Nucleic Acids
Research,
Vol.18,
No. 10 2861Regulatory elements in the first intron contribute
to
transcriptional
regulation
of
the
f33
tubulin
gene
by
20-hydroxyecdysone
in
Drosophila Kc cells
A.Bruhat,
S.Tourmente, S.Chapel,
M.L.Sobrier,
J.L.Couderc
and
B.Dastugue
Laboratoire
de Biochimie
Medicale,
CJF INSERM 88.06, UFR de
Medecine
BP
38,
63001
Clermont-Ferrand,
Cedex
France
Received March
9,
1990;Revised
andAccepted
April 19,
1990ABSTRACT
We have studied the
transcriptional regulation
of the(3
tubulin
gene bythe
steroidhormone
20-hydroxyecdysone
(20-OH-E)
inDrosophila
Kccells.
A series ofhybrid
genes with varyingtubulin
gene lengthsdriving
thebacterial
chloramphenicol acetyl
transferase(CAT)
gene were constructed.The
promoteractivity
wasassayed
after transient
expression
inKc
cells,
inthe
presence orabsence
of20-OH-E. We find that
0.911Kb
upstreamfrom the
transcription
startsite contain
one orseveralhormone
independent positive cis-acting elements, responsible
for
theconstitutive
expression of the
O3
tubulin
gene. In thelarge (4.5
Kb) first intronof
this
gene, we identifiedadditionnal hormone dependent
negative andpositive
regulatory elements, which
can act inboth
directions and in aposition-independence
manner.Then,
thenegative intron element(s), which repress thetranscription
in theabsence
of20-OH-E has
characteristics
ofsilencer.
INTRODUCTION
Many eukaryotic genes require cis acting regulatory elements
for transcription regulation. These elements appear to act as
binding sites fortrans-acting factors which stimulate (positive regulatory elements) or repress (negative regulatory elements)
promoter activity (1). In mammalians, some of them, called
Hormone Regulatory Elements (HRE) are steroid receptors
binding sites involvedinthetranscription regulation by steroid
hormones. These HREarepalindromic sequencesand
generally
exhibit the properties ofinducible promoter enhancers(2)(3).
20-hydroxyecdysone (20-OH-E) is an hormonal steroid ofcrucial importance for insectdevelopment. In
Drosophila,
the20-OH-E responseseems to be similar tothat of other steroid
response systems, and require cis
regulatory
elements calledEcdysone Responsive Elements (EcRE). These EcRE are the
presumed binding sites ofthe ecdysone receptor which is not
actuallycompletely characterized
(4).
Infact the mechanismby
which 20-OH-E induces
transcription
isnot aswell understoodasthe mammalian steroidmechanisms. The four small heat-shock
genes ofDrosophilarepresent the mostextensive studies of genes
whichrespond to 20-OH-E. Indeed, sequences found inhsp22,
hsp23, hsp26 and hsp27 promoters have been shown to be
necessary andsufficientforthe 20-OH-E induction of these genes
(5-8). Promoter sequence analysis of these heat-shock genes
and other20-OH-E regulated genes such asthe Sgs4 gene (9)
does not reveal a consensus sequence for the binding of the
hormone receptor. FurthermoreDrosophila EcRE binding site
sequencesonlyshowpartial sequence homology whencompared
to mammalian steroid receptor binding sites sequences.
Kc cultured Drosophila cells constitute a very convenient
systemfor studying the molecular mechanism of transcription
regulationby20-OH-E. Inthese cells,20-OH-Einduces dramatic
changesatthemorphological level that reflect changesatthegene
expression level (10). 20-OH-E regulates in Kc cells the
expression of one of the four (3 tubulin genes (1 l).The 60C (3
tubulin gene encodes, after hormonal treatment, for the
03
subunit (12). Previous work in Kc cells hasdescribedthe structure
of the gene anddemonstrated that its regulation is at least, in
part, at the level oftranscription (13).
Inorder to study themolecularmechanism of the transcription
activation triggered by 20-OH-E in Kccells,wedescribed in this
work,ourattempts to map andidentifycis-actingelement(s)that areinvolved in 20-OH-E regulated expression of the
03
tubulingene. Wehaveconstructed aseriesofhybridgeneswithvarying
gene lengths driving the bacterial chloramphenicol acetyl
transferase (CAT)gene. Inthe presence or absence of
20-OH-E, the promoteractivity following transfection in Kc cells was
assayed.
We show that elements responsible for the constitutive expression lie within 0.22 and 0.91 Kb upstream from the
transcriptionstartsite. We also showthat thesequences necessary
for 20-OH-E regulation of theexpressionofthe(3tubulin gene
are located downstream from the initiation site of the
transcription. The first intron particularly includes negative regulatory element(s) (NRE) intervening in the transcription
inhibition in the absence of 20-OH-E and positive regulatory
element(s) stimulating the promoter activity after hormonal
treatment. Thesecis-acting intronelementscan actupstreamor
2862 Nucleic Acids Research, Vol. 18, No. 10
MATERIALS AND METHODS Cell culture and hormone treatment
The Drosophila cell line used in this studywasderived from the
linesestablished by Echalier and Ohanessian (14). This Kc 167
linewas grown at 23°C in D22 medium with 2% foetal calf
serum. 20-hydroxyecdysone (SIMES)wasaddedtocell cultures
froma 10mMstock in 95 % ethanoltoa final concentration of
IAM.
The hormonal response was checked by observing themorphological modifications of the cells (15).
Construction of CAT fusion genes
To constructchimeric CAT expressionconstructs, the plasmid
pBLCAT3 (16) wasused. Fusions between the CATgene and
the
03
5' flanking region were performed within the 5'untranslatedregion, atthe AvaIl siteinthe (3tubulin firstexon.
A 0.36 Kb BamHI-AvaII fragment, a 1.03 Kb PstI-AvaII and
a6.1 KbEcoRI-AvaLI fragment containing respectively0.22Kb,
0.91Kband6 Kb of the 335'flanking regionwerepurified and
subcloned inpBLCAT3 to create pT-0.22, pT-0.91 and pT-6.
Forthat, these fragmentswererespectivelysubcloned in
BamHI-BglIH, PstI-BglIl and HindIH-BglIl sites of pBLCAT3, after
conversion of cohesive ends intobluntends andligation. Inorder
toobtainpTI-4.5,a6.25KbPstI-BamHIfragmentincluding 0.91
Kb of the 5' flanking region, the firstexon (0.26Kb) and the
beginningofthe first intron(1.64 Kb), a2.8 KbBamHI fragment
including the centralpartofthefirst intron anda0.13 Kb
BamHI-PvuIl fragment includingthe 3' part of the first intron(0.06 Kb)
and apartof the second exon (0.07 Kb) were used for the in
frame fusion via PstI and BglIl sites inpBLCAT3. From this
construct, a 3.4 KbHindIII-XbaIinternal intronfragment was
removed and the deleted plasmid was ligated afterconversion
of cohesive ends into blunt endsto createpTI-1.1. To construct
pTDI-1.1(+)and(-),aXhoI 1.1 Kbfragment, which contains
most of the (3 first deleted intron, was removed from the
previousconstruct,blunt ended and insertedinbothorientations,
downstream from the CATgene into theSmaIsite ofpT-0.91.
Thisfragment wasalso subcloned into theSmaI siteofpUC18
toobtainpUCI-1.1.UsingtheHindIHandEcoRIpolylinkersites
ofpUC 18, itwasremoved and insertedin the natural orientation
into theHindI site ofpT-0.91, upstream from the 5' flanking
region,to createpTUI-1.1(+).pUCI-l.1 wasusedtocleave the
1.1 Kb deleted intron fragmentinto twofragments of 0.6 and
0.5 Kbbyusingthe Sallsites within the intron and thepUC18
polylinker. These0.6 and 0.5 Kb SalI fragments wereinserted
downstream from the CATgeneinto theSmaI siteofpT-0.91,
respectivelyinthe natural orientationto createpTDI-0.6(+),and
inboth orientations to create pTDI-0.5(+) and (-).
Cell transfection and CAT assays
Cells were transfected using the calcium phosphate DNA
coprecipitation method (17) with the following additional
modifications. Solutionswere asdescribedpreviously (18). Each
assayused 5x106-107cellsin6mlof mediumtowhich 40/g
ofplasmidDNA were addedper90mm petridish . Afteran
incubation time of 4hrs,the cellswerewashed with6mlofTBS
(25 mM Tris HClpH 7.4, 0.75mM Na2HPO4, 5mMKcl, 15
mM NaCI) for 2mn, changed with 6 ml medium and then
incubated for 24 hours with or without a physiological
concentration of 20-OH-E(14M).Cellswereharvestedand crude extracts wereprepared by sonication in0.25 M Tris HCI, pH
7.8.Assaysforchloramphenicolacetyltransferase(CAT) activity
were performedaccording to the methodofGorman et al(19)
with modifications: the proteinconcentration of the cell extracts
was measured and identical proteinamounts persample were used for the enzymatic reaction.
In Kc 167 cells, 40 mg ofDNA are necessary to obtain a
significative CAT or ( galactosidase response. Indeed
co-transfections with a standardconstructcontaining the E.coli(
galactosidasegene,give riseto verylowactivities whicharenot
interpretable.As acontrol for theefficiencyof transfection,CAT
activityofeachconstruct wascompared with thatof the plasmid pAc(G-CAT(20),which hasthe5C actinpromoterfused to the
CAT gene. The 5C actingene is well expressedin Kc cellsin
the absenceorpresenceof20-OH-E.Inbothcasesthisconstruct
generates a very important activity which represents for our
experiments, 100% ofrelative CAT activity, respectively for
treated and untreated samples.
RESULTS
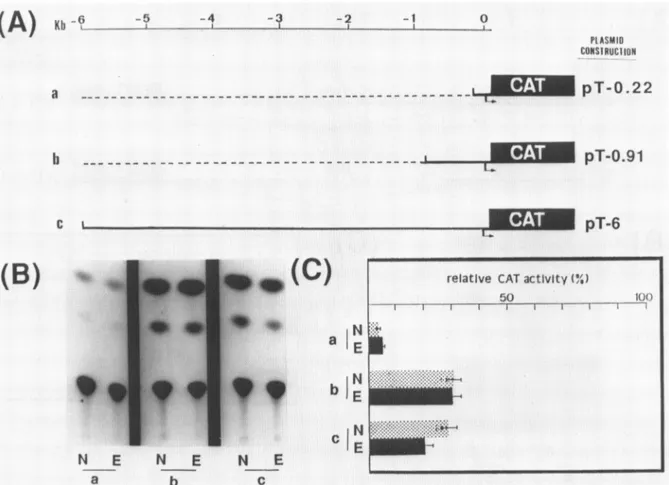
5' flanking region contains cis elements responsible for the expressionof the
03
tubulin gene in Kc cellsToidentifyecdysoneregulatory elementswithin the5' flanking region ofthe
(3
tubulin gene, different lengths ofthis regionwerefusedtothecodingregion of thebacterialchloramphenicol acetyl transferase (CAT)gene.Theseconstructs weretransiently transfccted into Kccultured Drosophila melanogastercells and
the response to hormone treatment was determined by CAT
assay, in untreated and treated cells. Control expression is
mediated by the plasmid
pAc(3G-CAT
asdescribedinmaterial and methods.When transfection iscarriedoutwiththe construction including
0.22 Kb upstream from the transcription start site (pT-0.22,
fig.
lA
lane a)thesameweakexpression( < 10%
of thecontrolexpression)wasobtainedin untreated and treatedcells
(fig.
1B,C).
Whenthe5'flankingregionwasextendedto0.91 Kb(pT-0.91, fig.1lane b) the CAT expressionwasincreased(30%) (fig. lB,C) but remained constitutivei.e. inpresenceorabsence of
20-OH-E. In order to identify ecdysone regulatory elements further
upstreamfrom thetranscriptionstartsite, wefusedalarge 6 Kb
flanking regiontoobtainpT-6 (fig.lAlane c). Theexpression
ofthisconstruct wasslightly repressed by 20-OH-E andthe CAT activity level in untreated cellswas similartotheoneobtained with theconstitutively expressedconstructpT-0.91 (fig.lB,C). Theseresults show thatone or severalcispositive elements
arepresent between 0.22 Kb and 0.91 Kb upstream from the
transcription
startsite, andareonly responsible
for the constitutive expression ofthe(3
tubulingene in Kc cells. The 5' flanking region extending from 0.91 over 6 Kb is not able to confer20-OH-E regulation to the CAT fusion gene. The promoter
strength is almost identical in the correspondingconstructs. Thefist intron isessential for the
(3
tubulin
generegulation
by 20-OH-E in Kc cells
Since the experiments described above have not permitted to
identifythe elementsresponsible for 20-OH-E regulation in the 5' flanking region,wehypothesizedthat these elementsshould
belocalized inanotherpart ofthe gene. A structural
particularity
of the
03
tubulin gene is a first large intron of 4.5 Kb. Thisintron isnotfound in the otherDrosophila ( tubulin genes and
what is more this gene is theonly one to be 20-OH-E regulated.
We decidedto construct aseries of fusiongenes containingthe
Nucleic Acids Research, Vol. 18, No. 10 2863 (A)
Kb
i1 -5 -4 -3I -2 -Ia
U
pT-0.22
b
I_-L
I
pT-0.91
C I,
pT-6
,.(C)
aIN
Eb
IN
CN
N E N E N E a b cFigure1: CATactivity of tubulin-CAT fusiongeneconstructstransfectedinKccells. A) Shematic representation ofplasmids: Thebacterialchloramphenicolacetyl transferase(CAT)genewasfusedtorespectively 0.22 Kb (lanea),0.91 Kb (lane b) and 6 Kb (lane c)upstreamfrom thestartsite of the 13tubulingene. (L,)
transcriptionstartsite.The scale indicatesthe position ofthe 33tubulinsequencesused intheconstructs,withrespecttothetranscriptionstartsite. B)Autoradiogram ofCATassaysillustrating activitiesfrom thesamethreepromoterfusionconstructsa)pT-0.22 without (N) and with (E) 20-OH-E. b) pT-0.91 without (N) and with(E)20-OH-E.c) pT-6without (N) and with (E) 20-OH-E. C) Relative CAT activity of these threeconstructsnormalizedwiththe plasmidpAc(3G-CAT(see
mat. andmethods). N: without 20-OH-E;E: with20-OH-E.
pTI-4.5 (fig.2A lane a) contains thesame 5' flanking region
as pT-0.91, the firstexon,the first large intron andpartofthe
sequence ofthe second exon. For a correctexpression of the
CATgene,itwasimportanttoconservethesequencesinvolved
inthesplicingprocessandlocalizedatthe 5' and 3' sides of the
intron. Asseeninfigure 2B and2C, in the absence of hormone,
pTI-4.5 yieldedaweakCATactivity. After 20-OH-Etreatment,
the expression ofthis construct including the first intron was
strongly induced. The activity varied intreatedcells, from 30
to 40% ofthat produced by pAc(3G-CAT.
Theseresults showfirst that the addition of the first intronto
aconstructcontaining 0.91Kb of5'upstreamflankingsequences allows the repression ofthe expression ofthe
O3
tubulin gene inuntreated cells. Moreover, inagreementwiththe known effect of 20-OH-E onO3
tubulingene expression inKc cells, properregulation ofthe gene was recovered with an induction level
varying from 4 to 10.
To better define the regions responsible for 20-OH-E
regulation, thelengthof the first intron wasreduced. Forthat,
the twouniquerestriction sites, Hind III and XbaI, wereused
todeleteafragmentof 3.4 Kb.pTI-1.1 (fig.2Alaneb)includes
the sameregions aspTI-4.5 exceptthe deletionof 3.4Kb in the
middle of the intron. Splicing sitesareconservedatthe 5' and
3'sides. Thisconstructshows thesamefeaturesastheprevious
one,withcomparableCAT activities and inductionlevelaverage
as seen infigure 2B and 2C. Thus, a 3.4 Kb internal deletion
ofthe first intron does not affect the expression level and the
regulation ofthe CAT fusion gene.
These results indicate that 1.1 Kb of the
03
first introncontainssequencesactingas anegative regulatory elementwhich
mediates the repression of pTI-4.5and pTI-1.1 expression in the
absence of20-OH-E. This repressionwasreleviedbythe addition
ofthe hormone.
However, it isverydifficulttocompareCATactivities obtained
with pT-0.91 in the absence of the intron with pTI-4.5 and
pTI-1.1 including the intron. Indeed, the first construct is a
transcriptional fusion, withoutatranslated (3tubulin region, the
second and thethirdconstruct,translational fusions with the first
exon of the (3 tubulin gene. We found that adding several
exogeneousadditionalamino-acidstothe CATgeneimpairs the
CATprotein activity (data not shown).
Regulatory sequences within the first intron can either
regulateorstimulate the promoter activity in bothdirections
and in a position independence manner
Because many eukaryotic transcriptional regulatory elements
function efficiently at considerable distances upstream or
downstream from promoters, irrespective of theirorientation,
weinvestigated whether the negative regulatorysequencespresent
in the first intron show any of these properties.
0
PtASMIO CONSIRUJCIION
(B)
..
...-._...
relative...
CATactivity0,0_ ~~~50 _ __ _ e ... ...,t...
_~~~
~~~~~~~~~~~~~~.
.1i
...i2864 Nucleic Acids Research, Vol. 18, No. 10 Lx I.
~~~~~~~~~~~~~~~~~~~~~~~~~~~~.
... . ..;i
§ *
o
w
-_.=
~~~~~~~pTl
4.5
pTI-1
.1
,¾, s~~~~~~~~P r(iy(oEIIIIIE
N
E
a & IN14E
b
Figure 2:CATactivityoftwotubulin-CATconstructs,includingthe 5'flanking regionand the first intron of the033tubulingene. A)The CATgenewasfused to0.91 Kbupstream from thestartsite,the firstexonandthelargefirstintron(IVS;lanea)orthe first deleted intron(lane b)of thej33tubulingene.(Q)transcription
startsite;hatched boxes:codingsequence of the gene;(---)deleted sequence. B) Autoradigram correspondingtoCATassays fromthesetwoconstructs:a)pTI-4.5 without (N)orwith (E) 20-OH-E.b) pTI-1.1 without(N)orwith(E)20-OH-E.C)Normalized relative CATactivityofpTI-4.5andpTI-1.1(seemat.andmethods)
N: without20-OH-E, E: with 20-OH-E.
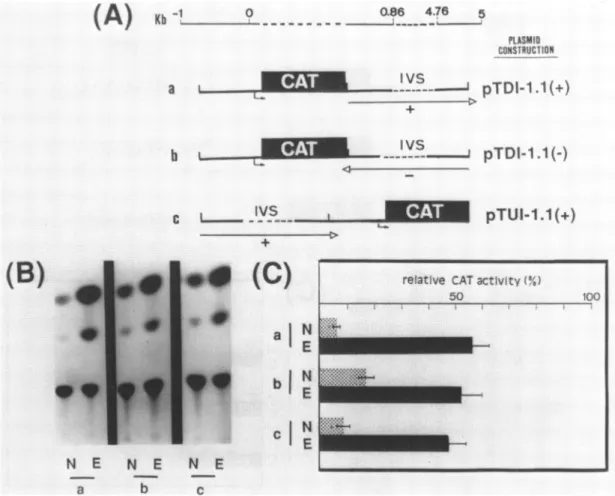
Since the 0.91 Kb
flanking
region
presentinpT-O.91,
was notefficientat
repressing
thetranscription
inthe absence of20-OH-E,
we used this construct to test the
properties
of the intronicsequences.Asseenin
figure
3A threeplasmids
wereconstructedbyinsertingatvarious
positions
the first deleted introninpT-0.91:
pTDI-1.1(+)
(lane a) contains the intronic sequences fused in the natural orientation downstream from the CAT gene;pTDI-1.1(-) (lane b) is thesameconstructwith thedeletedintron
inserted in the
opposite
orientation;
inpTUI-1. 1(+)
(lane
c)thedeleted intron is fusedinthe natural
orientation,
upstreamfromthe 5'flanking region.Inall thecases, we seethat CAT
activity
is low in untreated cells and
high
in treated cells(fig.3B,3C)
therefore showing that the
regulation
ofthe[3
tubulin gene ispresent. IndeedwithpTDI-1.1(+),the CAT generegulationleads
to anaverageinduction level of7,which issimilartotheprevious data,obtained when the fulllenghtintron is in its
physiological
position. However, when comparing to this plasmid,pTDI-1.
l(-)
showsalittlehigherCATactivityinuntreated cellswith a lower induction average of about 3. This indicates that
therepression isless efficient when the intronic sequencesare
in the opposite orientation. Analysis of CAT activity level
obtained from pTUI-1.1(+) did not reveal significative
differences with these ofpTDI-1.
1(+)
althoughthe average levelof induction of about 6 is a little lower.
So, in Kccells,the first intron of the
[3
tubulingene containsnegative regulatoryelements whichcan actineitherorientation,
and upstream or downstream from the gene, to repress the
transcription in absence of20-OH-E.
Becauseallthethreeplasmids aretranscriptional fusion, CAT
activitiescanbecompared with these of
the
constitutiveexpressedconstructpT-0.91. It appearsthatexpression levels aregreater
(about2 fold more) afterhormonaltreatmentinthepresencethan
inthe absenceof the intronic sequences. This suggests
that
the intron is also involved inthe transcription activation of the[3
tubulin gene under20-OH-E treatment.
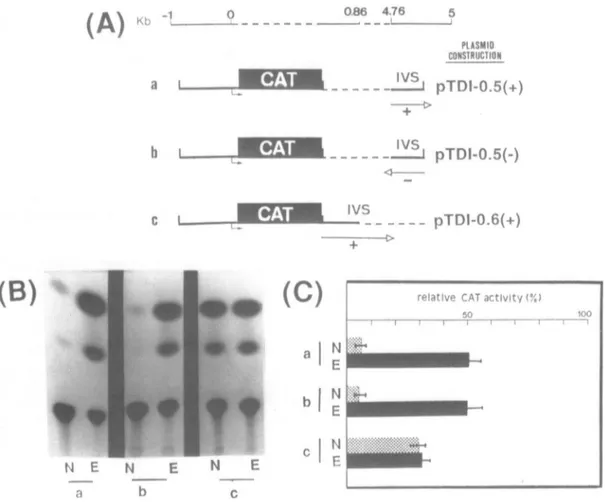
A 3' 0.5 Kb fragment of the first intron is sufficient for 20-OH-E regulation
Inordertoidentifyecdysoneregulatory elements responsible for
bothtranscriptioninhibition without 20-OH-E and transcription activation under hormonal treatment, the first deleted intron of
1.1
Kbwascleaved at a SalI site into two fragments of 0.6 and0.5 Kb. Afterwards these fragments were inserted downstream
from the CAT gene.
Asseeninfigure4A, in theconstitutively expressed plasmid
pT-0.91
either 0.5 Kb of the intron 3' side or 0.6Kbof the 5'sidewere inserted downstream from the CAT sequence in the
natural orientation: this creates respectively plasmids
pTDI-0.5(+)(lane a) and pTDI-0.6(+) (lane c). When cells are
transfected with pTDI-0.5(+), transcription inhibition and
activation are similar to those observed with pTDI-1.1(+), in
the absence or presence of 20-OH-E
(fig.4B,4C).
When cells1 V
X,. i
S.,i ,.
I v
Nucleic Acids Research, Vol. 18, No. 10 2865 Kb 0.86 4.76 5 ___ ____ _ PLASMIO .NSIRU.I. .N a I---...11-!Im ~ m - - -vst---. b c _ l IVspT I 'vs
(B)
C
al bi cI N E N E N E N E N E N E a b cFigure 3:CATactivity of tubulin-CAT fusiongenesconstructsincluding 5' and intronsequences. A)TheCATgene wasfusedto0.91 Kbupstreamfrom the startsite,and thedeleted intron(1.1 Kb)wasaddedrespectively downstream from the CATgene,in positive orientation (lanea);downstream from the CATgene, inopposite orientation (lane b);upstreamformthe CATgene,in positive orientation (lanec).(L)transcriptionstartsite;(---)deletedsequences.B)Autoradiogram correspondingtoCATassaysfor these threeconstructs. a)pTDI-1.l(+)without (N) and with(E)20-OH-E.b) pTDI-1. 1(-)without (N) and with (E) 20-OH-E.
c)pTUI-1. I(+) without(N) and with (E) 20-OH-E. C) Relative CAT activityof these three tubulin-CATconstructs(seemat.and methods). N: without 20-OH-E,
E:with20-OH-E.
aretransfected withpTDI-0.6(+) which contains only the first
part ofthe intron sequence, no difference in CAT activity in
untreated andtreated cells is detected. Activity levelsaresimilar
to these obtain withpT-0.91, which does notcontain any first
intron sequence.
These results show that negative and positivesequences,which
are20-OH-Edependent,arelocalized within0.5 Kb of theintron
3'side.Toverify theaction ineither orientation of theseelements,
intronic sequences downstream from the CAT gene of
pTDI-0.5(+)wereinvertedtoobtain pTDI-0.5(-)(laneb).With
these two constructs, CAT activities are almost equivalent,
respectively in untreated and treated cells. Thus, inaccordance
withtheprevious results,20-OH-Edependent elements,present
in the 3' 0.5 Kb fragment ofthe intron are functional in the
positive as well as in the opposite orientation.
DISCUSSION
We haveexamined the mechanism regulating thetranscription
ofthe 20-OH-E inducible, (3 tubulin gene, in Drosophila Kc
cells.
Using transient DNA transfer experiments, we have shown
that 0.22 kbupstream from thetranscription start siteleadsto
averyweakexpressionof this gene,inthepresenceorabsence
of20-OH-E. Sequence data didnot revealanyclassical TATA
box consensus (TATAAAA) at -30. The sequence found
between -28 and -35(GAACATC) (13) isverysimilartothat
ofthe (32tubulingene(GAACATT),localised between -26 and
-32 (21). The position and theconservation of this sequence
areinagreementwith theseofaTATA-like element.On the other
hand,wehave also shown that the 0.91kbregionupstreamfrom
thetranscriptionstartsite,contains hormoneindependent positive
element(s), leadingtoastrongconstitutiveexpressionof the
03
tubulin gene. Indeed, with this partof the 5' flanking region,
the regulation of this gene expression by 20-OH-E is absent.
These resultsareinaccordance withsequencing data which do
notreveal neither HREnorEcREsequences(3)(6)inthisregion
(13). Moreover, evenwith 6 kb of the 5' flanking region, the
20-OH-Eregulationis absent. The weakrepressionof thegene
expression, observed after hormonal treatment, suggests the
presenceof20-OH-Edependent negative regulatoryelements far
upstream from the start site.
DNA sequences responsible for regulation by steroid
hormones,havegenerallybeen foundinthe5' flankingregions
of genes (22 -24). For several years, many reports have
described the critical role of introns in the expression of
eukaryoticgenes,sincetheymaycarrytranscriptional regulatory
elements within their introns(25-28). However,suchelements
haveonlybeen described forafewhormonally regulatedgenes
(A)
relative CATactivity1%)
50 100
.
II
Im
hI-IpTDI-1
.1(+)
--I,, -L--.oldA
pTUI-1.1(+)
2866 Nucleic Acids Research, Vol. 18, No. 10 - f,.! .i...I ';'. 2: I .
...
...~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ N.Figure4: CATactivityoftubulin-CAT constructsincludingthe 5'flanking regionandrespectivelythe 5'or3'regionof the first intron.A)The CATgenewas
fusedto0.91 Kbupstream from the startsite,andwereadded: the 3' 500bpof the first intron in thepositiveorientation(lane a);the 3' 500bpof the firstintron
in theoppositeorientation(lane b);the 5' 600bpof the firstintron inpositiveorientation(lane c). (L*) transcriptionstartsite;(---)deletedsequence.B)Autoradiogram correspondingtoCATassaysfrom these three constructs:a)pTDI-0.5(+)without(N)and with(E)20-OH-E.b)pTDI-0.5(-)without(N)and with(E)20-OH-E. c) pTDI-0.6(+) without(N)and with (E)20-OH-E. C)Normalized relative CATactivityof these threeconstructs(seemat.andmethods) N:without20-OH-E, E: with 20-OH-E.
i.e. uteroglobingene(29), human(30-31)andrat(32) growth
hormonegene. In thesetwolastcases, the introncontains GRE
(glucocorticoid response element) and TRE (thyroid response
element), respectively. The (33 tubulin gene belongs to this
category.Indeed,addition ofthefirst intronto0.91 Kbupstream
fromthestartsite, allows 20-OH-E regulation. Uptonow, the
shortestregulatedconstruct includes the 3' 500pb fragmentof this intron. However,theabilityof thisfragmenttoregulatethe
expressionofthegene,doesnotexclude thepresenceofadditional
EcRE in otherparts ofthe intron.
Transcription activation ofgenesinducedby steroid hormones
involves binding of the hormone to the receptor, followed by
aspecificbindingtoHRE, whichexhibit theproperties ofpositive
regulatory elements. Insomecases, asfor thechicken ovalbumin
gene, thepromoteractivity isrepressed by negativeregulatory
elements andtherepressionisrelievedby steroid hormones (22).
In the case of the
03
tubulin gene, the 5' flanking regioncontains20-OH-E independent constitutive element(s), and the
first intronincludes negative regulatoryelement(s) (NRE)
20-OH-E dependent. Distantly located eukaryotic cis-repressor
sequencesoftenworkthroughanindirect mechanism, whereby
they interfere withthe binding of positive control factors to a
neighboring enhancer (33)(34). The 33 tubulin NRE does not
appear to function in this way. It appears to act over a long
distancetoinfluence theactivityof thebasalpromoter, similar
totheactivityofayeastsilencersequence(35). Theexactspacing
of the
(33
NRE relative to the transcription start site does not appeartobe criticalbecause itcanmediatethe repression of theactivityofa 1Kbpromoterbybeing placedupstream,downstream
and in both orientations. Moreoverthe activity of the NRE in
represssingtranscription fromabasalpromotermakes it similar
totheactivity ofayeast 'silencer' sequence asdefined in (35).
Besides the negative effect, thefirst intron is also involved in
the activation ofthe transcription. Indeed, 20-OH-E does not
simply relieve the transcription repression because higher CAT
activitiesareobtained afterhormonaltreatment,withconstructs
carryingintronsequences, comparedtotheconstitutiveone(see
Results).
The model that we propose for the
03
tubulin gene consistsofa repressor-silencer system localized in the first intron and
lockingthetranscription without E. Addition of
20-OH-E would relieve this repression, resulting in activated
transcription. This is mediated by positiveelements localized in
intronic regions and possibly by cooperation between these
intronicelements andthe 5'cispositive elements.Furtherstudies
will allow to precise these different regulatory elements. So,
20-OH-Enotonlyderepresses the transcription of thisgenebut
also activates it. A similar kind of regulation ofa hormone
(A)
(B)
4 E: I. ..1-I-,.
.= R. -' J niNucleicAcids Research, Vol. 18, No. 10 2867
responsive
promoterbeing repressed in abscence of hormone andbeing derepressed
in presence ofhormone has been recentlydescribed
(36)(37).
Thethyroid-hormone
a-receptors (TRa)regulate
the expression of several thyroid-hormone-responsivegenes in vertebrates
by binding
to the TREs and functionningas
TRE-specific
repressors in the abscence of hormone. Inthepresenceof hormonetheyact asTRE-specific activators. Sowe
can
speculate,
inthe caseof the regulation by 20-OH-E of the03
tubulin gene in Kccells,
that unfilled 20-OH-E receptorscouldact as repressors. Although we havenot yet proved that
the
repression
mediatedby
the NRE of the(3
tubulin geneinvolved an 20-OH-E receptor, it has been reported that
ecdysteroid
receptorsarepermanently located inthenuclei (38).Moreoverwe cantestthishypothesis by purifying therepressor
ofthe
(3
tubulin gene in Kc cells in absence of hormone andlooking
ifit can be a steroid binding protein.The
negative regulation
ofthe(3
tubulin promoter byacis-acting negative regulatory
element(NRE) isprobably mediatedatthe level ofDNA-protein and protein-protein interactions. It
is not due to a simple steric hindrance mechanism preventing
transcription complex
fromgoing
further downstream this element in absence ofhormone like in(39). Moreoverthese intronsequences couldinteract with 5'flankingsequencesthroughDNA
binding proteins
as described for the Human a 1 collagen gene(40).
These interactions between distantproteinscanbeexplainedby
thelooping
model proposed by Ptashne (41).During Drosophila
development, this03
tubulin gene isexpressed
atmidembryogenesis
andatthe end of the third instarlarvae-early
pupae(42)(43) and is also 20-OH-Eregulated (44).The
03
tubulin gene is a good marker for mesodermdevelopment (45)(46).
Concerning the regulationofthis gene inDrosophila,
Gashetal(47) have shown thatsequences upstreamfrom the cap site contain distinct cis-acting elements, for
constitutive and tissue specific expression. In addition, some
intronsequencesarealsoinvolvedintissue specific expression.
Although
the03
tubulingenesequence is almost identical in Kccells andin
Drosophila,
the differentcellularcontexts of thesetwomodels involve differences in therequirement of cis acting
elementsused for theregulationofthe expression of the gene.
In this paper, we have described cis-acting elements, which
canactinboth directions and inaposition-independancemanner,
involved in20-OH-E regulated expressionof the
03
tubulin gene inDrosophila
Kccells. Precise determinationof these cis-actingelements andtrans-acting factors involvedin20-OH-E regulation of the
(3
tubulin gene expression is in progress in ourlaboratory.
ACKNOWLEDGEMENTS
Theauthors thank
P.Couble,
H. DeThM,
M.RaymondjeanandC.Besmond for
helpful
discussions and criticalreading
ofthemanuscript.
We alsothankProfessorD.Beytoutand ProfessorB.Joly
forlaboratory
facilities forDrosophila
cellsandbacterialcultures.
A.Bruhat is a
recipient
of a french MRTpre-doctoral
scholarship.
S.Tourmente andS.Chapel
arerespectively
supported by
afellowship
fromthe Fondation pour la RechercheMedicale and from the Association pour la Recherche sur le
Cancer.
REFERENCES
3. Beato, M. (1989) Cell56, 335-344.
4. Handler,A.M.andMaroy, P.(1989). Molecularandcellularendocrinology
63,103-109.
5. Klemenz, R. and Gehring, W.J. (1986) Mol. CellBiol. 6, 2011-2019. 6. Mestril, R., Schiller, P., Amin, J.,Klapper,H.,Ananthan,J.andVoellmy,
R. (1986) Embo J. 5, 1667-1673.
7. Riddihough, G. and Pelham, R.B. (1987)EmnboJ. 6, 3729-3734. 8. Riddihough, G. andPelham, H.R.B. (1986)EmnboJ. 5, 1653-1658. 9. Mc. Ginnis, W., Shermoen, A.W., Heemskerk J. and Beckendorf, S.K.
(1983) Proc. Natl. Acad. Sci. 80, 1063-1067.
10. Couderc, J.L.,Becker,J.L.,Sobrier, M.L., Dastugue, B.,Best-Belpomme, M.,Lepesant,J.A.and Pardue, H.L. (1984) Chromosomna89,338-342. 11. Montpied, P.,Sobrier, M-L., Chapel, S., Couderc, J.L. and Dastugue B.
(1988)Biochem. Biophys. Acta. 949, 79-86.
12. Sobrier, M.L.,Couderc, J.L., Chapel, S. and DastugueB.(1986) Biochem. Biophys. Res. Comm. 134, 191-200.
13. Chapel, S., Montpied, P.,Bruhat, A., Sobrier, M.L.,Micard, D.,Couderc,
J.L. and Dastugue, B. (1990) submitted.
14. Echalier, G. and Ohanessian, A. (1969) CR. Acad. Sc. Paris 268,
1771-1773.
15. Courgeon, A.M. (1972) Exp. Cell. Res. 74, 327-336. 16. Luckow, B. and Schuitz, G. (1987) Nucl. Acids. Res. 15, 5490 17. Bourouis, M. and Jarry, B. (1983) EmboJ. 2, 1099-1104.
18. Di Nocera, P.P. and Dawid, J.B. (1983) Proc. Natl. Acad. Sci. 80,
7095-7098.
19. Gorman, C.M., Moffat, L.F. and Howard, B.H. (1982)Mol. Cell. Biol. 2, 1044-1051.
20. Martin, M. and Jarry, B. (1988) J. InsectPhysio. 34, 691-699. 21. Michiels,F.,Gash,A.,Kaltschmidt,B.andRenkawitz-Pohl,R.(1989)Embo.
J. 8, 1559-1565.
22. Gaub, M.P. Dierich, A., Astinotti, D., Touitou, I. and Chambon, P.
(1987)EmboJ. 6, 2313-2320.
23. Sakai, D.D., Helms,S., Carlstedt-Dike, J., Gustafson, J.A., Rottman,F.M. and Yamamoto, K.R. (19887 Genesand Development2, 1144-1154. 24. Luckow, B. and Shutz, G. (1989)Nucl. Acids Res. 17, 8451-8462. 25. Rippe,A.R., Lorenzen, S.I., Brenner D.A.andBreindl, M. (1989) Mol.
Cell. Biol. 9, 2224-2227.
26. Evans, M J. and Scarpulla, R.C. (1988) Mol. Cell. Biol. 8, 35-41. 27. Geyer, P.K. and Corces, V.G. (1987) Genes andDevelopment1, 996-1004. 28. Neuberger, M.S. and Williams, G.T. (1988) Nucl. Acids Res. 16,
6713-6724.
29. Bailly, A., Le Page, C., Rauch, M. and Milgrom, E.(1986)Embo J. 5, 3235-3241.
30. Moore, D.D., Marks, A.R., Buckley, D.I.,Kapler, G., Payvar, F. and
Goodman, H.M. (1985)Proc. Natl. Acad. Sci. 82, 699-702.
31. Slater, E.P., Rabenau,O.,Karin, M.,Baxter, J.D. and Beato, M(1985)Mol.
Cell Biol. 5,2984-2992.
32. Sap, J., De Magistris, L., Stunnenberg, H. andVennstrom, B. (1990)Emnbo
J. 9, 887-896.
33. Jones, P.B.C., Galeazzi, D.R., Fisher,J.M., Whitlock,J.R. (1985) Science
227, 1499-1502.
34. Maniatis,T. Goodbourn, S., Fisher,J.A.(1987)Science 236, 1237-1244. 35. Brand, H., Breeden,L.,Abraham,J., Sternglanz, R. and Nasmyth K.(1985)
Cell41,41-48.
36. Damm, K., Thompson, C.C. and Evans, R.M. (1989) Nature 339, 593-597. 37. Graupner, G., Wills, K.N., Tzukerman, M., Zhang, X.K. andPfhal,M.
(1989) Nature 340, 653-656.
38. Sage, B.A., Tanis,M., and O'Connor,J.D. (1982) J. Biol. Chem. 257, 6373-6379.
39. Oro,A.E.,Hollenberg,S.M.and Evans, R.M.(1988) Cell 55,1109-1114. 40. Bornstein,P., McKay, J., Liska, D.J., Apone, S. and Devarayalu,S.(1988)
Mol. Cell. Biol. 8, 4851-4857.
41. Ptashne, M. (1986) Nature 322, 697-701.
42. Bialojan, S., Falkenburg, D.and Renkawitz-Pohl, R. (1984) Embo. J. 3, 2543-2548.
43. Natzle,J. and McCarthy, B. (1984) Dev. Biol. 104, 187-198. 44. Sobrier,M.L., Chapel, S. Couderc, J.L., Micard,D.,Lecher,P.,
Somme-Martin, G. andDastugue, B. (1989)Exp CellRes. 184, 241-249. 45. Gasch,A.,Hinz,U.,andRenkawitz-Pohl, R.(1988)Mol.Gen. Genet.211,
8-16.
46. Leiss, D.,Hinz, U., Gasch, A.,Mertz, R.andRenkawitz-Pohl,R.(1988) Development 104, 525-531.
47. Gasch, A., Hinz, U. and Renkawitz-Pohl, R. (1989) Proc. Natl. Sci. 86, 3215-3218.
1. Dynan, W.S.(1989) Cell58, 1-4. 2. Berg,J.M. (1989) Cell 57, 1065-1068.