were the target of RANKL as well as osteoclasts/osteoclast precursors. Thus, we present for the first time that RANKL can directly and significantly modulate gene expression of RANK-expressing Saos-2 cells. RANKL-modulated genes included genes that were implicated in protein metabolism, nucleic acid metabolism, intracellular transport, cytoskeleton organization and biogenesis, apoptosis and signaling cascade. Our results strengthen the involvement of the RANK/RANKL/OPG axis in osteosarcoma biology and capability to identify novel therapeutic approaches targeting RANK-positive osteo-sarcomas.
Introduction
Osteosarcoma is the most frequent malignant primary bone tumor. Some important pathogenetic roles of p53, RB and
mdm2 have been already reported (1); however the
patho-genesis of osteosarcoma and mainly the role of the bone microenvironment in cancer cell biology are not fully under-stood. Therefore, understanding the biological mechanisms that govern osteosarcoma development at the molecular level should lead to the determination of new potential therapeutic targets.
The discovery of key factors involved in the control of osteoclastogenesis has moved bone research into a new era. The most notable of these factors belong to the tumor necrosis factor (TNF)/TNF receptor family: receptor activator of nuclear factor κB (RANK/TNFRSF11A), its ligand RANKL/TNFSF11 and decoy receptor for RANKL, osteoprotegerin (OPG/TNFRSF11B) (2-4). Consequently, RANKL has been shown both to mediate osteoclastogenesis and activate mature osteoclasts, whereas OPG negatively regulates RANKL binding to RANK, reduces the half-life of membranous RANKL, therefore inhibiting bone resorption induced by osteoclasts (5). RANK/RANKL/OPG axis is the key regulator of bone metabolism not only in normal but also pathological conditions. Indeed, bone-related tumors including osteosarcoma are very often associated with dysregulated RANK/RANKL/OPG axis leading to altered bone remodeling (6,7). RANK has also attracted special attention because a functional RANK expression has been reported in several bone-associated tumors (8-11). Interestingly, RANKL triggered migration of human prostate cancer cells (8,9), breast cancer cells and melanoma cells that express RANK (9). Recently, we have reported functional RANK expression in a mouse (POS-1 cells) (10) and several human osteosarcoma cell lines (11). All these findings suggest the major involvement of RANK/RANKL/OPG axis in osteosarcoma which appears to be one of the most relevant and confidential therapeutic targets.
In the present study, we analyzed the RANKL effect on RANK-positive human osteosarcoma cells using Saos-2 cells that expresses functional RANK.
Receptor activator of nuclear factor-
κκ
B ligand
(RANKL) directly modulates the gene expression profile
of RANK-positive Saos-2 human osteosarcoma cells
KANJI MORI1,5, MARTINE BERREUR1,2, FRÉDERIC BLANCHARD1,2,
CATHERINE CHEVALIER3, ISABELLE GUISLE-MARSOLLIER3, MARTIAL MASSON4, FRANÇOISE RÉDINI1,2 and DOMINIQUE HEYMANN1,2,5
1Université de Nantes, Nantes Atlantique Universités, Laboratoire de Physiopathologie de la Résorption Osseuse
et Thérapie des Tumeurs Osseuses Primitives, EA3822; 2INSERM, ERI 7; 3INSERM U533;
4INSERM U791; 5University Hospital of Nantes, Nantes, F-44035, France
Received June 28, 2007; Accepted August 2, 2007
_________________________________________
Correspondence to:Dr D. Heymann or Dr K. Mori, Université de Nantes, Nantes Atlantique Universités, Laboratoire de Physio-pathologie de la Résorption Osseuse et Thérapie des Tumeurs Osseuses Primitives, EA3822, Nantes, F-44035, France
E-mail: dominique.heymann@univ-nantes.fr kanchi@belle.shiga-med.ac.jp
Key words:RANK, RANKL, osteosarcoma, bone tumor, bone microenvironment, gene modulation
Cell culture. The human osteosarcoma cell line Saos-2 was
purchased from the American Tissue Cell Collection (LGC Promochem, Molsheim, France). Saos-2 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Bio Whittaker, Verviers, Belgium) supplemented with 10% fetal calf serum (FCS) (Hyclone-Perbio, Brebières, France) at 37˚C in a humidified atmosphere (5% CO2and 95% air).
Cell proliferation and viability assays. Cell proliferation was
determined as previously reported with slight modifications (8). Briefly, Saos-2 cells were seeded at the density of 2x103
cells per well into a 96-multiwell plate, and cultured for 72 h in DMEM supplemented with 0.5% FCS in the absence or presence of recombinant human RANKL (rhRANKL) kindly provided by Amgen Inc. (Thousand Oaks, CA, USA) (5, 50, 100 ng/ml). The medium was changed every 24 h and the cell proliferation was determined by an XTT based method, using Cell Proliferation Kit II (Sigma, Saint-Quentin Fallavier, France) following to the supplier's recommendations. In addition, trypan-blue exclusion was used to quantify the viable and dead cells. Saos-2 cells were seeded into a 24-multiwell plate (5x103cells/well) and cultured in DMEM supplemented
with several FCS concentrations (0.5-10%) in the absence or presence of 5 to 100 ng/ml rhRANKL, and then the viable and dead cell number was counted at days 1, 4 and 7 under a light microscope.
Cell migration analyses by slit assay. Cell migration analyses
were performed as previously described with slight modifi-cations (8). Briefly, Saos-2 cells were seeded at the density of 40x103cells per well into a 24-multiwell plate, and cultured
in DMEM supplemented with 1% FCS. At the time of confluence, cells were incubated in the absence or presence of rhRANKL (5, 50 and 100 ng/ml) for 24 h. Then, a slit was made horizontally with a white tip at the centre of each confluent well, the medium was changed after gentle rinse and cells were cultured for 24 h with or without rhRANKL (5, 50 and 100 ng/ml). Cell invasion on the slit of the confluent well was assessed in each condition by light micro-scope.
cDNA microarray. Total RNAs were extracted using TRIzol
reagent (Invitrogen, Eragny, France) from Saos-2 osteosarcoma cell line cultured in the absence (control condition, n=3) or presence of 50 ng/ml rhRANKL (treatment condition, n=3) for 24 h. One microgram of total RNA was amplified using the Amino Allyl MessageAmp™ II-aRNA amplification Kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions: 2 amplifications were performed from each control condition (n=6), 3 amplifications from each treatment condition (n=9). Cy3- and Cy5-labeled Amino-allyl RNA (aaRNA) samples were prepared using the CyDye Post Labeling Reactive Dye Pack (Amersham BioSciences, Uppsala, Sweden). aaRNA samples from treatment condition were labeled with Cy3. The same amount of control aaRNA was pooled, labeled with Cy5 and used as the reference. For each step of the preparation, sample quality was assessed by capillary electrophoresis with Bioanalyser 2100 (Agilent
sample was mixed with an equal amount of Cy5-labeled reference and then hybridized to the microarrays. Micro-arrays were prepared by INSERM U533 Laboratory (Dr J. Léger, Nantes, France) using 50-mer oligonucleotide probes (MWG Biotech, Hebersberg, Germany). The 6864 genes, spotted in triplicate on the microarrays, were composed of relevant gene collections already selected by teams from the West of France specialized in studies of various types of cancer or their related mechanisms (for further infor-mation see: http://cardioserve.nantes.inserm.fr/ptf-puce/ cancerochips_en.php) (12). Hybridized arrays were scanned at 10-μm resolution with a Scanarray 3000 (Packard Bio-sciences, Wellesley). Data were analyzed with GenePix Pro 4.0 (Axon Union City, CA, USA). Raw data were normalized with the Madscan application (12,13) according to the instructions. Genes lacking valid expression values for more than one array were eliminated for all conditions. Two-class impaired Significance Analysis of Microarrays (SAM) was used to identify genes with statistically significant dif-ferential expression between control and treatment conditions (14).
Quantitative real-time RT-PCR. Quantitative real-time
RT-PCR (qRT-PCR) was performed in the Stratagene Mx3000p (Stratagene, CA, USA) using SYBR Green I dye and Sure Start Taq Polymerase (Stratagene), according to the manufacture's recommendations. The primers were designed with Beacon Designer 5.0 (Premier Biosoft). Validation of primers was done by optimization of each couple of primers between 50-900 nM each. Fluorescence was measured at the end of extension period by ramping from 60 to 95˚C (0.2˚C step) to generate a melting curve for each set of primers. The lowest Ctfound for a set of primer
was used the determination in qPCR and checked in a 2% agarose gel. qPCRs were performed with the ‘Comparative qPCR’ program of the MxPro software (Stratagene). Reaction was achieved with 1X BrillantR SYBERRGreen Master Mix
(Stratagene), appropriated volumes of each primer, 30 nM final concentration of SYBERRGreen, 5 μl of cDNA diluted
1:20. Amplification and detection were performed using the Mx3000P system (Stratagene), with ß-actin as normalizer and controls as calibrators. The following profile: 1 cycle of 95˚C for 10 min and 40 cycles each of 95˚C for 30 sec, 60˚C for 1 min, and 72˚C for 30 sec was applied for all experiments. Fluorescence was measured at the end of annealing period of each cycle to monitor the amplification and was plotted in real-time manner. qPCR analysis was automatically done by the software. Comparative quantification was calculated with the formula of Pfaffl et al (15). Discrepancies were corrected with serial dilutions generating a standard curve, then if an efficiency of amplification between 95 and 105% was found, the difference between the Genes of Interest (GOI) and the Normalizer (housekeeping) genes fold change results can be obtained using Comparative Quantification algorithms.
Statistical analyses. Mann-Whitney's U test was employed
when appropriate. Results with p<0.05 were considered significant.
Results
Cell proliferation, viability and cell migration assays. Neither
XTT assay nor manual cell counting could demonstrate any significant difference of Saos-2 cell proliferation and viability after rhRANKL treatment (data not shown). Moreover, rhRANKL did not modulate Saos-2 cell migration in the experimental conditions used (data not shown).
RANKL-induced gene modulations in RANK-positive Saos-2 cells. Three independent cDNA microarray experiments clearly
demonstrated that RANKL was a powerful modulator of genes expressed by osteosarcoma cells (Fig. 1A). Thus, 69 genes out of 6,864 genes analyzed, showed significantly different levels of expression in rhRANKL-treated Saos-2 cells compared to the control group; 48 were down-regulated whereas the remaining 21 were up-regulated (Fig. 1B and Table I). The down-regulated group involved some genes implicated in protein
metabolism, nucleic acid metabolism, intracellular transport, cytoskeleton organization and biogenesis and apoptosis and signaling cascade. In the up-regulated group, the main genes affected by RANKL as referred to ontology biological pro-cesses were nucleic acid and protein metabolisms (Table I).
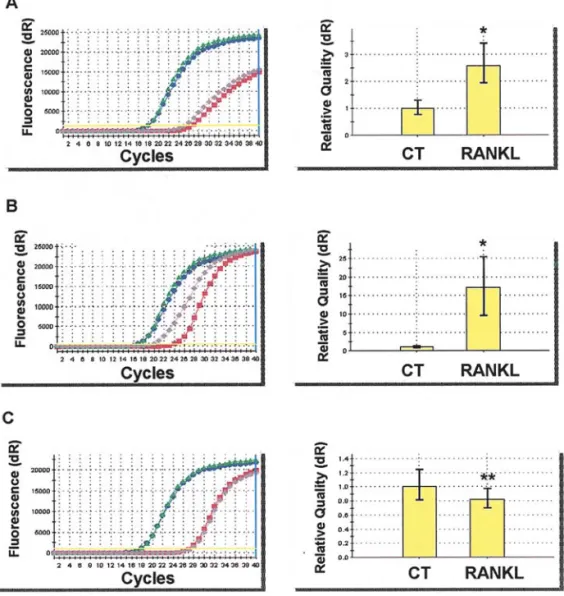
To confirm the results of cDNA microarray analysis, 10% of rhRANKL-modulated genes were measured by qRT-PCR. For instance, ROCK1 (Rho associated, coiled-coil containing protein kinase 1) and SEMA3A (Semaphorin 3A), significantly up-regulated by rhRANKL as revealed by microarray analysis (1.21- and 1.43-fold respectively) were also markedly up-regulated in qRT-PCR assay (2.59- and 17.1-fold, respectively, p<0.001) (Fig. 2). Furthermore, the expression of GDF15 (growth differentiation factor 15) was down-regulated 0.78-fold in microarray analysis and around 0.8-fold in qRT-PCR (p<0.05) (Fig. 2). Correspondingly, other results obtained through cDNA microarray analysis were also confirmed by qRT-PCR.
Figure 1. RANKL modulated the expression of 69 genes in human osteosarcoma Saos-2 cell line. (A) Two-way hierarchical clustering of the human osteosarcoma Saos-2 cell line and the 69 genes differentially expressed between control and RANKL-stimulated condition. The position of each is mentioned on the right. Expression values are indicated by color coding: red > grey > green. For each gene, the expression values were median centered and therefore represent relative expression ratios. (B) Gene Ontology classification of the genes differentially regulated by RANKL in Saos-2 cells. Representation of the genes up- and down-regulated by RANKL is according to the biological process categories (as defined by the Gene Ontology Consortium).
–––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––
Full name Genebank Gene Map Fold
accession symbol location change no.
––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––– 1 Aggrecan 1 M55172 AGC1 15q26.1 0.82 2 Adenylate kinase 5 BC036666 AK5 1p31 0.82 3 Apolipoprotein B mRNA editing enzyme NM_004900 APOBEC3B 22q13.1-q13.2 0.76 4 Attractin NM_139321 ATRN 20p13 0.83 5 Complement component 1, s subcomponent NM_001734 C1S 12p13 0.84 6 Chromosome 20 open reading frame 64 NM_033550 C20orf64 20q13.2 0.85 7 Calumenin NM_001219 CALU 7q32 0.82 8 Chaperonin containing TCP1, subunit 7 (eta) NM_006429 CCT7 2p13.2 0.85 9 Chromosome 19, cosmid R32469, complete sequence AC005197 chr19cosmidR324 19p12 0.86 10 CSE1 chromosome segregation 1-like (yeast) AF053640 CSE1L 20q13 0.81 11 Dystonia 1, torsion (autosomal dominant; torsin A) AF007871 DYT1 9q34 0.81 12 Enolase 1, (alpha) X84907 ENO1 1p36.3-p36.2 0.83 13 F-box only protein 32 NM_058229 FBXO32 8q24.13 0.86 14 Hypothetical protein FLJ23467 AF271774 FLJ23467 1q21.3 0.87 15 Fusion [involved in t(12;16) in malignant liposarcoma] NM_004960 FUS 16p11.2 0.87 16 Growth differentiation factor 15 BC000529 GDF15 19p13.1-13.2 0.78 17 Glutamate-ammonia ligase (glutamine synthase) BC031964 GLUL 1q31 0.83 18 Growth factor receptor-bound protein 2 NM_002086 GRB2 17q24-q25 0.82 19 Heterogeneous nuclear ribonucleoprotein A/B NM_031266 HNRPAB 5q35.3 0.87 20 Keratin 8 NM_002273 KRT8 12q13 0.84 21 Tubulin, beta polypeptide paralog BC001352 MGC8685 6p25 0.78 22 Myopalladin AK027343 MYPN 10q22.1 0.80 23 Nucleosome assembly protein 1-like 4 NM_005969 NAP1L4 11p15.5 0.82 24 NADH dehydrogenase (ubiquinone) 1 alpha NM_004542 NDUFA3 19q13.42 0.86
subcomplex, 3, 9 kDa
25 2',5'-oligoadenylate synthetase 1, 40/46 kDa NM_002534 OAS1 12q24.1 0.78 26 Ornithine decarboxylase antizyme 2 NM_002537 OAZ2 15q22.1 0.86 27 Beta 5-tubulin NM_178014 OK/SW-cl.56 6p21.32 0.78 28 Procollagen-proline, 2-oxoglutarate 4-dioxygenase
(proline 4-hydroxylase), alpha polypeptide I M24486 P4HA1 10q21.3-q23.1 0.85 29 Protein phosphatase 1, catalytic subunit, alpha isoform NM_002708 PPP1CA 11q13 0.83 30 Hypothetical protein PRO1855 NM_018509 PRO1855 17q21.33 0.79 31 Proteasome 26S subunit, non-ATPase, 4 NM_002810 PSMD4 1q21.3 0.85 32 Proteasome 26S subunit, non-ATPase, 8 NM_002812 PSMD8 19q13.13 0.82 33 Ribophorin I NM_002950 RPN1 3q21.3-q25.2 0.85 34 SeryltRNA synthetase NM_006513 SARS 1p13.3-p13.1 0.86 35 Small EDRK-rich factor 2 NM_005770 SERF2 15q15.1 0.84 36 GMP synthase (glutamine-hydrolyzing), mRNA XM_167338 siGMPsLOC222152 7 0.84 37 Importin alpha-2 subunit, mRNA XM_070941 siImportinalpha 9 0.84 38 Similar to tropomyosin 4, mRNA XM_088391 simtotropomyosi 8 0.74 39 Solute carrier family 2 (facilitated glucose transporter), K03195 SLC2A1 1p35-p31.3 0.77
member 1
40 Solute carrier family 2 (facilitated glucose transporter), AF481879 SLC2A14 12p13.31 0.81 member 14
41 Stress-induced-phosphoprotein 1 (Hsp70/Hsp90- NM_006819 STIP1 11q13 0.86 organizing protein)
Discussion
This is the first report on direct gene modulations by RANKL in RANK-positive human osteosarcoma cells, Saos-2. RANK expressed by osteoclasts/osteoclast precursors is recognized as the key molecule involved in osteoclastogenesis and mature osteoclast activation (4). However, its expression is not restricted to the osteoclastic lineage as it is also demonstrated
in other tissues including mammary gland, heart, lung and skeletal muscle (4). Furthermore, recent finding clearly suggested a pivotal role of RANK in bone-associated tumors by presenting RANKL-triggered RANK-positive cell migration (8,9). Moreover, a positive correlation has been reported between constant expressions of RANK with decreased/absent expression of RANKL and a high metastatic phenotype in breast carcinoma (16). We have recently demonstrated Table I. Continued.
–––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––
Full name Genebank Gene Map Fold
accession symbol location change no.
––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––– 42 Synaptogyrin 3 NM_004209 SYNGR3 16p13 0.74 43 Testis enhanced gene transcript (BAX inhibitor 1) NM_003217 TEGT 12q12-q13 0.84 44 Thymosin, beta 4, Y-linked NM_004202 TMSB4Y Yq11.221 0.75 45 Translocase of outer mitochondrial membrane 34 NM_006809 TOMM34 20q12-q13.1 0.77 46 Tropomyosin 3 AB062125 TPM3 1q21.2 0.81 47 Tripartite motif-containing 5 NM_033034N TRIM5 11p15 0.82 48 ZW10 interactor NM_032997 ZWINT 10q21-q22 0.85 49 ASF1 anti-silencing function 1 homolog A NM_014034 ASF1A 6q22.31 1.21
(S. cerevisiae)
50 Axotrophin AK022973 AXOT 2q24.2 1.18 51 Carnitine deficiency-associated gene expressed in AF078932 CDV-1 12q24.13 1.15
ventricle 1
52 Kinesin family member 18A AB062483 DKFZP434G2226 11p14.1 1.33 53 Ets variant gene 5 (ets-related molecule) NM_004454 ETV5 3q28 1.18 54 Hypothetical protein FLJ20249 NM_015590 FLJ20249 1q22 1.18 55 Histamine N-methyltransferase NM_006895 HNMT 2q22.1 1.18 56 IDN3 protein NM_133433 IDN3 5p13.2 1.23 57 Likely ortholog of mouse immediate early BC021102 LEREPO4 2q32.2 1.21
response, erythropoietin 4
58 Neurogenic differentiation 6 NM_022728 NEUROD6 7p15.1 1.21 59 Nuclear factor IB U70862 NFIB 9p24.1 1.24 60 Oculocerebrorenal syndrome of Lowe NM_001587 OCRL Xq25-q26.1 1.21 61 Protein kinase, cAMP-dependent, regulatory, NM_002736 PRKAR2B 7q22 1.17
type II, beta
62 Rho-associated, coiled-coil containing protein NM_005406 ROCK1 18q11.2 1.21 kinase 1
63 Semaphorin 3A L26081 SEMA3A 7p12.1 1.43 64 C-myc purine-binding transcription factor (PUF), XM_070869 siNuclLOC138342 9 1.16
mRNA
65 Solute carrier family 7 (cationic amino acid AB040875 SLC7A11 4q28-q32 1.35 transporter)
66 Sterol Oacyltransferase 2 AF099031 SOAT2 12q13.13 1.22 67 Translocation protein 1 AB024586 TLOC1 3q26.2-q27 1.19 68 Tetratricopeptide repeat domain 3 NM_003316 TTC3 21q22.2 1.26 69 Zinc finger protein 281 NM_012482 ZNF281 1q32.1 1.27 –––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––––
functional RANK expression in several human osteosarcoma cell lines (11). RANK expression in bone-associated tumors is therefore very hot spot of tumor-bone biology.
Alternatively, as bone environment is rich in RANKL, RANKL can bind to RANK expressed on osteosarcoma cells in a paracrine (soluble RANKL) and/or juxtacrine (membrane RANKL) manner. Therefore, RANKL can act as activator of two targets in this tumoral bone environment: one is osteo-clasts/osteoclast precursors and the other is RANK-positive osteosarcoma cells. RANKL activates osteoclasts/osteoclast precursors and increases osteoclastic activity leading to disturbed bone remodeling and then releases several tumor-supportive growth factors. This interaction resides between pathologic bone remodeling and osteosarcoma development creates a vicious cycle (17). In high grade osteosarcoma, this vicious cycle should be more accelerated because of its increased RANKL/OPG ratio (7). Such phenomenon has been well documented in osteolytic bone tumors (6-8); however recent findings suggested the importance of osteoclast functions in osteoblastic bone metastasis (18,19). RANK also activates RANK-positive osteosarcoma cells and induce gene modulations. In Saos-2 cells, RANKL up-regulated the expression of genes such as SEMA3A and axotrophin that
exert immunosuppressive activity (20,21). Interestingly, knockout of the SEMA3A gene induces abnormal bone and cartilage development (22). It has been also reported that SEMA3A signaling molecules are in a position to modulate the vascularization of bone, and the innervation of osteoblasts and osteoclasts during bone development and remodeling (23). In this respect, RANKL appears as a deleterious factor thereby facilitating tumor progression. Namely, RANKL might have a propensity for osteosarcoma development by synergistic effect of osteoclasts activity acting as a ‘soil’ factor in bone environment suggested in other bone-associated tumors (8,9). In addition, RANKL-induced SEMA3A modulation might play, at least in part, the osteoblastic profile of osteosarcoma.
On the contrary, RANKL appears as a protective factor against osteosarcoma development by modulating other genes involved in the cellular metabolism. For instance, RANKL down-regulated genes encoding proteasome 26S and ribophorin I, known to reduce the proteasomal degradation machinery (24,25) and GDF-15 that is associated with early prostate carcinogenesis (26). In addition, RANKL up-regulated NF-IB which is potentially implicated in cell morphology and susceptibility to nuclear oncogenes (27).
Figure 2. Representative results of quantitative real-time RT-PCR (qRT-PCR). To confirm the results of cDNA microarray analysis, 10% of RANKL-modulated genes were measured by qRT-PCR as described in Materials and methods. (A) ROCK1 (Rho associated, coiled-coil containing protein kinase 1), (B) SEMA3A (Semaphorin 3A) and (C) GDF15 (growth differentiation factor 15).*p<0.0001, **p<0.05, by Mann-Whitney's U test.
Moreover, except for these direct RANKL-induced gene modulations in RANK-positive osteosarcoma cells, RANKL could be involved as a tumor development protector, as RANKL can act as a potent immune activator by inhibiting dendritic cell apoptosis (28).
Further experiments are needed to determine the balance between pro- and anti-tumor activities of RANKL in osteo-sarcoma that could provide new therapeutic approaches targeting RANK-positive osteosarcoma.
Acknowledgements
This work was supported by The Région des Pays de la Loire and by a grant from the West Committee of the Ligue Contre le Cancer. Kanji Mori received a personal fellowship from the Ligue Nationale Contre le Cancer. We thank Dr J. Léger (INSERM U533) and Dr J. Guicheux (INSERM U791) for their discussions concerning the design and the interpretation of the cDNA microarrays and the qRT-PCR.
References
1. Miller CW, Aslo A, Won A, Tan M, Lampkin B and Koeffler HP: Alterations of the p53, Rb and MDM2 genes in osteosarcoma. J Cancer Res Clin Oncol 122: 559-565, 1996.
2. Simonet WS, Lacey DL, Dunstan CR, et al: Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell 89: 309-319, 1997.
3. Lacey DL, Timms E, Tan HL et al: Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93: 165-176, 1998.
4. Theoleyre S, Wittrant Y, Tat SK, Fortun Y, Redini F and Heymann D: The molecular triad OPG/RANK/RANKL: involvement in the orchestration of pathophysiological bone remodeling. Cytokine Growth Factor Rev 15: 457-475, 2004. 5. Tat SK, Padrines M, Theoleyre S, Couillaud-Battaglia S,
Heymann D, Redini F and Fortun Y: OPG/membranous -RANKL complex is internalized via the clathrin pathway before a lysosomal and a proteasomal degradation. Bone 39: 706-715, 2006.
6. Terpos E, Szydlo R, Apperley JF, et al: Soluble receptor activator of nuclear factor kappaB ligand-osteoprotegerin ratio predicts survival in multiple myeloma: proposal for a novel prognostic index. Blood 102: 1064-1069, 2003.
7. Grimaud E, Soubigou L, Couillaud S, et al: Receptor activator of nuclear factor kappaB ligand (RANKL)/osteoprotegerin (OPG) ratio is increased in severe osteolysis. Am J Pathol 163: 2021-2031, 2003.
8. Mori K, Le Goff B, Charrier C, Battaglia S, Heymann D and Redini F: DU145 human prostate cancer cells express functional receptor activator of NFkappaB: new insights in the prostate cancer bone metastasis process. Bone 40: 981-990, 2007. 9. Jones DH, Nakashima T, Sanchez OH, et al: Regulation of
cancer cell migration and bone metastasis by RANKL. Nature 440: 692-696, 2006.
10. Wittrant Y, Lamoureux F, Mori K, Riet A, Kamijo A, Heymann D and Redini F: RANKL directly induces bone morphogenetic protein-2 expression in RANK-expressing POS-1 osteosarcoma cells. Int J Oncol 28: 261-269, 2006.
11. Mori K, Le Goff B, Berreur M, et al: Human osteosarcoma cells express functional receptor activator of nuclear factor-kappa B. J Pathol 211: 555-562, 2007.
12. Le Meur N, Lamirault G, Bihouee A, et al: A dynamic, web-accessible resource to process raw microarray scan data into consolidated gene expression values: importance of replication. Nucleic Acids Res 32: 5349-5358, 2004.
13. Tseng GC, Oh MK, Rohlin L, Liao JC and Wong WH: Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res 29: 2549-2557, 2001.
14. Tusher VG, Tibshirani R and Chu G: Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116-5121, 2001.
15. Pfaffl MW, Horgan GW and Dempfle L: Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36, 2002.
16. Bhatia P, Sanders MM and Hansen MF: Expression of receptor activator of nuclear factor-kappaB is inversely correlated with metastatic phenotype in breast carcinoma. Clin Cancer Res 11: 162-165, 2005.
17. Guise TA: The vicious cycle of bone metastases. J Musculoskelet Neuronal Interact 2: 570-572, 2002.
18. Garnero P, Buchs N, Zekri J, Rizzoli R, Coleman RE and Delmas PD: Markers of bone turnover for the management of patients with bone metastases from prostate cancer. Br J Cancer 82: 858-864, 2000.
19. Whang PG, Schwarz EM, Gamradt SC, Dougall WC and Lieberman JR: The effects of RANK blockade and osteoclast depletion in a model of pure osteoblastic prostate cancer metastasis in bone. J Orthop Res 23: 1475-1483, 2005.
20. Lepelletier Y, Moura IC, Hadj-Slimane R, et al: Immuno-suppressive role of semaphorin-3A on T cell proliferation is mediated by inhibition of actin cytoskeleton reorganization. Eur J Immunol 36: 1782-1793, 2006.
21. Metcalfe SM: Axotrophin and leukaemia inhibitory factor (LIF) in transplantation tolerance. Philos Trans R Soc Lond B Biol Sci 360: 1687-1694, 2005.
22. Behar O, Golden JA, Mashimo H, Schoen FJ and Fishman MC: Semaphorin III is needed for normal patterning and growth of nerves, bones and heart. Nature 383: 525-528, 1996.
23. Gomez C, Burt-Pichat B, Mallein-Gerin F, et al: Expression of Semaphorin-3A and its receptors in endochondral ossification: potential role in skeletal development and innervation. Dev Dyn 234: 393-403, 2005.
24. Mani A and Gelmann EP: The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol 23: 4776-4789, 2005. 25. Elsasser S, Gali RR, Schwickart M, et al: Proteasome subunit
Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol 4: 725-730, 2002.
26. Cheung PK, Woolcock B, Adomat H, et al: Protein profiling of microdissected prostate tissue links growth differentiation factor 15 to prostate carcinogenesis. Cancer Res 64: 5929-5933, 2004. 27. Schuur ER, Kruse U, Iacovoni JS and Vogt PK: Nuclear factor I interferes with transformation induced by nuclear oncogenes. Cell Growth Differ 6: 219-227, 1995.
28. Wong BR, Josien R, Lee SY, Sauter B, Li HL, Steinman RM and Choi Y: TRANCE [tumor necrosis factor (TNF)-related activation-induced cytokine], a new TNF family member predominantly expressed in T cells, is a dendritic cell-specific survival factor. J Exp Med 186: 2075-2080, 1997.