HAL Id: inserm-00349146
https://www.hal.inserm.fr/inserm-00349146
Submitted on 21 May 2014
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
country living, and asthma in adults.
Lidwien Smit, Valérie Siroux, Emmanuelle Bouzigon, Marie-Pierre Oryszczyn,
Mark Lathrop, Florence Desmenais, Francine Kauffmann
To cite this version:
Lidwien Smit, Valérie Siroux, Emmanuelle Bouzigon, Marie-Pierre Oryszczyn, Mark Lathrop, et al..
CD14 and toll-like receptor gene polymorphisms, country living, and asthma in adults.: CD14, TLR2,
rural living and asthma. American Journal of Respiratory and Critical Care Medicine, American
Thoracic Society, 2009, 179 (5), pp.363-8. �10.1164/rccm.200810-1533OC�. �inserm-00349146�
Am J Respir Crit Care Med – in press
CD14 and Toll-like receptor gene polymorphisms, country living, and asthma in adults
Lidwien A. M. Smit1,2, Valérie Siroux3,4, Emmanuelle Bouzigon5,6,7, Marie-Pierre Oryszczyn1,8, Mark Lathrop9, Florence Demenais5,6,7, Francine Kauffmann1,8, on behalf of the EGEA cooperative group
1 INSERM, U780, Villejuif, France
2 Division of Environmental Epidemiology, Institute for Risk Assessment Sciences, Utrecht, The Netherlands 3 INSERM, U823, Centre de Recherche Albert Bonniot, Epidémiologie des cancers et des affections graves, La
Tronche, France
4 Université Joseph Fourier, Grenoble, France 5 INSERM, U794, Paris, France
6 Université d'Evry, Evry, France 7
Fondation Jean Dausset-Centre d'Etude du Polymorphisme Humain (CEPH), Paris, France
8
Université Paris-Sud, IFR69, Villejuif, France
9 Commissariat à l'Energie Atomique, Institut de Génomique, Centre National de Génotypage, Evry, France
Correspondence: Francine Kauffmann
INSERM U780, 16 Avenue Paul Vaillant Couturier 94807 Villejuif Cedex, France
Tel + 33 145595072, Fax +33 145595169 Email: francine.kauffmann@.inserm.fr Funded by ANR-SEST, AFSSET.
Lidwien Smit is supported by an EAACI-GA2LEN exchange fellowship award.
Running head: CD14, TLR2, rural living and asthma Subject category: Asthma: epidemiology (57) Manuscript: 3304 words
This article has an online data supplement, which is accessible from this issue's table of content online at www.atsjournals.org
AT A GLANCE COMMENTARY Scientific Knowledge on the Subject
Several studies have suggested that gene-environment interactions play an important role in the occurrence of asthma and allergy. Genes dependent on the innate immunity pathway are obvious candidates for the understanding of the protective effects of exposure to microbial agents on allergy and asthma.
What This Study Adds to the Field
TLR2 and CD14 SNPs were associated with asthma and atopic asthma respectively. Variation in CD14, TLR2, TLR4, and TLR9 genes modified associations between country living during childhood and asthma, in particular among atopic subjects.
ABSTRACT
Rationale: It has been shown that country living protects against asthma, which may be explained by microbial
exposures. Objectives: To study whether single nucleotide polymorphisms (SNPs) in CD14 and Toll-like receptor (TLR) 2, TLR4 and TLR9 genes are associated with asthma in adults, and whether these SNPs modify associations between country living and asthma. Methods: Twenty-five SNPs in CD14, TLR2, TLR4, and TLR9 genes were genotyped in adult subjects from the French Epidemiological study on the Genetics and Environment of Asthma, Bronchial Hyperresponsiveness, and Atopy (EGEA). We conducted a case-control analysis on unrelated subjects (239 asthmatics and 596 non asthmatics), and a family based association test (FBAT) in 192 families ascertained through asthmatic probands. Measurements and Main Results: The TLR2/+596 C allele was associated with an increased risk for asthma in both case-control and family-based analyses (under a dominant model, OR [95%CI]=1.91 [1.34-2.72], P=.0003; z statistics from FBAT=2.48, P=.01). In skin prick test (SPT) positive subjects, the CD14/-260 C allele was negatively associated with asthma (additive model, OR[95%CI]=0.66 [0.48-0.91]). Significant gene-environment interactions between variation in CD14 and TLR genes and country living during childhood were found for ten SNPs. In SPT positive subjects carrying CD14/-260 CC, country living protected against asthma (OR=0.32 [0.12-0.85]), whereas country living was not associated with asthma in atopics carrying CD14/-260 T (OR=1.11 [0.65-1.90]) (gene-environment
interaction P<.05). Conclusions: TLR2 and CD14 SNPs were associated with asthma and atopic asthma respectively. In addition, CD14, TLR2, TLR4, and TLR9 SNPs modified associations between country living and asthma.
Abstract: 249 words
Keywords: asthma; atopy; epidemiology; gene-environment interaction; hygiene hypothesis INTRODUCTION
Epidemiological studies have shown a lower prevalence of asthma and allergic sensitization in children and adults who lived on a farm or in a rural area during childhood (1-6). According to the hygiene hypothesis, the development of allergic disease may be influenced by bacterial and viral infections, and environmental exposure to noninfectious microbial agents such as endotoxin – which are abundantly present in the farm environment (7). Both exposure to environmental factors and genetic susceptibility play a role in the occurrence of asthma and allergy. Therefore, genes dependent of the innate immunity pathway are obvious candidates for the understanding of the protective effects of exposure to microbial agents on allergy and asthma. Indeed, several single nucleotide polymorphisms (SNPs) in genes encoding pattern recognition receptors such as CD14 and Toll-like receptors (TLR) have been associated with atopic sensitization and asthma (8, 9). The first report by Baldini et al. (10) on the functional (11) CD14 C -159 T SNP (C -260 T when counting from the translation start site) has shown increased levels of circulating soluble CD14 (sCD14) in TT homozygotes, and a lower number of positive skin prick tests in atopic children carrying this genotype. Although subsequent studies have often replicated the association between CD14/-260 T and increased sCD14, results on atopic sensitization have been conflicting and results for asthma were mainly negative (9, 12-14). Several studies have suggested that gene-environment interactions play an important role, since associations between CD14/-260 genotype and allergy and asthma-related outcomes have been shown to depend on the level of microbial exposures (15). A few studies have also shown gene-environment interactions in relation to SNPs in TLR2 and TLR4 genes, and other SNPs in the CD14 gene (-1721 showing a high linkage disequilibrium with CD14/-260) (16-18).
In the French Epidemiological study on the Genetics and Environment of Asthma, Bronchial Hyperresponsiveness, and Atopy (EGEA) a lower prevalence of asthma has been shown in adults who lived in the country during childhood (3). The purpose of the present study was to investigate whether SNPs in the candidate genes CD14, TLR2, TLR4, and TLR9 are associated with asthma in adults from the EGEA study by conducting a case-control analysis on unrelated subjects and a family based association test (19). We further investigated whether these SNPs modify associations between living in the country and asthma. Studies in children and adults have shown that exposure to endotoxin and other microbial agents may protect against asthma or wheeze in atopic subjects, whereas increased exposure may be a risk factor for nonatopic asthma or wheeze (20-22). Therefore, the role of atopy was evaluated by conducting separate analyses for atopic and nonatopic subjects. Some of the results presented here have been previously reported in the form of an abstract (23).
METHODS
Study population and design
The design of EGEA combines a case-control study and a family study of adult and childhood asthma. The design, ascertainment of asthmatic cases, and descriptive characteristics have been described earlier (24-27). Briefly, asthmatic patients (age 7-70 years) were recruited from six chest clinics in five French cities. Family members of asthmatic probands were included, either by including the proband’s parents and siblings, or by including the proband’s spouse and children. Controls were population-based. Inclusion criteria used to define asthma in probands were based on self-reported answers to the four questions “Have you ever had attacks of breathlessness at rest with wheezing?”, “Have you ever had asthma attacks?”, “Was this diagnosis confirmed by a physician?”, and “Have you had an asthma attack in the last 12 months?”, or a positive response to at least two questions and a positive expertise of their medical record (25). Asthma in relatives of probands was defined as a positive answer to the question “Have you ever had attacks of breathlessness at rest with wheezing?” or “Have you ever had asthma attacks?” (25). In the present study, “asthmatics” were defined as probands and relatives with asthma, “nonasthmatics” as population-based controls and relatives without asthma. Atopy was defined as a positive skin prick test (SPT) to one of the 11 common allergens tested (28). Country living during childhood was defined as a positive answer to the question “Have you ever lived in the country for at least one year?”, and being 16 years of age or younger when beginning to live in the country (3). Age 16 was chosen as a cut-off because in the initial study (3), living in the country before age 16 was most strongly associated with a reduced prevalence of asthma. Sensitivity analyses were conducted using country living before being 1 year of age, and additional information on the definition of country living is given in the online data supplement.
A case-control analysis was conducted in 825 genetically unrelated adults from the parental generation (aged 27-70 years), i.e. adult probands and their spouses, parents of probands from families where the proband is offspring, and population-based controls from the same generation. In addition, a family-based analysis was
conducted in 708 adult subjects from 192 families, comprising 378 parents who were included in the case-control analysis as well, and 330 adult offspring subjects who were not included in the analysis among unrelated subjects. All participants gave written informed consent. Additional information is given in the online data supplement.
Genotyping
Twenty-five SNPs belonging to the four candidate genes considered were selected using a tagging approach: CD14 (5 SNPs), TLR2 (5 SNPs), TLR4 (10 SNPs) and TLR9 (5 SNPs). This set of SNPs allowed us to capture the majority of common haplotype variation of these four genes (i.e., haplotype with a frequency > 5%). Details on genotyping are given in the online data supplement.
Statistical analysis
In the 825 genetically unrelated adults, odds ratios (ORs) for associations between genotype and asthma were calculated by generalized estimating equations (GEE model; Proc GENMOD using SAS statistical software) to adjust for dependence between subjects sharing the same household (probands and their spouses, and mothers and fathers of probands). ORs were adjusted for age and sex. The effect of each SNP on disease was tested under a general genetic model (2-degrees-of-freedom-test) and ORs were calculated using homozygotes for the most frequent allele as reference group. In addition, dominant, recessive, and additive genetic models were considered and the best-fitting model was selected (likelihood ratio test). We tested the null hypothesis that effects of genotype and country living on asthma were independent by including an interaction term in the model (gene-environment interaction) and testing the significance (GEE model, Z-score). False Discovery Rate (FDR) adjusted P-values were calculated to take multiple comparisons (n=25 SNPs) into account (29).
In order to confirm associations among unrelated subjects, a family-based association test, as implemented in FBAT (30) was performed as a complementary method to test for association between asthma and genetic polymorphisms. Details on the FBAT method are given in the online data supplement.
RESULTS
Identification numbers, position, and minor allele frequencies of studied SNPs are shown in Table E2 in the online data supplement. All SNPs were in Hardy-Weinberg equilibrium (P>.05). Tables E3-E6 in the online data supplement show linkage disequilibrium (LD) between polymorphisms for the four genes under study.
Characteristics of nonasthmatic (n=586) and asthmatic (n=239) subjects are shown in Table I. Living in the country during childhood was associated with a lower prevalence of asthma with an OR of 0.67 (0.48-0.92) (before 16y) and 0.72 (0.52-1.01) (before 1y, P=.06), which confirmed the earlier report in a different subpopulation of the EGEA study (3). Adjusting for age, sex, and smoking habits did not change the association.
Association between CD14, TLR2, TLR4, and TLR9 genetic polymorphisms and asthma
The genotypic distribution of CD14, TLR2, TLR4, and TLR9 SNPs and the associations between these polymorphisms and asthma in the 825 unrelated subjects are shown in Tables E7-E10 in the online data supplement. Carrying the TLR2/+596 C allele was associated with an increased risk for asthma (OR (95%CI) = 1.91 (1.34-2.72); P=.0003, P value adjusted for multiple comparisons =.008). Two other TLR2 SNPs, TLR2/-24438 and rs2289318, and two TLR9 SNPs, rs353547 and TLR9/+2848, were also associated with asthma, but not significantly after adjusting for multiple comparisons. Since low LD was observed among the three TLR2 SNPs on the one hand (LD coefficient r2≤0.34; Table E4) and between the two TLR9 SNPs on the other hand (LD coefficient r2=0.53; Table E6), we conducted two separate multivariate-SNP analyses by including these polymorphisms in the regression model. TLR2/+596 was the only SNP significantly associated with asthma in the model testing for three TLR2 markers. Estimates for each TLR9 SNP in the multiple regression model were similar to those in the univariate model. However, none of them was significant after mutual adjustment. Associations between genotype and asthma were not different for men and women (interaction P>.10 for all SNPs). Similar results were obtained when either population-based controls (n=117) or non-asthmatic family members (n=469) were excluded from the control group (data not shown).The positive association between the TLR2/+596 C allele and asthma under a dominant model was confirmed by a family-based association test in 192 families (Z=2.48; P=.01, FBAT). In the family-based analysis, asthma outcome of 330 offspring subjects determined the magnitude of the test statistic, whereas the case-control analyses in the 825 genetically unrelated subjects as described above included only the parental generation. For other SNPs no significant association was found using FBAT (P>.05; data not shown).
In the 825 genetically unrelated subjects, the influence of TLR2/+596 on associations between other SNPs and asthma (i.e. gene-gene interactions) was assessed by including an interaction term in the model and testing the significance (GEE model, Z-score). For three SNPs (CD14/+1188 and two TLR9 SNPs), a significant interaction with TLR2/+596 genotype was found (Table II), showing significant associations between these SNPs and asthma in TLR2/+596 TT homozygotes, but ORs around 1 in subjects carrying TLR2/+596 C.
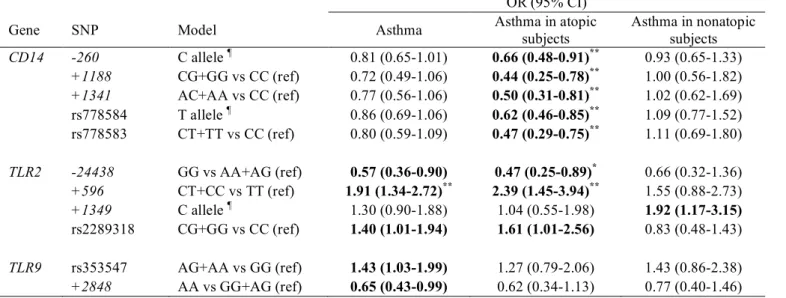
Separate analyses were conducted for atopic and nonatopic subjects. Associations between genotype and asthma outcomes for 11 SNPs associated with at least one of the asthma outcomes are presented in Table III (unadjusted P<.05 for 11 SNPs, FDR-adjusted P<.05 for 6 SNPs). Carrying the TLR2/+596 C allele was a significant risk factor for asthma in atopic subjects (OR 2.39, P=.0006), whereas a positive but not significant association was found in nonatopic subjects (OR 1.55, P=.13). The CD14/-260 C allele protected against asthma in atopic subjects with an OR of 0.66 (0.48-0.91). Four other CD14 SNPs were also significantly associated with a lower risk of asthma in atopic subjects, and for these SNPs, associations differed significantly from those observed in non-atopic subjects, who had ORs around 1 (gene-atopy interaction P<.05). Atopic subjects carrying a protective allele for each CD14 SNP had significantly less asthma compared with subjects carrying none of the protective alleles, with an OR of 0.34 (0.18-0.67). Carrying a protective allele for one to four CD14 SNPs showed an OR of 0.64 (0.37-1.16) compared with subjects carrying none. Multiple regression models including more than one CD14 SNP showed independent effects that were similar in size for each different SNP, thus the effect was not dominated by one particular SNP.
Role of CD14, TLR2, TLR4, and TLR9 genetic polymorphisms in the associations between living in the country and asthma
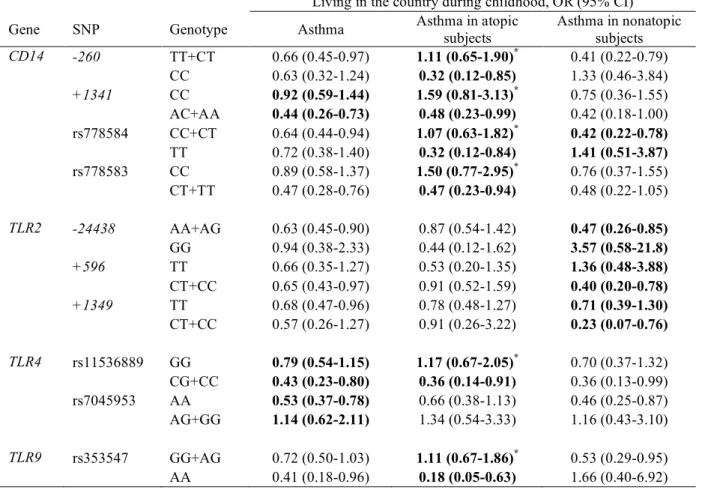
Gene-environment interactions between 10 SNPs and living in the country during childhood were found for at least one of the asthma outcomes (unadjusted P-value for interaction <.05 for 10 SNPs, FDR-adjusted P<.10 for 6 SNPs; Table IV). In atopic subjects with CD14/-260 CC genotype, country living protected against asthma (OR 0.32), whereas country living was not associated with asthma in atopics carrying the CD14/-260 T allele (OR 1.11). Conversely, in non-atopic subjects carrying CD14/-260 CC, country living was not associated with asthma (OR 1.33), whereas country living was associated with a lower risk of asthma in non-atopic subjects carrying the CD14/-260 T allele (OR 0.41). For this SNP, a significant second-order interaction term (gene x environment x atopy) was found (P=.005).
In atopics, gene-environment interactions were found as well for three other CD14 SNPs, one TLR4 SNP, and one TLR9 SNP (all FDR-adjusted P-values for interaction were borderline significant). In nonatopic subjects, gene-environment interactions were found for four SNPs, including TLR2/+596. In nonatopic carriers of the TLR2/+596 C allele, country life protected against asthma. Alternatively, when the results were stratified for living in the country, carrying at least one TLR2/+596 C allele (CC/CT vs TT) was a risk factor for asthma in nonatopic subjects who had not grown up in the country with an OR of 2.94 (1.16-7.41), but not in those who had grown up in the country (OR 0.77 (0.35-1.70)).
Sensitivity analyses using 1 year of age as a cut-off for beginning to live in the country showed very similar results, but the P-values for interaction were less often statistically significant, especially for atopic asthma (Table E11 in the online data supplement).
DISCUSSION
In the present study, an association was found between the TLR2/+596 polymorphism and asthma among French adults from the EGEA study. The well-studied CD14/-260 SNP was associated with asthma in SPT positive subjects – once again confirming the importance of this SNP in the occurrence of allergy and asthma related phenotypes. Gene-environment interactions were found between ten SNPs in candidate genes CD14, TLR4, TLR2, and TLR9 and living in the country during childhood, which was presumed to represent higher exposures to various microbial agents. Main effects and gene-environment interactions were stronger in atopic subjects than in non-atopic subjects. Significant interactions between CD14 SNPs and atopy were found in the association with asthma, moreover for CD14/-260, a significant interaction between genotype, atopy, and country living was found.
Eder et al. found that a SNP in TLR2 (A -16934 T) was associated with asthma, atopy, and hay fever in children of farmers (16). On the other hand, two studies investigating genetic variation in TLR2 reported no association with childhood-onset atopic asthma in a Japanese population (31), and no association with atopy or new-onset asthma in young Danish farmers (32). In the present study, we observed replication for the association between TLR2/+596 and asthma in case-control and family-based analyses. In the case-control analysis only the parental generation was included, whereas in the family-based analysis, genotype and asthma outcome of offspring determined the magnitude of the test statistic. Thus, results of the two different analyses were based on a different population within the EGEA study, which substantiates findings on TLR2. Moreover, although we tested a large number of SNPs in four candidate genes which increases the risk for false positive results, the findings on TLR2/+596 were still statistically significant after adjusting for multiple comparisons. TLR2 is involved in the recognition of microbial motifs of a wide range of Gram-positive microorganisms, mycobacteria and yeast, and exposures to these micoorganisms are likely to be higher in rural than in urban areas. We can only speculate on a possible mechanism explaining our findings, as the functional significance of TLR2/+596 is so far unknown. A possible explanation could be a lower expression of TLR2 on the surface of innate immune cells in carriers of the TLR2/+596C allele. As a consequence, carriers of TLR2/+596C would
benefit less from the protective effects of environmental exposures to TLR2 ligands on asthma. Further analyses of different asthma outcomes showed that TLR2/+596C was in particular associated with asthma in SPT positive subjects, whereas in nonatopic subjects, a significant interaction with childhood environment was found, showing that TLR2/+596C was only a risk factor for asthma in nonatopic subjects who had not grown up in the country. Thus, variation in TLR2 appears to be associated with asthma, but environmental exposure, asthma subphenotype, and also interactions with other genes such as CD14 and TLR9 were shown to play a role. TLR9 is a receptor for bacterial CpG DNA motifs, which have been found in increased levels in farm barn dust and dust from rural homes (33). Only few studies have investigated TLR9 SNPs in relation to allergy or asthma, showing inconsistent results (31, 34, 35). In the present study, only minor associations with asthma were found for TLR9 SNPs when other factors were not taken into account. However, significant gene-gene interactions with the TLR2/+596 SNP were found, showing effects of two TLR9 SNPs on asthma in TLR2/+596 TT subjects. Since TLR9 and TLR2 have different ligands, this finding might reflect an interactive effect of multiple microbial exposures.
In contrast to TLR2 and TLR9, which have not been studied widely in relation to asthma or allergic outcomes, CD14 is one of the best established susceptibility genes for asthma-related phenotypes such as allergic sensitization and total IgE (36). Studies investigating asthma in relation to CD14/-260 have often been negative, whereas either the C allele or T allele were alternately found to be a risk factor for allergic sensitization, number of positive SPT, or increased total IgE (9, 15). In the present study, CD14 polymorphisms protected against asthma, but only in atopic subjects, showing significant gene-atopy interactions.
In various studies, inverse associations between house dust endotoxin and allergic sensitization, total serum IgE, and asthma have been shown to be strongest in CD14/-260 CC homozygotes (15, 37, 38). In the present study, the inverse relationship between a rural childhood and asthma in atopic adults was also strongest in CD14/-260 CC subjects, and a significant gene-environment interaction was found. This apparent replication of a gene-environment interaction is remarkable, given the different phenotypes, age of study subjects, timing of exposure, study region, and exposure definitions across these studies. Conversely, in three other studies, the CD14/-260 TT genotype protected against atopic dermatitis, atopy, and increased IgE, especially in children with a dog in the home (39, 40), and in adults or young farmers who lived on a farm during childhood (32, 41). Gene-environment interactions are therefore likely to depend on qualitative and quantitative characteristics of environmental exposure, but also on phenotype, interactions with other genes, and on the age of study subjects (9, 36). The importance of atopy as an effect modifier in the present study underlines the complex interrelationships between environmental exposures, genetic variation in CD14, and asthma.
The fact that interactions were found between a rural childhood and polymorphisms in genes encoding four different pattern recognition receptors that become activated by a wide array of ligands, suggests that the protective effect of living in the country is – at least partly – a result of increased microbial exposures. In the present study, living in the country was used as a proxy for high exposure to microbial agents during childhood, since measured domestic endotoxin levels or more specific information regarding farm (animal) exposures was not available. Nevertheless, in the EGEA study it has been demonstrated that several lifestyle factors differed substantially between subjects who ever lived in rural communities and subjects who had always lived in urban regions: for instance, rural life was associated with factors such as using wood for heating, household size, and the presence of pets (3). Self-reported country living during childhood was also associated with parental farming and residential history (see Table E1 in the online data supplement). One could argue that concentrations of microbial agents in dust collected in rural homes are lower compared with dust from farm homes (42), and in our study only few subjects had farming parents. However, levels of microbial exposures in rural households have been found to be higher than exposure levels in urban homes (33). Moreover, considering the fact that most of the present study subjects were born in the 1940s/50s, it is likely that other relevant factors such as raw milk consumption, and exposure to stables and livestock were relatively common among those living in the country during their childhood compared with those in urban areas.
For country living during childhood, a 16 year cut-off was used. When we considered subjects with early-life rural exposures (before 1 year), very similar results were obtained, although gene-environment interactions were less often statistically significant. This may partly be due to a decreased power because of the lower number of early-life rural exposed subjects. It is also possible that rural exposures later in childhood may have a protective effect on asthma as well, which is suggested by the observed stronger relationship between country living and asthma when using 16 years as a cut-off.
In conclusion, TLR2 and CD14 are important genes for asthma in adults. Variation in CD14, TLR2, TLR4, and TLR9 genes modified associations between country living during childhood and asthma, in particular for asthma in atopic subjects.
EGEA cooperative group
Coordination: F Kauffmann; F Demenais (genetics); I Pin (clinical aspects).
Respiratory epidemiology: Inserm U 700, Paris M Korobaeff (EGEA1), F Neukirch (EGEA1); Inserm U 707,
Paris: I Annesi-Maesano; Inserm U 780, Villejuif: F Kauffmann, N Le Moual, R Nadif, MP Oryszczyn; Inserm U 823, Grenoble: V Siroux.
Genetics: Inserm U 393, Paris: J Feingold; Inserm U 535, Villejuif: MH Dizier; Inserm U 794, Paris: E
Bouzigon, F Demenais; CNG, Evry: I Gut, M Lathrop.
Clinical centers: Grenoble: I Pin, C Pison; Lyon: D Ecochard (EGEA1), F Gormand, Y Pacheco; Marseille: D
Charpin (EGEA1), D Vervloet; Montpellier: J Bousquet; Paris Cochin: A Lockhart (EGEA1), R Matran (now in Lille); Paris Necker: E Paty, P Scheinmann; Paris-Trousseau: A Grimfeld, J Just.
Data and quality management: Inserm ex-U155 (EGEA1): J Hochez; Inserm U 780, Villejuif: N Le Moual, C
Ravault; Inserm U 794: N Chateigner; Grenoble: J Ferran
REFERENCES
1. Riedler J, Braun-Fahrlander C, Eder W, Schreuer M, Waser M, Maisch S, Carr D, Schierl R, Nowak D, von Mutius E. Exposure to farming in early life and development of asthma and allergy: A cross-sectional survey. Lancet 2001;358:1129-1133.
2. Von Ehrenstein OS, Von Mutius E, Illi S, Baumann L, Bohm O, von Kries R. Reduced risk of hay fever and asthma among children of farmers. Clin Exp Allergy 2000;30:187-193.
3. Kauffmann F, Oryszczyn MP, Maccario J. The protective role of country living on skin prick tests, immunoglobulin e and asthma in adults from the epidemiological study on the genetics and environment of asthma, bronchial hyper-responsiveness and atopy. Clin Exp Allergy 2002;32:379-386.
4. Kilpelainen M, Terho EO, Helenius H, Koskenvuo M. Childhood farm environment and asthma and sensitization in young adulthood. Allergy 2002;57:1130-1135.
5. Leynaert B, Neukirch C, Jarvis D, Chinn S, Burney P, Neukirch F. Does living on a farm during childhood protect against asthma, allergic rhinitis, and atopy in adulthood? Am J Respir Crit Care Med 2001;164:1829-1834.
6. Portengen L, Sigsgaard T, Omland O, Hjort C, Heederik D, Doekes G. Low prevalence of atopy in young danish farmers and farming students born and raised on a farm. Clin Exp Allergy 2002;32:247-253.
7. von Mutius E. Allergies, infections and the hygiene hypothesis--the epidemiological evidence. Immunobiology 2007;212:433-439.
8. Yang IA, Fong KM, Holgate ST, Holloway JW. The role of Toll-like receptors and related receptors of the innate immune system in asthma. Curr Opin Allergy Clin Immunol 2006;6:23-28.
9. Zhang G, Goldblatt J, LeSouef PN. Does the relationship between IgE and the CD14 gene depend on ethnicity? Allergy 2008;63:1411-7.
10. Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A polymorphism* in the 5' flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol 1999;20:976-983.
11. LeVan TD, Bloom JW, Bailey TJ, Karp CL, Halonen M, Martinez FD, Vercelli D. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J Immunol 2001;167:5838-5844.
12. Kedda MA, Lose F, Duffy D, Bell E, Thompson PJ, Upham J. The CD14 C-159T polymorphism is not associated with asthma or asthma severity in an Australian adult population. Thorax 2005;60:211-214. 13. Koppelman GH, Reijmerink NE, Colin Stine O, Howard TD, Whittaker PA, Meyers DA, Postma DS,
Bleecker ER. Association of a promoter polymorphism of the CD14 gene and atopy. Am J Respir Crit Care Med 2001;163:965-969.
14. Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet 2000;67:1154-1162.
15. Martinez FD. CD14, endotoxin, and asthma risk: actions and interactions. Proc Am Thorac Soc 2007;4:221-225.
16. Eder W, Klimecki W, Yu L, Von Mutius E, Riedler J, Braun-Fahrlander C, Nowak D, Martinez FD. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol 2004;113:482-488.
17. Bieli C, Eder W, Frei R, Braun-Fahrlander C, Klimecki W, Waser M, Riedler J, von Mutius E, Scheynius A, Pershagen G, et al. A polymorphism in CD14 modifies the effect of farm milk consumption on allergic diseases and CD14 gene expression. J Allergy Clin Immunol 2007;120:1308-1315.
18. Werner M, Topp R, Wimmer K, Richter K, Bischof W, Wjst M, Heinrich J. TLR4 gene variants modify endotoxin effects on asthma. J Allergy Clin Immunol 2003;112:323-330.
19. Cordell HJ, Clayton DG. Genetic association studies. Lancet 2005;366:1121-31.
20. Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, Maisch S, Carr D, Gerlach F, Bufe A, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med 2002;347:869-877.
21. Douwes J, Pearce N, Heederik D. Does environmental endotoxin exposure prevent asthma? Thorax 2002;57:86-90.
22. Smit LA, Heederik D, Doekes G, Blom C, van Zweden I, Wouters IM. Exposure-response analysis of allergy and respiratory symptoms in endotoxin-exposed adults. Eur Respir J 2008;31:1241-1248.
23. Smit LAM, Siroux V, Bouzigon E, Oryszczyn MP, Demenais F, Kauffmann F. CD14 and Toll-like receptor gene polymorphisms, country living during childhood, and asthma in adults of the French EGEA study [abstract]. Eur Respir J 2008: 481s.
24. Kauffmann F, Dizier MH. EGEA (Epidemiological study on the Genetics and Environment of Asthma, bronchial hyperresponsiveness and atopy)--design issues. EGEA Co-operative group. Clin Exp Allergy 1995;25 Suppl 2:19-22.
25. Kauffmann F, Dizier MH, Pin I, Paty E, Gormand F, Vervloet D, Bousquet J, Neukirch F, Annesi I, Oryszczyn MP, et al. Epidemiological study of the Genetics and Environment of Asthma, bronchial hyperresponsiveness, and atopy: Phenotype issues. Am J Respir Crit Care Med 1997;156:S123-129. 26. Kauffmann F, Dizier MH, Annesi-Maesano I, Bousquet J, Charpin D, Demenais F, Ecochard D,
Feingold J, Gormand F, Grimfeld A, et al. EGEA (Epidemiological study on the Genetics and Environment of Asthma, bronchial hyperresponsiveness and atopy)-- descriptive characteristics. Clin Exp Allergy 1999;29 Suppl 4:17-21.
27. Kauffmann F, Dizier MH, Annesi-Maesano I, Bousquet J, Charpin D, Demenais F, Ecochard D, Feingold J, Gormand F, Grimfeld A, et al. [Epidemiological study of genetic and environmental factors in asthma, bronchial hyperresponsiveness and atopy. Protocol and potential selection bias]. Rev Epidemiol Sante Publique 2001;49:343-356.
28. Maccario J, Oryszczyn MP, Charpin D, Kauffmann F. Methodologic aspects of the quantification of skin prick test responses: The EGEA study. J Allergy Clin Immunol 2003;111:750-756.
29. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995;57:289-300.
30. Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet 2001;9:301-306.
31. Noguchi E, Nishimura F, Fukai H, Kim J, Ichikawa K, Shibasaki M, Arinami T. An association study of asthma and total serum Immunoglobin E levels for Toll-like receptor polymorphisms in a Japanese population. Clin Exp Allergy 2004;34:177-183.
32. Smit LA, Bongers SI, Ruven HJ, Rijkers GT, Wouters IM, Heederik D, Omland O, Sigsgaard T. Atopy and new-onset asthma in young Danish farmers and CD14, TLR2, and TLR4 genetic polymorphisms: a nested case-control study. Clin Exp Allergy 2007;37:1602-1608.
33. Roy SR, Schiltz AM, Marotta A, Shen Y, Liu AH. Bacterial DNA in house and farm barn dust. J Allergy Clin Immunol 2003;112:571-578.
34. Lazarus R, Klimecki WT, Raby BA, Vercelli D, Palmer LJ, Kwiatkowski DJ, Silverman EK, Martinez F, Weiss ST. Single-nucleotide polymorphisms in the Toll-like receptor 9 gene (TLR9): Frequencies, pairwise linkage disequilibrium, and haplotypes in three U.S. ethnic groups and exploratory case-control disease association studies. Genomics 2003;81:85-91.
35. Berghofer B, Frommer T, Konig IR, Ziegler A, Chakraborty T, Bein G, Hackstein H. Common human Toll-like receptor 9 polymorphisms and haplotypes: association with atopy and functional relevance. Clin Exp Allergy 2005;35:1147-1154.
36. Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol 2008;8:169-182. 37. Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, Nowak D, Martinez FD, The
Allergy and Endotoxin Alex Study Team. Opposite effects of CD14/-260 on serum IgE levels in children raised in different environments. J Allergy Clin Immunol 2005;116:601-607.
38. Simpson A, John SL, Jury F, Niven R, Woodcock A, Ollier WE, Custovic A. Endotoxin exposure, CD14, and allergic disease: an interaction between genes and the environment. Am J Respir Crit Care Med 2006;174:386-392.
39. Gern JE, Reardon CL, Hoffjan S, Nicolae D, Li Z, Roberg KA, Neaville WA, Carlson-Dakes K, Adler K, Hamilton R, et al. Effects of dog ownership and genotype on immune development and atopy in infancy. J Allergy Clin Immunol 2004;113:307-314.
40. Bottema RW, Reijmerink NE, Kerkhof M, Koppelman GH, Stelma FF, Gerritsen J, Thijs C, Brunekreef B, van Schayck CP, Postma DS. Interleukin 13, CD14, pet and tobacco smoke influence atopy in three Dutch cohorts: the allergenic study. Eur Respir J 2008;32:593-602.
41. Leynaert B, Guilloud-Bataille M, Soussan D, Benessiano J, Guenegou A, Pin I, Neukirch F. Association between farm exposure and atopy, according to the CD14 C-159T polymorphism. J Allergy Clin Immunol 2006;118:658-665.
42. von Mutius E, Braun-Fahrlander C, Schierl R, Riedler J, Ehlermann S, Maisch S, Waser M, Nowak D. Exposure to endotoxin or other bacterial components might protect against the development of atopy. Clin Exp Allergy 2000;30:1230-1234.
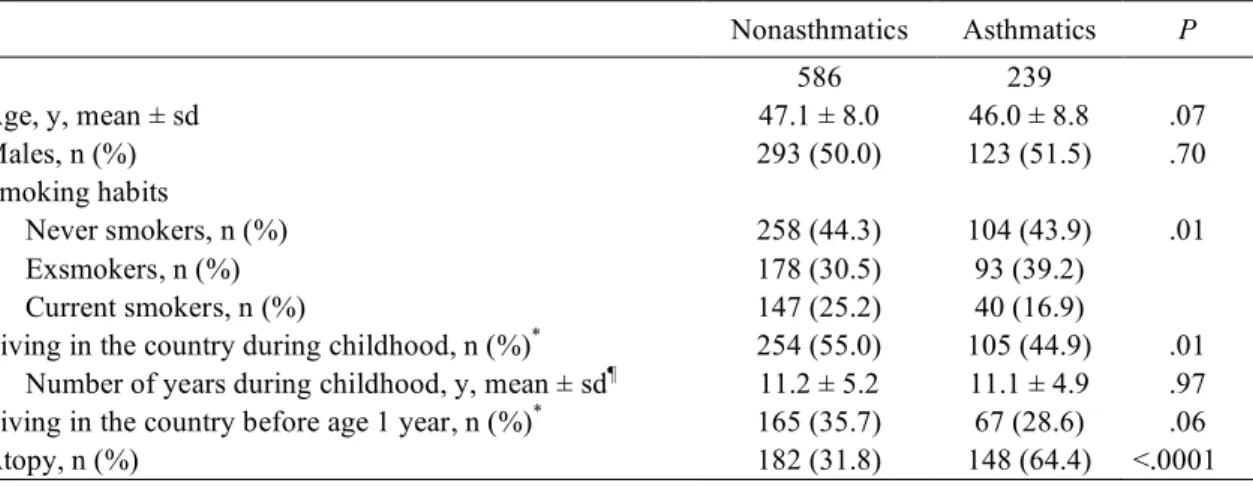
TABLE I. Characteristics of the study population
Nonasthmatics Asthmatics P n 586 239 Age, y, mean ± sd 47.1 ± 8.0 46.0 ± 8.8 .07 Males, n (%) 293 (50.0) 123 (51.5) .70 Smoking habits Never smokers, n (%) 258 (44.3) 104 (43.9) .01 Exsmokers, n (%) 178 (30.5) 93 (39.2) Current smokers, n (%) 147 (25.2) 40 (16.9)
Living in the country during childhood, n (%)* 254 (55.0) 105 (44.9) .01 Number of years during childhood, y, mean ± sd¶ 11.2 ± 5.2 11.1 ± 4.9 .97 Living in the country before age 1 year, n (%)* 165 (35.7) 67 (28.6) .06
Atopy, n (%) 182 (31.8) 148 (64.4) <.0001
* population-based controls(n=117) excluded ¶
TABLE II. Associations between CD14 and TLR9 SNPs and asthma for SNPs showing significant gene-gene interactions with TLR2/+596 Asthma (OR (95% CI)) P interaction SNP Model All subjects (n=825)# TLR2/+596 TT
(n=259)# TLR2/+596 CT/CC (n=529)# Unadjusted FDR adjusted CD14/+1188 CG+GG vs CC (ref) 0.72 (0.49-1.06) 0.30 (0.11-0.81) 0.93 (0.60-1.45) .03 .16 TLR9 rs352143 AG+GG vs AA (ref) 1.15 (0.85-1.55) 2.31 (1.24-4.30) 0.95 (0.66-1.38) .02 .15 TLR9 rs352163 T allele ¶ 0.87 (0.70-1.09) 0.48 (0.30-0.74) 1.04 (0.80-1.35) .002 .04
¶ Additive genetic model, the genotype was categorized into a three level variable for the number of minor alleles (0,1,2).
Ref: reference genotype coded as 0. Bold: p<0.05.
# Exact numbers of genotyped subjects for each SNP are given in Table E2. TLR2/+596 was genotyped in 788 subjects.
TABLE III. Associations between CD14, TLR2, TLR4, and TLR9 SNPs and asthma for SNPs significantly associated with at least one of the asthma outcomes OR (95% CI)
Gene SNP Model Asthma Asthma in atopic
subjects Asthma in nonatopic subjects CD14 -260 C allele ¶ 0.81 (0.65-1.01) 0.66 (0.48-0.91)** 0.93 (0.65-1.33) +1188 CG+GG vs CC (ref) 0.72 (0.49-1.06) 0.44 (0.25-0.78)** 1.00 (0.56-1.82) +1341 AC+AA vs CC (ref) 0.77 (0.56-1.06) 0.50 (0.31-0.81)** 1.02 (0.62-1.69) rs778584 T allele ¶ 0.86 (0.69-1.06) 0.62 (0.46-0.85)** 1.09 (0.77-1.52) rs778583 CT+TT vs CC (ref) 0.80 (0.59-1.09) 0.47 (0.29-0.75)** 1.11 (0.69-1.80) TLR2 -24438 GG vs AA+AG (ref) 0.57 (0.36-0.90) 0.47 (0.25-0.89)* 0.66 (0.32-1.36) +596 CT+CC vs TT (ref) 1.91 (1.34-2.72)** 2.39 (1.45-3.94)** 1.55 (0.88-2.73) +1349 C allele ¶ 1.30 (0.90-1.88) 1.04 (0.55-1.98) 1.92 (1.17-3.15) rs2289318 CG+GG vs CC (ref) 1.40 (1.01-1.94) 1.61 (1.01-2.56) 0.83 (0.48-1.43) TLR9 rs353547 AG+AA vs GG (ref) 1.43 (1.03-1.99) 1.27 (0.79-2.06) 1.43 (0.86-2.38) +2848 AA vs GG+AG (ref) 0.65 (0.43-0.99) 0.62 (0.34-1.13) 0.77 (0.40-1.46) Bold: p<0.05, not adjusted for multiple comparisons; * p<0.10, FDR adjusted p value; ** p<0.05, FDR adjusted p value
¶ Additive genetic model, the genotype was categorized into a three level variable for the number of minor alleles (0,1,2).
TABLE IV. Gene-environment interactions between CD14, TLR2, TLR4, and TLR9 SNPs and country living during childhood in asthma outcomes, shown as associations between country living and asthma, stratified by genotype
Living in the country during childhood, OR (95% CI) Gene SNP Genotype Asthma Asthma in atopic
subjects Asthma in nonatopic subjects CD14 -260 TT+CT 0.66 (0.45-0.97) 1.11 (0.65-1.90)* 0.41 (0.22-0.79) CC 0.63 (0.32-1.24) 0.32 (0.12-0.85) 1.33 (0.46-3.84) +1341 CC 0.92 (0.59-1.44) 1.59 (0.81-3.13)* 0.75 (0.36-1.55) AC+AA 0.44 (0.26-0.73) 0.48 (0.23-0.99) 0.42 (0.18-1.00) rs778584 CC+CT 0.64 (0.44-0.94) 1.07 (0.63-1.82)* 0.42 (0.22-0.78) TT 0.72 (0.38-1.40) 0.32 (0.12-0.84) 1.41 (0.51-3.87) rs778583 CC 0.89 (0.58-1.37) 1.50 (0.77-2.95)* 0.76 (0.37-1.55) CT+TT 0.47 (0.28-0.76) 0.47 (0.23-0.94) 0.48 (0.22-1.05) TLR2 -24438 AA+AG 0.63 (0.45-0.90) 0.87 (0.54-1.42) 0.47 (0.26-0.85) GG 0.94 (0.38-2.33) 0.44 (0.12-1.62) 3.57 (0.58-21.8) +596 TT 0.66 (0.35-1.27) 0.53 (0.20-1.35) 1.36 (0.48-3.88) CT+CC 0.65 (0.43-0.97) 0.91 (0.52-1.59) 0.40 (0.20-0.78) +1349 TT 0.68 (0.47-0.96) 0.78 (0.48-1.27) 0.71 (0.39-1.30) CT+CC 0.57 (0.26-1.27) 0.91 (0.26-3.22) 0.23 (0.07-0.76) TLR4 rs11536889 GG 0.79 (0.54-1.15) 1.17 (0.67-2.05)* 0.70 (0.37-1.32) CG+CC 0.43 (0.23-0.80) 0.36 (0.14-0.91) 0.36 (0.13-0.99) rs7045953 AA 0.53 (0.37-0.78) 0.66 (0.38-1.13) 0.46 (0.25-0.87) AG+GG 1.14 (0.62-2.11) 1.34 (0.54-3.33) 1.16 (0.43-3.10) TLR9 rs353547 GG+AG 0.72 (0.50-1.03) 1.11 (0.67-1.86)* 0.53 (0.29-0.95) AA 0.41 (0.18-0.96) 0.18 (0.05-0.63) 1.66 (0.40-6.92) Bold: interaction p<0.05, not adjusted for multiple comparisons; * interaction p<0.10, FDR adjusted p value
ONLINE DATA SUPPLEMENT
CD14 and Toll-like receptor gene polymorphisms, country living, and asthma in adults
Lidwien A. M. Smit, Valérie Siroux, Emmanuelle Bouzigon, Marie-Pierre Oryszczyn, Mark Lathrop, Florence Demenais, Francine Kauffmann, on behalf of the EGEA cooperative group
Study population and design
Subjects were enrolled in six chest clinics from five French cities (Grenoble, Lyon, Marseille, Montpellier, Paris) between 1991 and 1995. Inclusion criteria used to define asthma in probands were based on self-reported answers to the four questions “Have you ever had attacks of breathlessness at rest with wheezing?”, “Have you ever had asthma attacks?”, “Was this diagnosis confirmed by a physician?”, and “Have you had an asthma attack in the last 12 months?”, or a positive response to at least two questions and a positive expertise of their medical record (E1). All probands and their two parents were Caucasian and were born in France. All family members answered a detailed questionnaire based on British Medical Research Council (BMRC)/European Coal and Steel Community (ECSC), American Thoracic Society (ATS), and European Community Respiratory Health Survey (ECRHS) questionnaires. Based on the BMRC/ECSC questionnaire, asthma in relatives of probands was defined as a positive answer to the question “Have you ever had attacks of breathlessness at rest with wheezing?” or “Have you ever had asthma attacks?” (E1). Population-based controls were selected from electoral rolls, a surgery department, and a social security check-up center. In the present study, “asthmatics” were defined as probands and relatives with asthma, “nonasthmatics” as population-based controls and relatives without asthma. Population-based controls with asthma (n=15) were excluded from data analysis.
Figure E1 represents a diagram describing the population used in the present analysis. As shown in Figure E1, a case-control analysis was conducted in genetically unrelated adults from the parental generation. Population-based controls were assigned to the parental generation (controls aged 40-64 yrs) or offspring generation (controls aged 18-47 yrs) on the basis of age in order to obtain an optimal age distribution between cases and controls. The study protocol was approved by the institutional ethics committee and all participants gave written informed consent.
Country living
The present study was restricted to adult subjects of the EGEA study because information on country living was only available for adults. Country living was not taken into account during recruitment, as this was not considered of importance when the study was designed in 1990/1991. As described earlier, the area of residence for asthmatic probands and their relatives was broader than for most population-based controls, reflecting the area of attraction of the chest clinics, and significantly more asthmatic probands were living in the country than population-based controls (E2). Analyses that included country living were therefore conducted in asthmatic probands and relatives only, excluding controls (n=117).
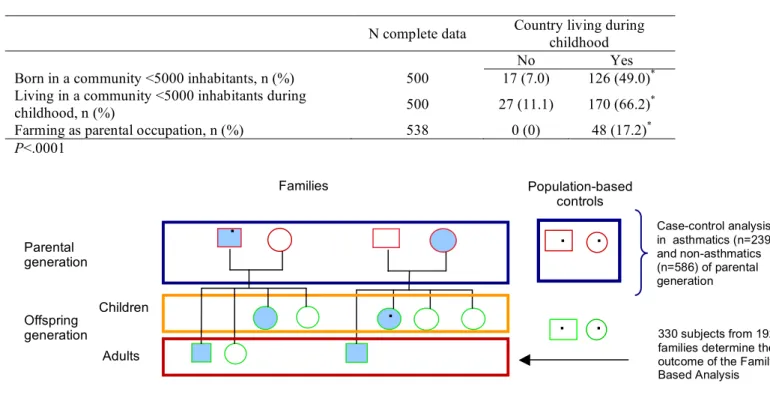
Country living during childhood was defined as living in a rural area for at least one year in a row, and being 16 years of age or younger when beginning to live in the country. In EGEA2, the 12 year follow-up of the first EGEA survey which has been conducted recently (between 2003 and 2006) (E3), more questions on residential history and farming were asked. In EGEA2 the complete residential history (at the French ‘commune’ level) was obtained and these data were combined with demographic statistics indicating the number of inhabitants of the ‘commune’. Table E1 shows that subjects with and without a rural childhood are significantly different with regard to farming parents and residential history.
Genotyping
Genotyping was performed using either Taqman Probes (Applied Biosystems, Foster City, CA) on an ABI7900HT Sequence Detection System or 1536-plex Illumina Golden Gate assay (Illumina, San Diego, CA) at the Centre National de Génotypage (CNG, Evry, France).
FBAT
A family-based association test, as implemented in FBAT (E4) was performed as a complementary method to test for association between asthma and genetic polymorphisms in the adult offspring sample. This approach tests for the association between the phenotype and excess transmission of a specific allele from parent to offspring. The advantage of this approach is that populationstratification is avoided. We used the –o option in FBAT program allowing to take into account affected and unaffected offspring in the analysis. Association was initially tested under a general model and subsequently under additive, dominant and recessive models.
TABLE E1. Factors related to living in the country during childhood
N complete data Country living during childhood
No Yes
Born in a community <5000 inhabitants, n (%) 500 17 (7.0) 126 (49.0)* Living in a community <5000 inhabitants during
childhood, n (%) 500 27 (11.1) 170 (66.2)
*
Farming as parental occupation, n (%) 538 0 (0) 48 (17.2)* P<.0001
Figure E1. Diagram describing the study population used in the case-control and family-based analysis.
Asthmatic subjects are represented by shaded shapes. Asthmatic probands and population-based controls are represented by a dot.
TABLE E2. SNP identification numbers, position, and minor allele frequencies
dbSNP ID Gene Genomic location Position from translation start site Alleles Minor allele frequency N rs2569190 CD14 5 :139993100 -260 T/C 0.50 777 rs4914 CD14 5 :139991652 +1188 C/G 0.12 783 rs2563298 CD14 5 :139991499 +1341 C/A 0.27 781 rs778584 CD14 5 :139985396 C/T 0.49 819 rs778583 CD14 5 :139985294 C/T 0.28 822 rs13150331 TLR2 4 :154819072 -24438 A/G 0.40 817 rs3804099 TLR2 4 :154844106 +596 T/C 0.43 788 rs3804100 TLR2 4 :154844859 +1349 T/C 0.08 822 rs5743700 TLR2 4 :154845132 +1622 C/T 0.04 786 rs2289318 TLR2, RNF175 4 :154853184 C/G 0.17 822 rs10759930 TLR4 9 :119501442 C/T 0.41 822 rs2737191 TLR4 9 :119502536 A/G 0.27 821 rs4986791 TLR4 9 :119515423 +1196, Thr399Ile C/T 0.06 792 rs7869402 TLR4 9 :119517853 C/T 0.02 790 rs11536889 TLR4 9 :119517952 G/C 0.17 820 rs1927906 TLR4 9 :119519936 A/G 0.09 821 rs1554973 TLR4 9 :119520633 T/C 0.24 817 rs913930 TLR4 9 :119523830 T/C 0.34 820 Population-based controls Families Parental generation
.
.
.
.
.
.
x
.
Offspring generation Case-control analysis in asthmatics (n=239) and non-asthmatics (n=586) of parental generation Children Adults 330 subjects from 192 families determine the outcome of the Family Based Analysisrs1927905 TLR4 9 :119525129 A/G 0.06 822 rs7045953 TLR4 9 :119525616 A/G 0.15 821 rs353547 TLR9, PTK9L 3 :52243906 G/A 0.39 821 rs11717574 TLR9, PTK9L 3 :52243286 T/C 0.15 815 rs352143 TLR9, PTK9L 3 :52239947 A/G 0.21 822 rs352140 TLR9 3 :52231737 +2848 G/A 0.47 778 rs352163 TLR9, ALAS1 3 :52222150 C/T 0.47 817
TABLE E3. Linkage disequilibrium (r2) between CD14 SNPs -260 +1188 +1341 rs778584 rs778583 -260 0.14 0.37 0.95 0.37 +1188 0.37 0.14 0.35 +1341 0.38 0.98 rs778584 0.39 rs778583
TABLE E4. Linkage disequilibrium (r2) between TLR2 SNPs -24438 +596 +1349 +1622 rs2289318 -24438 0.34 0.05 0.03 0.05 +596 0.11 0.06 0.26 +1349 0 0.02 +1622 0.23 rs2289318
TABLE E5. Linkage disequilibrium (r2) between TLR4 SNPs
rs10759930 rs2737191 Thr399Ile rs7869402 rs11536889 rs1927906 rs1554973 rs913930 rs1927905 rs7045953 rs10759930 0.26 0.05 0.02 0.26 0.07 0.21 0.34 0.04 0.12 rs2737191 0.03 0.01 0.06 0.04 0.11 0.65 0.02 0.06 Thr399Ile 0 0.01 0.70 0.22 0.03 0 0.01 rs7869402 0.01 0.25 0.08 0.01 0 0 rs11536889 0.02 0.07 0.11 0.01 0.04 rs1927906 0.31 0.05 0 0.01 rs1554973 0.16 0.21 0.57 rs913930 0.03 0.09 rs1927905 0.37 rs7045953
TABLE E6. Linkage disequilibrium (r2) between TLR9 SNPs
rs353547 rs11717574 rs352143 +2848 rs352163 rs353547 0.11 0.01 0.53 0.45 rs11717574 0.65 0.13 0.13 rs352143 0.21 0.21 +2848 0.89 rs352163
TABLE E7. Association between CD14 genotype and asthma
SNP Nonasthmatics, n (%) Asthmatics, n (%) OR (95%CI) CD14/-260
TT 133 (24.0) 67 (30.0) 1.00
CT 276 (49.8) 107 (48.0) 0.75 (0.51-1.10)
C allele ¶ 0.81 (0.65-1.01) CD14/+1188 CC 428 (76.3) 181 (81.5) 1.00 CG 122 (21.8) 37 (16.7) 0.71 (0.48-1.06) GG 11 (2.0) 4 (1.8) 0.85 (0.27-2.69) CG+GG vs CC (ref) 0.72 (0.49-1.06) CD14/+1341 CC 292 (52.4) 131 (58.5) 1.00 AC 214 (38.4) 76 (33.9) 0.78 (0.56-1.09) AA 51 (9.2) 17 (7.6) 0.74 (0.42-1.30) AC+AA vs CC (ref) 0.77 (0.56-1.06) rs778584 CC 146 (25.1) 69 (29.0) 1.00 CT 286 (49.2) 117 (49.2) 0.85 (0.59-1.23) TT 149 (25.7) 52 (21.9) 0.73 (0.48-1.12) T allele ¶ 0.86 (0.69-1.06) rs778583 CC 303 (52.0) 137 (57.3) 1.00 CT 226 (38.8) 85 (35.6) 0.82 (0.60-1.14) TT 54 (9.3) 17 (7.1) 0.70 (0.39-1.22) CT+TT vs CC (ref) 0.80 (0.59-1.09)
¶ Additive genetic model, the genotype was categorized into a three level variable for the number of minor
alleles (0,1,2).
Ref: reference genotype coded as 0.
TABLE E8. Association between TLR2 genotype and asthma
SNP Nonasthmatics, n (%) Asthmatics, n (%) OR (95%CI) TLR2/-24438 AA 192 (33.2) 94 (39.3) 1.00 AG 286 (49.5) 120 (50.2) 0.86 (0.62-1.21) GG 100 (17.3) 25 (10.5) 0.52 (0.31-0.86) GG vs AA+AG (ref) 0.57 (0.36-0.90) TLR2/+596 TT 207 (36.6) 52 (23.3) 1.00 CT 259 (45.8) 122 (54.7) 1.88 (1.30-2.71) CC 99 (17.5) 49 (22.0) 1.99 (1.24-3.19) CT+CC vs TT (ref) 1.91 (1.34-2.72) TLR2/+1349 TT 502 (86.1) 199 (83.3) 1.00 CT 77 (13.2) 36 (15.1) 1.20 (0.79-1.85) CC 4 (0.7) 4 (1.7) 2.58 (0.64-10.39) C allele ¶ 1.30 (0.90-1.88) TLR2/+1622 CC 515 (91.3) 201 (90.5) 1.00 CT 49 (8.7) 21 (9.5) 1.10 (0.64-1.92) rs2289318 CC 419 (71.9) 154 (64.4) 1.00 CG 145 (24.9) 81 (33.9) 1.51 (1.08-2.10) GG 19 (3.3) 4 (1.7) 0.57 (0.19-1.72) CG+GG vs CC (ref) 1.40 (1.01-1.94) ¶
Additive genetic model, the genotype was categorized into a three level variable for the number of minor alleles (0,1,2).
TABLE E9. Association between TLR4 genotype and asthma
SNP Nonasthmatics, n (%) Asthmatics, n (%) OR (95%CI) rs10759930 CC 192 (32.9) 88 (36.8) 1.00 CT 305 (52.3) 107 (44.8) 0.76 (0.55-1.06) TT 86 (14.8) 44 (18.4) 1.14 (0.73-1.78) TT vs CC+CT (ref) 1.34 (0.88-2.03) rs2737191 AA 299 (51.4) 123 (51.5) 1.00 AG 248 (42.6) 101 (42.3) 0.99 (0.73-1.34) GG 35 (6.0) 15 (6.3) 1.00 (0.52-1.89) AG+GG vs AA (ref) 0.99 (0.74-1.32) TLR4 Thr399Ile CC 492 (86.6) 198 (88.4) 1.00 CT 76 (13.4) 26 (11.6) 0.85 (0.52-1.39) rs7869402 CC 544 (96.3) 211 (93.8) 1.00 CT 21 (3.7) 13 (5.8) 1.56 (0.78-3.12) TT 0 (0.0) 1 (0.4) n.e. rs11536889 GG 385 (66.2) 174 (73.1) 1.00 CG 183 (31.4) 55 (23.1) 0.69 (0.48-0.99) CC 14 (2.4) 9 (3.8) 1.63 (0.64-4.11) CG+CC vs GG (ref) 0.72 (0.51-1.00) rs1927906 AA 482 (82.7) 196 (82.4) 1.00 AG 99 (17.0) 39 (16.4) 0.97 (0.65-1.47) GG 2 (0.3) 3 (1.3) 3.33 (0.58-19.08) AG+GG vs AA (ref) 1.03 (0.69-1.53) rs1554973 TT 332 (57.4) 140 (58.6) 1.00 CT 219 (37.9) 83 (34.7) 0.92 (0.67-1.26) CC 27 (4.7) 16 (6.7) 1.39 (0.75-2.59) CC vs TT+CT (ref) 1.44 (0.79-2.64) rs913930 TT 252 (43.3) 99 (41.6) 1.00 CT 270 (46.4) 113 (47.5) 1.06 (0.78-1.45) CC 60 (10.3) 26 (10.9) 1.07 (0.64-1.78) CT+CC vs TT (ref) 1.06 (0.79-1.43) rs1927905 AA 513 (88.0) 208 (87.0) 1.00 AG 69 (11.8) 29 (12.1) 1.06 (0.67-1.66) GG 1 (0.2) 2 (0.8) 4.62 (0.44-49.07) AG+GG vs AA (ref) 1.11 (0.72-1.72) rs7045953 AA 415 (71.3) 174 (72.8) 1.00 AG 157 (27.0) 60 (25.1) 0.92 (0.66-1.29) GG 10 (1.7) 5 (2.1) 1.21 (0.45-3.21) GG vs AA+AG (ref) 1.23 (0.47-3.26)
Ref: reference genotype coded as 0 n.e. not estimable
TABLE E10. Association between TLR9 genotype and asthma
SNP Nonasthmatics, n (%) Asthmatics, n (%) OR (95%CI) rs353547 GG 233 (40.0) 76 (31.9) 1.00 AG 263 (45.1) 124 (52.1) 1.46 (1.04-2.07) AA 87 (14.9) 38 (16.0) 1.35 (0.85-2.15) AG+AA vs GG (ref) 1.43 (1.03-1.99) rs11717574 TT 419 (72.5) 171 (72.2) 1.00 CT 140 (24.2) 62 (26.2) 1.08 (0.77-1.53) CC 19 (3.3) 4 (1.7) 0.52 (0.17-1.55) CC vs TT+CT (ref) 0.51 (0.17-1.51) rs352143 AA 370 (63.5) 144 (60.3) 1.00 AG 180 (30.9) 83 (34.7) 1.19 (0.87-1.64) GG 33 (5.7) 12 (5.0) 0.92 (0.47-1.83) AG+GG vs AA (ref) 1.15 (0.85-1.55) TLR9/+2848 GG 154 (27.6) 65 (29.6) 1.00 AG 275 (49.3) 118 (53.6) 1.00 (0.70-1.44) AA 129 (23.1) 37 (16.8) 0.65 (0.40-1.07) AA vs GG+AG (ref) 0.65 (0.43-0.99) rs352163 CC 159 (27.4) 67 (28.3) 1.00 CT 286 (49.3) 128 (54.0) 1.06 (0.75-1.50) TT 135 (23.3) 42 (17.7) 0.73 (0.46-1.16) T allele ¶ 0.87 (0.70-1.09) ¶
Additive genetic model, the genotype was categorized into a three level variable for the number of minor alleles (0,1,2).
Ref: reference genotype coded as 0
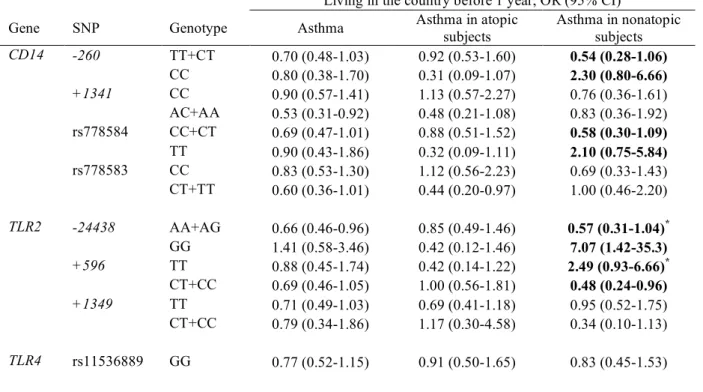
TABLE E11. Gene-environment interactions between CD14, TLR2, TLR4, and TLR9 SNPs and country living
before 1 year in asthma outcomes, shown as associations between country living before 1 year and asthma, stratified by genotype
Living in the country before 1 year, OR (95% CI) Gene SNP Genotype Asthma Asthma in atopic
subjects Asthma in nonatopic subjects CD14 -260 TT+CT 0.70 (0.48-1.03) 0.92 (0.53-1.60) 0.54 (0.28-1.06) CC 0.80 (0.38-1.70) 0.31 (0.09-1.07) 2.30 (0.80-6.66) +1341 CC 0.90 (0.57-1.41) 1.13 (0.57-2.27) 0.76 (0.36-1.61) AC+AA 0.53 (0.31-0.92) 0.48 (0.21-1.08) 0.83 (0.36-1.92) rs778584 CC+CT 0.69 (0.47-1.01) 0.88 (0.51-1.52) 0.58 (0.30-1.09) TT 0.90 (0.43-1.86) 0.32 (0.09-1.11) 2.10 (0.75-5.84) rs778583 CC 0.83 (0.53-1.30) 1.12 (0.56-2.23) 0.69 (0.33-1.43) CT+TT 0.60 (0.36-1.01) 0.44 (0.20-0.97) 1.00 (0.46-2.20) TLR2 -24438 AA+AG 0.66 (0.46-0.96) 0.85 (0.49-1.46) 0.57 (0.31-1.04)* GG 1.41 (0.58-3.46) 0.42 (0.12-1.46) 7.07 (1.42-35.3) +596 TT 0.88 (0.45-1.74) 0.42 (0.14-1.22) 2.49 (0.93-6.66)* CT+CC 0.69 (0.46-1.05) 1.00 (0.56-1.81) 0.48 (0.24-0.96) +1349 TT 0.71 (0.49-1.03) 0.69 (0.41-1.18) 0.95 (0.52-1.75) CT+CC 0.79 (0.34-1.86) 1.17 (0.30-4.58) 0.34 (0.10-1.13) TLR4 rs11536889 GG 0.77 (0.52-1.15) 0.91 (0.50-1.65) 0.83 (0.45-1.53)
CG+CC 0.60 (0.30-1.18) 0.52 (0.20-1.39) 0.66 (0.22-2.01) rs7045953 AA 0.56 (0.38-0.85) 0.64 (0.36-1.15) 0.56 (0.29-1.09)
AG+GG 1.34 (0.71-2.55) 1.00 (0.37-2.66) 1.73 (0.66-4.57)
TLR9 rs353547 GG+AG 0.84 (0.59-1.21) 1.09 (0.62-1.92)* 0.82 (0.47-1.45) AA 0.28 (0.11-0.73) 0.12 (0.03-0.48) 0.80 (0.19-3.30) Bold: interaction p<0.05, not adjusted for multiple comparisons; * interaction p<0.10, FDR adjusted p value
REFERENCES
E1. Kauffmann F, Dizier MH, Pin I, Paty E, Gormand F, Vervloet D, Bousquet J, Neukirch F, Annesi I, Oryszczyn MP, et al. Epidemiological study of the Genetics and Environment of Asthma, bronchial hyperresponsiveness, and atopy: Phenotype issues. Am J Respir Crit Care Med 1997;156:S123-129. E2. Kauffmann F, Oryszczyn MP, Maccario J. The protective role of country living on skin prick tests,
immunoglobulin e and asthma in adults from the epidemiological study on the genetics and environment of asthma, bronchial hyper-responsiveness and atopy. Clin Exp Allergy 2002;32:379-386.
E3. Siroux V, Le Moual N, Rage E, Ferran J, Bousquet J, Gormand F, Just J, Nadif R, Oryszczyn MP, Ravault C, et al. Twelve-year follow-up of asthma, FEV1 and bronchial-hyperresponsiveness in children and adults in the Epidemiological study on Genetics and Environment of asthma, bronchial hyperresponsiveness and atopy – Preliminary results [abstract]. Am J Respir Crit Care Med 2007;175:A828.
E4. Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet 2001;9:301-306.