HAL Id: hal-02653040
https://hal.inrae.fr/hal-02653040
Submitted on 29 May 2020
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Permanent genetic resources added to molecular ecology
resources database 1 august 2011-30 september 2011
S.W. A’Hara, Paul Amouroux, Emily.E. Argo, Arman Avand-Faghih,
Ashoktaru Barat, Luiz Barbieri, Theresa M. Bert, R. Blatrix, Aurelie Blin, D.
Bouktila, et al.
To cite this version:
S.W. A’Hara, Paul Amouroux, Emily.E. Argo, Arman Avand-Faghih, Ashoktaru Barat, et al..
Per-manent genetic resources added to molecular ecology resources database 1 august 2011-30 september
2011. Molecular Ecology Resources, Wiley/Blackwell, 2012, 12 (1), pp.185-189.
�10.1111/j.1755-0998.2011.03088.x�. �hal-02653040�
P E R M A N E N T G E N E T I C R E S O U R C E S N O T E
Permanent Genetic Resources added to Molecular Ecology
Resources Database 1 August 2011–30 September 2011
MOLECULARECOLOGYRESOURCESPRIMERDEVELOPMENTCONSORTIUM,1S. W. A’HARA,2P. AMOUROUX,3,4EMILY E. ARGO,5 A. AVAND-FAGHIH,6ASHOKTARU BARAT,7LUIZ BARBIERI,8THERESA M. BERT,8R. BLATRIX,9AURE´ LIE BLIN,10D. BOUKTILA,11,12A. BROOME,2C. BURBAN,13C. CAPDEVIELLE-DULAC,14N. CASSE,15SURESH CHANDRA,7KYUNG JIN CHO,16J. E. COTTRELL,2CHARLES R. CRAWFORD,8MICHELLE C. DAVIS,8H. DELATTE,17NICOLAS DESNEUX,18 C. DJIETO-LORDON,19M. P. DUBOIS,9R. A. A. M. EL-MERGAWY,20C. GALLARDO-ESCA´RATE,21M. GARCIA,9MARY M. GARDINER,22THOMAS GUILLEMAUD,10P. A. HAYE,23B. HELLEMANS,24P. HINRICHSEN,25JI HYUN JEON,26
C. KERDELHUE´ ,27I. KHARRAT,11KI HWAN KIM,26YONG YUL KIM,16YE-SEUL KWAN,28ELLEN M. LABBE,29ERIC LAHOOD,30KYUNG MI LEE,16WAN-OK LEE,31YAT-HUNG LEE,32ISABELLE LEGOFF,10H. LI,33CHUNG-PING LIN,32S. S. LIU,34Y. G. LIU,35D. LONG,36G. E. MAES,24E. MAGNOUX,37PRABIN CHANDRA MAHANTA,7H. MAKNI,11,38
M. MAKNI,11THIBAUT MALAUSA,10RAKESH MATURA,7D. MCKEY,9ANNE L. MCMILLEN-JACKSON,8
M. A. ME´ NDEZ,39M. MEZGHANI-KHEMAKHEM,11ANDY P. MICHEL,22MORAN PAUL,30JANICE MURIEL-CUNHA,40 S. NIBOUCHE,17F. NORMAND,3ERIC P. PALKOVACS,5VEENA PANDE,41K. PARMENTIER,42J. PECCOUD,9F. PIATS-CHECK,9CECILIA PUCHULUTEGUI,8R. RAMOS,25,43G. RAVEST,25HEINZ RICHNER,44J. ROBBENS,42D. ROCHAT,45 J. ROUSSELET,37VERENA SALADIN,44M. SAUVE,9ORA SCHLEI,46THOMAS F. SCHULTZ,5A. R. SCOBIE,47
N. I. SEGOVIA,23SEIFU SEYOUM,8J.-F. SILVAIN,14ELISABETH TABONE,48J. K. J. VAN HOUDT,24,49
S. G. VANDAMME,42,24F. A. M. VOLCKAERT,24JOHN WENBURG,46THEODORE V. WILLIS,50YONG-JIN WON,28 N. H. YE,51W. ZHANG22and Y. X. ZHANG35
1
Molecular Ecology Resources Editorial Office, 6270 University Blvd, Vancouver, British Columbia, Canada V6T 1Z4,2Forest Research, Northern Research Station, Roslin, Midlothian, Scotland EH25 9SY, UK,3CIRAD, UPR HortSys, Station de Bassin Plat, BP180, F-97455 Saint-Pierre, La Re´union, France,4Universite´ de la Re´union, 15 avenue Rene´ Cassin BP 7151, F-97715 Saint-Denis Messag, Cedex 9, La Re´union, France,5Marine Conservation Molecular Facility, Duke University Marine Laboratory, Nicholas School of the Environment, 135 Duke Marine Lab Road, Beaufort, NC 28516, USA,6Plant Pests & Diseases Research Institute, PO box 1454, 19395 Tehran, Iran,7Molecular Genetics Laboratory, Directorate of Coldwater Fisheries Research, Indian Council of Agri-cultural Research, Bhimtal-263136, Nainital, Uttarakhand, India,8Florida Fish and Wildlife Research Institute, 100 Eighth Avenue S.E., Saint Petersburg, FL 33701-5095, USA,9Centre d’Ecologie Fonctionnelle et Evolutive (CEFE), UMR 5175 (CNRS, Universite´ Montpellier 2), 1919 route de Mende, 34293 Montpellier Cedex 5, France,10Equipe ‘Biologie des populations en interaction’, UMR 1301 IBSV INRA-CNRS-Universite´ de Nice-Sophia Antipolis 400 route des Chappes, 06903 Sophia-Antipolis Cedex, France,
11Unite´ ge´nomique des insectes ravageurs des cultures d’inte´reˆt agronomique, Faculte´ des Sciences de Tunis, Universite´ de
Tunis-El-Manar, Tunisia,12Institut Supe´rieur de Biotechnologie Be´ja, Universite´ de Jendouba, Tunisia,13INRA, UMR1202 BIOGECO
(INRA ⁄ Universite´ de Bordeaux), F-33610 Cestas, France,14IRD, UR 072, Laboratoire Evolution, Ge´nomes et Spe´ciation, UPR
9034, CNRS, 91198 Gif-sur-Yvette, France and Universite´ Paris Sud 11, 91405 Orsay Cedex, France,15Laboratoire Mer, Mole´cules,
Sante´ (MMS), Universite´ du Maine, Le Mans, France,16Division of Seed & Seedling Management, Korea Forest Seed and Variety
Center, 72, Suhoeri-ro, Chungju-si, Chungcheongbuk-do, 380-941, Korea,17CIRAD, UMR PVBMT, 7 chemin de l’IRAT, Ligne Paradis, F-97410 Saint-Pierre, La Re´union, France,18Unite´ de recherche inte´gre´e en horticulture, INRA, 400 route des Chappes, 06903 Sophia-Antipolis Cedex, France,19Laboratory of Zoology, University of Yaounde´ I, Faculty of Science, PO Box 812, Yaounde´, Cameroun,20Department of Molecular Biology, Genetic Engineering and Biotechnology Research Institute (GEBRI), Minoufia Uni-versity, El-Sadat City, Minoufia, Egypt,21Departamento de Oceanografı´a, Facultad de Ciencias Naturales y Oceanogra´ficas, Centro de Biotecnologı´a, Universidad de Concepcio´n, Casilla 160-C, Concepcio´n, Chile,22Department of Entomology, The Ohio Agricul-tural Research and Development Center, The Ohio State University, 1680 Madison Avenue, Wooster, OH 44691, USA,23 Departa-mento de Biologı´a Marina, Universidad Cato´lica del Norte & Centro de Estudios Avanzados en Zonas A´ ridas (CEAZA), Larrondo 1281, Coquimbo, Chile,24Katholieke Universiteit Leuven (KULeuven), Laboratory of Animal Diversity and Systematics, BioGenom-ics Division, Charles Deberiotstraat 32, 3000 Leuven, Belgium,25Laboratorio de Biotecnologı´a, Centro de Investigacio´n La Platina,
Instituto de Investigaciones Agropecuarias, INIA, Santa Rosa 11,610, P.O. Box 439-3, Santiago, Chile,26Biomedic, 1143-3,
Joong-dong, Wonmi-gu, Bucheon-si, Gyeonggi-do, 420-020, Korea,27INRA, UMR CBGP (INRA ⁄ IRD ⁄ CIRAD ⁄ Montpellier Supagro),
F-34988 Montferrier-sur-Lez, France,28Division of EcoScience, Ewha Womans University, Seoul, Korea,29Department of Biology,
University of Southern Maine, 96 Falmouth St, Portland, ME 04102, USA,30Conservation Biology Division, Northwest Fisheries
Science Center, 2725 Montlake East, Seattle, WA 98112, USA,31Inland Fisheries Research Institute, National Fisheries Research &
Development Institute, Gapyeong, Gyeonggi-do, Korea,32Department of Life Science & Center for Tropical Ecology and Biodiversity, Tunghai University, Taichung, 40704, Taiwan,33Jiangsu Provincial Key Laboratory of Coastal Wetland Bioresources and
Environ-mental Protection, Yancheng Teachers University, Yancheng 224002, China,34Ocean University of China, Qingdao 266003, China,
35
Shandong Entry-Exit Inspection and Quarantine Bureau, Qingdao 266002, China,36Plantlife Scotland, Balallan House, Allan Park, Stirling, Scotland FK8 2QG, UK,37INRA, UR633 Zoologie forestie`re, 45075 Orle´ans cedex 2, France,38Institut Supe´rieur de l’Animation pour la Jeunesse et la Culture, Bir El Bey, Universite´ de Tunis, Tunisia,39Laboratorio de Gene´tica y Evolucio´n, Facultad de Ciencias, Univ. de Chile, Las Palmeras 3425, N˜ un˜oa, Box 780-0024, Santiago, Chile,40Universidade Federal do Para´ - Altamira, Faculdade de Cieˆncias Biolo´gicas, Rua Coronel Jose´ Porfı´rio, N 2515, 68372-040 – Altamira, PA, Brasil,41Department of Biotechnol-ogy, Kumaon University, Bhimtal-263136, Uttarakhand, India,42Institute for Agricultural and Fisheries Research (ILVO-Fisheries),
Ankerstraat 1, 8400 Ostend, Belgium,43Syngenta-Chile, Av. Vitacura 2939 Of. 201, Santiago, Chile,44University of Bern, Institute
of Ecology and Evolution, Dept. Evolutionary Ecology, Baltzerstrasse 6, 3012 Bern, Switzerland,45UMR 1272, UPMC-INRA,
Physiologie de l’insecte: Signalisation et Communication, Route de Saint Cyr, 78026 Versailles Cedex, France,46Conservation
Genetics Laboratory, USFWS, 1011 East Tudor Rd., Anchorage, AK 99503, USA,47Cairngorms Rare Plants Project, Scottish
Natural Heritage, Achantoul, Aviemore, Inverness-shire, PH22 1QD,48Unite´ expe´rimentale de lutte biologique, INRA, 400 route
des Chappes, 06903 Sophia-Antipolis Cedex, France,49KULeuven, Laboratory for Cytogenetics and Genome Research, O&N, Herestraat 49, 3000 Leuven, Belgium,50Department of Environmental Science, University of Southern Maine, 37 College Ave, Gorham, ME 04038, USA,51Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao, 266071, China
Abstract
This article documents the addition of 299 microsatellite marker loci and nine pairs of single-nucleotide polymorphism (SNP) EPIC primers to the Molecular Ecology Resources (MER) Database. Loci were developed for the following species: Alosa pseudoharengus, Alosa aestivalis, Aphis spiraecola, Argopecten purpuratus, Coreoleuciscus splendidus, Garra gotyla, Hippodamia convergens, Linnaea borealis, Menippe mercenaria, Menippe adina, Parus major, Pinus densiflora, Portunus tritu-berculatus, Procontarinia mangiferae, Rhynchophorus ferrugineus, Schizothorax richardsonii, Scophthalmus rhombus, Tetrap-onera aethiops, Thaumetopoea pityocampa, Tuta absoluta and Ugni molinae. These loci were cross-tested on the following species:Barilius bendelisis, Chiromantes haematocheir, Eriocheir sinensis, Eucalyptus camaldulensis, Eucalyptus cladocalix, Eucalyptus globulus, Garra litaninsis vishwanath, Garra para lissorhynchus, Guindilla trinervis, Hemigrapsus sanguineus, Luma chequen. Guayaba, Myrceugenia colchagu¨ensis, Myrceugenia correifolia, Myrceugenia exsucca, Parasesarma plicatum, Parus major, Portunus pelagicus, Psidium guayaba, Schizothorax richardsonii, Scophthalmus maximus, Tetraponera latifrons, Thaumetopoea bonjeani, Thaumetopoea ispartensis, Thaumetopoea libanotica, Thaumetopoea pinivora, Thaumetopoea pityo-campa ena clade, Thaumetopoea solitaria, Thaumetopoea wilkinsoni and Tor putitora. This article also documents the addi-tion of nine EPIC primer pairs forEuphaea decorata, Euphaea formosa, Euphaea ornata and Euphaea yayeyamana.
This article documents the addition of 299 microsatellite marker loci and nine pairs of single-nucleotide polymor-phism (SNP) genotyping primers to the Molecular Ecol-ogy Resources Database. Table 1 contains information on the focal species, the number of loci developed, any other species the loci were tested in and the accession numbers for the loci in both the Molecular Ecology Resources Database and GenBank. The authors responsible for each set of loci are listed in the final column. Table 2 presents information on SNP genotyping resources added to the MER database and presents data on the focal species, the
number of sequencing primer pairs, the observed num-ber of SNPs, other species the loci were tested in, and the number of allele specific primers or probes. The MER database and GenBank accession numbers and the authors responsible are also listed. Table 3 outlines addi-tional permanent genetic resources that have been uploaded to the MER program wiki (http://tomato.biol. trinity.edu/programs/). A full description of the devel-opment protocol for the loci presented in Tables 1 & 2 can be found on the Molecular Ecology Resources Database (http://tomato.biol.trinity.edu/).
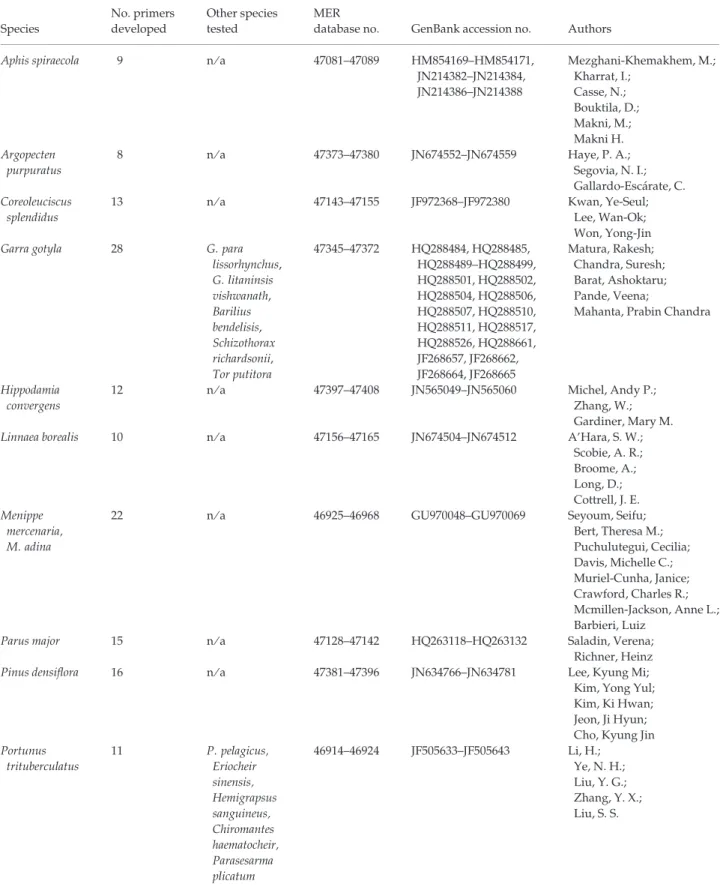
Table 1 Information on the focal species, the number of loci developed, any other species the loci were tested in and the accession numbers for the loci in both the Molecular Ecology Resources (MER) Database and GenBank. The authors responsible for each set of loci are listed in the final column
Species No. primers developed Other species tested MER
database no. GenBank accession no. Authors Alosa pseudoharengus,
Alosa aestivalis
18 n ⁄ a 47166–47201 JN383992–JN384009 Labbe, Ellen M.; Argo, Emily E.; Schultz, Thomas F.; Palkovacs, Eric P.;Willis, Theodore V. Correspondence: Molecular Ecology Resources Primer Development Consortium, E-mail: editorial.office@molecol.com
Table 1 (Continued) Species No. primers developed Other species tested MER
database no. GenBank accession no. Authors Aphis spiraecola 9 n ⁄ a 47081–47089 HM854169–HM854171, JN214382–JN214384, JN214386–JN214388 Mezghani-Khemakhem, M.; Kharrat, I.; Casse, N.; Bouktila, D.; Makni, M.; Makni H. Argopecten purpuratus 8 n ⁄ a 47373–47380 JN674552–JN674559 Haye, P. A.; Segovia, N. I.; Gallardo-Esca´rate, C. Coreoleuciscus splendidus 13 n ⁄ a 47143–47155 JF972368–JF972380 Kwan, Ye-Seul; Lee, Wan-Ok; Won, Yong-Jin Garra gotyla 28 G. para
lissorhynchus, G. litaninsis vishwanath, Barilius bendelisis, Schizothorax richardsonii, Tor putitora 47345–47372 HQ288484, HQ288485, HQ288489–HQ288499, HQ288501, HQ288502, HQ288504, HQ288506, HQ288507, HQ288510, HQ288511, HQ288517, HQ288526, HQ288661, JF268657, JF268662, JF268664, JF268665 Matura, Rakesh; Chandra, Suresh; Barat, Ashoktaru; Pande, Veena;
Mahanta, Prabin Chandra
Hippodamia convergens
12 n ⁄ a 47397–47408 JN565049–JN565060 Michel, Andy P.; Zhang, W.; Gardiner, Mary M. Linnaea borealis 10 n ⁄ a 47156–47165 JN674504–JN674512 A’Hara, S. W.;
Scobie, A. R.; Broome, A.; Long, D.; Cottrell, J. E. Menippe mercenaria, M. adina
22 n ⁄ a 46925–46968 GU970048–GU970069 Seyoum, Seifu; Bert, Theresa M.; Puchulutegui, Cecilia; Davis, Michelle C.; Muriel-Cunha, Janice; Crawford, Charles R.; Mcmillen-Jackson, Anne L.; Barbieri, Luiz
Parus major 15 n ⁄ a 47128–47142 HQ263118–HQ263132 Saladin, Verena; Richner, Heinz Pinus densiflora 16 n ⁄ a 47381–47396 JN634766–JN634781 Lee, Kyung Mi;
Kim, Yong Yul; Kim, Ki Hwan; Jeon, Ji Hyun; Cho, Kyung Jin Portunus trituberculatus 11 P. pelagicus, Eriocheir sinensis, Hemigrapsus sanguineus, Chiromantes haematocheir, Parasesarma plicatum 46914–46924 JF505633–JF505643 Li, H.; Ye, N. H.; Liu, Y. G.; Zhang, Y. X.; Liu, S. S.
Table 1 (Continued) Species No. primers developed Other species tested MER
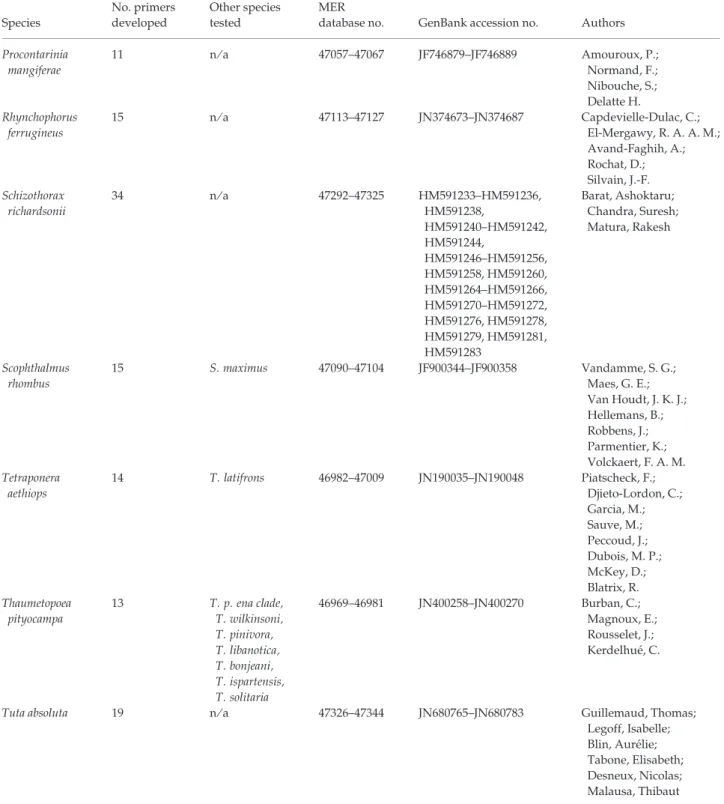
database no. GenBank accession no. Authors Procontarinia mangiferae 11 n ⁄ a 47057–47067 JF746879–JF746889 Amouroux, P.; Normand, F.; Nibouche, S.; Delatte H. Rhynchophorus ferrugineus 15 n ⁄ a 47113–47127 JN374673–JN374687 Capdevielle-Dulac, C.; El-Mergawy, R. A. A. M.; Avand-Faghih, A.; Rochat, D.; Silvain, J.-F. Schizothorax richardsonii 34 n ⁄ a 47292–47325 HM591233–HM591236, HM591238, HM591240–HM591242, HM591244, HM591246–HM591256, HM591258, HM591260, HM591264–HM591266, HM591270–HM591272, HM591276, HM591278, HM591279, HM591281, HM591283 Barat, Ashoktaru; Chandra, Suresh; Matura, Rakesh Scophthalmus rhombus 15 S. maximus 47090–47104 JF900344–JF900358 Vandamme, S. G.; Maes, G. E.; Van Houdt, J. K. J.; Hellemans, B.; Robbens, J.; Parmentier, K.; Volckaert, F. A. M. Tetraponera aethiops 14 T. latifrons 46982–47009 JN190035–JN190048 Piatscheck, F.; Djieto-Lordon, C.; Garcia, M.; Sauve, M.; Peccoud, J.; Dubois, M. P.; McKey, D.; Blatrix, R. Thaumetopoea pityocampa 13 T. p. ena clade, T. wilkinsoni, T. pinivora, T. libanotica, T. bonjeani, T. ispartensis, T. solitaria 46969–46981 JN400258–JN400270 Burban, C.; Magnoux, E.; Rousselet, J.; Kerdelhue´, C.
Tuta absoluta 19 n ⁄ a 47326–47344 JN680765–JN680783 Guillemaud, Thomas; Legoff, Isabelle; Blin, Aure´lie; Tabone, Elisabeth; Desneux, Nicolas; Malausa, Thibaut 188 P E R M A N E N T G E N E T I C R E S O U R C E S N O T E
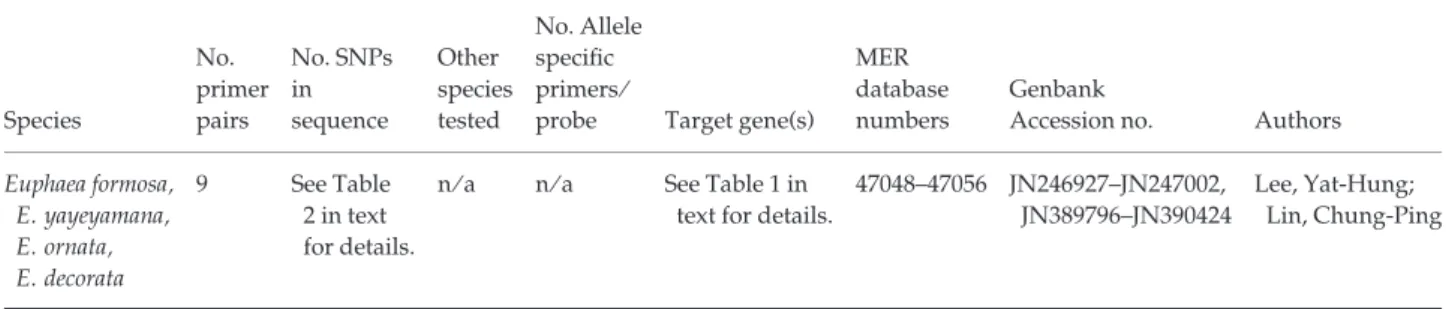
Table 2 Information on the focal species, the sequencing primer pairs developed, the number of single-nucleotide polymorphisms (SNPs) observed and any other species the loci were tested in. The next columns contain the number of allele specific primers and probes developed and the Molecular Ecology Resources (MER) database and GenBank accession numbers, respectively. The authors responsible for each set of loci are listed in the final column
Species No. primer pairs No. SNPs in sequence Other species tested No. Allele specific primers ⁄
probe Target gene(s) MER database numbers
Genbank
Accession no. Authors Euphaea formosa, E. yayeyamana, E. ornata, E. decorata 9 See Table 2 in text for details. n ⁄ a n ⁄ a See Table 1 in text for details.
47048–47056 JN246927–JN247002, JN389796–JN390424
Lee, Yat-Hung; Lin, Chung-Ping
Table 3 Information on other resources recently uploaded to the Molecular Ecology Resources program wiki (http:// tomato.biol.trinity.edu/programs/). The authors are listed in the final column
Species Category Type of resource Authors Oncorhynchus tshawytscha Technique Microsatellite allele ladder-based standardization LaHood, Eric; Schlei, Ora; Wenburg, John; Moran, Paul Table 1 (Continued) Species No. primers developed Other species tested MER
database no. GenBank accession no. Authors Ugni molinae 16 Myrceugenia correifolia,
M. colchagu¨ensis, M. exsucca, Guindilla trinervis, Luma chequen. Guayaba, Psidium guayaba, Eucalyptus cladocalix, E. camaldulensis, E. globulus 46809–46824 HQ917086–HQ917101 Ramos, R.; Ravest, G.; Me´ndez, M.A.; Hinrichsen, P.