HAL Id: hal-01269201
https://hal.archives-ouvertes.fr/hal-01269201
Submitted on 27 May 2020
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
A long photoperiod relaxes energy management in
[i]Arabidopsis[/i] leaf six
Katja Baerenfaller, Catherine Massonnet, Lars Hennig, Doris Russenberger,
Ronan Sulpice, Sean Walsh, Mark Stitt, Christine Granier, Wilhelm Gruissem
To cite this version:
Katja Baerenfaller, Catherine Massonnet, Lars Hennig, Doris Russenberger, Ronan Sulpice, et al..
A long photoperiod relaxes energy management in [i]Arabidopsis[/i] leaf six. Current Plant Biology,
Elsevier, 2015, 2, pp.34-45. �10.1016/j.cpb.2015.07.001�. �hal-01269201�
Contents lists available atScienceDirect
Current
Plant
Biology
j o u r n a l h o m e p a g e :w w w . e l s e v i e r . c o m / l o c a t e / c p b
A
long
photoperiod
relaxes
energy
management
in
Arabidopsis
leaf
six
Katja
Baerenfaller
a,∗,
Catherine
Massonnet
b,1,2,
Lars
Hennig
a,c,
Doris
Russenberger
a,
Ronan
Sulpice
d,3,
Sean
Walsh
a,4,
Mark
Stitt
d,
Christine
Granier
b,
Wilhelm
Gruissem
a,∗aDepartmentofBiology,ETHZurich,CH-8092Zurich,Switzerland
bLaboratoired’EcophysiologiedesPlantessousStressEnvironnementaux(LEPSE),INRA-AGRO-M,F-34060MontpellierCedex1,France cDepartmentofPlantBiology,SwedishUniversityofAgriculturalSciencesandLinneanCenterforPlantBiology,SE-75007Uppsala,Sweden dMaxPlanckInstituteofMolecularPlantPhysiology,D-14476Golm,Germany
a
r
t
i
c
l
e
i
n
f
o
Articlehistory:
Received18February2015 Receivedinrevisedform7May2015 Accepted10July2015 Keywords: Photoperiod Arabidopsisthaliana Leafgrowth Proteomics iTRAQ Transcriptomics Tilingarray Phenotyping
a
b
s
t
r
a
c
t
Plantsadapttotheprevailingphotoperiodbyadjustinggrowthandfloweringtotheavailabilityofenergy. Tounderstandthemolecularchangesinvolvedinadaptationtoalong-dayconditionwecomprehensively profiledleafsixattheendofthedayandtheendofthenightatfourdevelopmentalstagesonArabidopsis thalianaplantsgrownina16hphotoperiod,andcomparedtheprofilestothosefromleaf6ofplants grownina8hphotoperiod.WhenArabidopsisisgrowninalong-dayphotoperiodindividualleafgrowth isacceleratedbutwholeplantleafareaisdecreasedbecausetotalnumberofrosetteleavesisrestricted bytherapidtransitiontoflowering.Carbohydratemeasurementsinlong-andshort-dayphotoperiods revealedthatalongphotoperioddecreasestheextentofdiurnalturnoverofcarbonreservesatallleaf stages.Atthetranscriptlevelwefoundthatthelong-dayconditionhassignificantlyreduceddiurnal tran-scriptlevelchangesthaninshort-daycondition,andthatsometranscriptsshifttheirdiurnalexpression pattern.Functionalcategorisationofthetranscriptswithsignificantlydifferentlevelsinshortandlong dayconditionsrevealedphotoperiod-dependentdifferencesinRNAprocessingandlightandhormone signalling,increasedabundanceoftranscriptsforbioticstressresponseandflavonoidmetabolisminlong photoperiods,andforphotosynthesisandsugartransportinshortphotoperiods.Furthermore,wefound transcriptlevelchangesconsistentwithanearlyreleaseoffloweringrepressioninthelong-day condi-tion.Differencesinproteinlevelsbetweenlongandshortphotoperiodsmainlyreflectanadjustmentto thefastergrowthinlongphotoperiods.Insummary,theobserveddifferencesinthemolecularprofilesof leafsixgrowninlong-andshort-dayphotoperiodsrevealchangesintheregulationofmetabolismthat allowplantstoadjusttheirmetabolismtotheavailablelight.Thedataalsosuggestthatenergy manage-mentisinthetwophotoperiodsfundamentallydifferentasaconsequenceofphotoperiod-dependent energyconstraints.
©2015TheAuthors.PublishedbyElsevierB.V.ThisisanopenaccessarticleundertheCCBYlicense (http://creativecommons.org/licenses/by/4.0/).
Contents
1. Introduction...35
2. Materialandmethods...35
2.1. Plantmaterial,leaf6androsettegrowthmeasurements...35
2.2. Carbohydratedeterminations...36
2.3. TilingarraytranscriptdataandquantitativeiTRAQproteomicsdata...36
2.4. Statisticalanalysesoftheproteinandtranscriptchanges ... 36
2.5. GOfunctionalclassification...36
∗ Correspondingauthorsat:ETHZurich,LFWE18,Universitaetstrasse2,8092Zurich,Switzerland. E-mailaddresses:kbaerenfaller@ethz.ch(K.Baerenfaller),wgruissem@ethz.ch(W.Gruissem).
1 Currentaddress:INRA,UMREcologieetEcophysiologieForestière,F-54280Champenoux,France.
2 Currentaddress:UniversitédeLorraine,UMREcologieetEcophysiologieForestière,BP239,F-54506Vandoeuvre,France.
3 Currentaddress:NUIGalway,PlantSystemsBiologyLab,PlantandAgriBiosciencesResearchCentre,BotanyandPlantScience,Galway,Ireland. 4 Currentaddress:Albert-Ludwigs-UniversityofFreiburg,FacultyofBiology,D-79104Freiburg,Germany.
http://dx.doi.org/10.1016/j.cpb.2015.07.001
3. Resultsanddiscussion...36
3.1. LDacceleratesArabidopsisgrowthandincreasesindividualleafareabutdecreasesrosettearea...36
3.2. Successivecellularstagesofleaf6developmentareafunctionofphotoperiod...36
3.3. Photoperiodaffectsindividualleafexpansioninthecontextofwholerosettedevelopment...37
3.4. Experimentaldesignforassessingmolecularchangesduringleafdevelopment...37
3.5. Photoperiodaffectstheamountanddiurnalturnoverofcarbonreserves...37
3.6. DiurnaltranscriptlevelchangesarelesspronouncedinaLDphotoperiod...38
3.7. DiurnaltranscriptfluctuationsareshiftedinLDandmostpronouncedforstressresponse...38
3.8. Photoperiodandgrowthbehaviourhavespecifictranscriptsignatures...40
3.9. Transcriptsregulatedbyphotoperiodbelongtospecificfunctionalcategories...40
3.9.1. RNAprocessingmechanismsdifferdependingonphotoperiodlength...41
3.9.2. FlavonebiosynthesisisenhancedintheLDphotoperiod...41
3.9.3. LightandhormonesignallingdifferbetweenSDandLD...41
3.9.4. SDincreasestranscriptlevelsforsugartransportandphotosystemproteins...42
3.10. ProteinsthatdifferbetweenSDandLDcanmainlybeattributedtodifferencesingrowth...42
3.11. Floweringgeneshavephotoperiod-specifictranscriptsignaturesinleaves...42
3.12. AtGRP7protein,butnottranscript,ismorehighlyexpressedinLD...43
4. Conclusions...43
Conflictsofinterest...43
Acknowledgements ... 43
AppendixA. Supplementarydata...43
References...43
1. Introduction
Plantsaslight-dependent,autotrophicorganismshaveadapted
totheregularlight–darkcyclesresultingfromtherotationofthe
earth.Thelengthofthelightperiod,orphotoperiod,dependson
thelatitudeandtime oftheyear.Plantsmustadjusttochanges
inday-lengthtooptimizegrowthinvaryingphotoperiodlengths.
Althoughthisrequirestightcontrolofphysiologicalandmolecular
processes,theunderlyingregulatorymechanismsarestillpoorly
understood.It is now wellestablished that thecircadian clock
synchronizesmetabolismwiththechangingphotoperiods[1–4].
Photoperiodlength affects netdaily photosynthesis and starch
metabolism[5,6]andadjustsseasonalgrowth[7–9].However,the
molecularintegrationofphotoperiod,clockandmetaboliccontrol
duringleafdevelopmentremainsachallengingproblem.
Arabidopsis is a facultative long-day plant whose flowering
iscontrolledbythephotoperiodpathway[7,8,10,11]inconcert
withmolecular,hormonalandenvironmentalsignals[10].
Interac-tionsbetweenthecircadianclockandphotoperiodlengthduring
vegetative growthaffectleaf number and size,as wellas their
morphologicalandcellularproperties[12–16].Plantsinwhichthe
vegetativetofloralgrowthtransitionisacceleratedbyincreasing
day-lengthor repressionofregulatorygeneshave fewerleaves,
increasedsingleleafareas,anda higherepidermal cellnumber
inindividualleavescomparedtolatefloweringplants[12,15,16].
Whiletheseadaptationstophotoperiodarewelldocumentedat
thephenotypiclevel,littleisknownabouthowconcerted
regula-tionofphotoperiod-dependentgeneexpressionandproteinlevels
isachieved duringdiurnal cyclesandat differentstages ofleaf
development.
We therefore asked how phenotypic changes are related to
molecularprofilesinasingleleafofArabidopsisplantsgrowingin
along-day(LD;16hlight,8hdark)orshort-day(SD;8hlight,16h
dark)condition.Thesetwophotoperiodscauseconsistent
pheno-typicchangesinthenumberandmorphologyofsuccessiveleaves
ontherosette[12,16].Becausesizeandshapeofsuccessiveleaves
varyduringArabidopsisdevelopment[17]wedecidedtofocusthe
analysisonleafnumber6,whichisthefirstadultleafofthe
Ara-bidopsis(Col-4)rosetteinshort-dayconditions.Leaf6wasused
previouslytogeneratemoleculardataforArabidopsisgrowninSD
[18].Togain insightsintothemolecularpatternunderlyingthe
phenotypicchangesbetweenphotoperiods,wethereforeanalyzed
transcriptandproteinlevelsofleafnumber6growninLDatfour
developmentalstages,bothattheendoftheday(EOD)andendof
thenight(EON).Wethencomparedthedatawiththe
correspond-ingpreviouslyestablishedmoleculardataforleaf6ofArabidopsis
growninSDeitherunderoptimalwatering(SOW)ora40%water
deficit(SWD)[18].Integrationand comparativeanalysesof the
quantitative proteomicsand transcriptomics datarevealed that
fewergeneshavesignificantdiurnaltranscriptlevelfluctuations
inLDthanSD.Transcriptsandproteinswithsignificantlydifferent
levelsinSDandLDvalidatethehypothesisthatashort
photope-riodrequiresatightenergymanagement,whichisrelaxedinalong
photoperiod.
2. Materialandmethods
2.1. Plantmaterial,leaf6androsettegrowthmeasurements
ArabidopsisthalianaaccessionCol-4(N933)plantsweregrown
inagrowthchamberequippedwiththePHENOPSISautomaton[19]
asdescribedpreviously[18]withtheexceptionthatdaylengthin
thegrowthchamberwasfixedat16h.Inbrief,seedsweresownin
potsfilledwithamixture(1:1,v/v)ofaloamysoilandorganic
com-postatasoilwatercontentof0.3gwater/gdrysoilandjustbefore
sowing10mlofamodifiedone-tenth-strengthHoaglandsolution
wereaddedtothepotsurface.After2daysinthedark,daylength
inthegrowthchamberwasadjustedto16hat∼220mol/m2/s
incidentlightintensityatthecanopy.Plantsweregrownatanair
temperatureof21.1◦Cduringthelightperiodand20.5◦Cduring
thedarkperiodwithconstant70%humidity.Duringthe
germina-tionphasewater wassprayed onthesoiltomaintainsufficient
humidityatthesurface.Beginningatplantgermination,eachpost
wasweighedtwiceadaytocalculatethesoilwatercontent,which
wasadjustedto0.4gwater/gdry soilbytheadditionof
appro-priatevolumesofnutrientsolution.Theexperimentwasrepeated
independentlythreetimesandeachleaf6samplewaspreparedby
bulkingmaterialfromnumerousplants.Thefrozenplantmaterial
wassenttotheMPIinGolm,whereitwasgroundandaliquotted
usingacryogenicgrinder(GermanPatentNo.8146.0025U1).
Growth-relatedtraitsofleaf6atsingleleafandcellularscales
weremeasuredas described [20]. Fiverosettes wereharvested
anddissectedevery2–3daysduringeachexperiment.Leaf6area
(×160)glassforleavessmallerthan2mm2orwithascannerfor
largerones.Anegativefilmoftheadaxialepidermisofthesameleaf
6astheonemeasuredinsurfacewasobtainedafterevaporationof
avarnishspreadonitssurface.Theseimprintswereanalyzedusing
amicroscope(LeitzDMRB;Leica)supportedbytheimage-analysis
softwareOptimas.Meanepidermalcelldensity[cellsmm−2]was
estimatedbycountingthenumberofepidermalcellsintwozones
(atthetipandbase)ofeachleaf.Totalepidermalcellnumberinthe
leafwasestimatedfromepidermalcelldensityandleafarea.Mean
epidermalcellarea[m2]wasmeasuredfrom25epidermalcells
intwozones(atthetipandbase)ofeachleaf.
Forrosettegrowthmeasurements,ateachdateofharvestall
leaveswithanarealargerthan 2mm2 fromfive rosetteswere
imagedwithascanner.Thenumberofleaveswascountedandtotal
rosetteareawascalculatedasthesumofeachindividualleafarea
measuredonthescanwiththeImageJsoftware.
2.2. Carbohydratedeterminations
Starch,glucose,fructoseandsucrosecontentweredetermined
byenzymaticassaysinethanolextractsof20mgfrozenplant
mate-rialasdescribedinCrossetal.[21].Chemicalswerepurchasedas
inGibonetal.[22].Assayswereperformedin96wellmicroplates
usingaJanuspipettingrobot(PerkinElmer,Zaventem,Belgium).
AbsorbancesweredeterminedusingaSynergymicroplatereader
(Bio-Tek,BadFriedrichshall,Germany).Foralltheassays,two
tech-nicalreplicatesweredeterminedperbiologicalreplicate.
2.3. TilingarraytranscriptdataandquantitativeiTRAQ
proteomicsdata
Geneexpressioninleavesofthefourdevelopmentalstagesand
atthetwodiurnaltimepointsinthelongdayoptimalwater(LD)
experimentandinareferencemixedrosettesamplewasprofiled
asdescribedpreviously[18]usingAGRONOMICS1microarrays[23]
andanalyzedusinga TAIR10CDFfile[24].Alllog2-transformed
sample/referenceratios without p-value filtering were used in
theanalyses.Microarrayrawandprocesseddataareavailablevia
ArrayExpress(E-MTAB-2480).
Proteins in the same sampleswere quantified using the
8-plexiTRAQisobarictaggingreagent[25,26]asdescribedindetail
previously [18] according to the labelling scheme in
Support-ing Table S5. The resulting spectra were searched against the
TAIR10proteindatabase[27]withconcatenateddecoydatabase
andsupplementedwithcommoncontaminantswithMascot
(Mas-cotScience,London,UK).Thepeptidespectrumassignmentswere
filteredforpeptideunambiguityinthepep2prodatabase[28,29].
Acceptingonlyunambiguouspeptideswithanionscoregreater
than 24 and an expect value smallerthan 0.05 resulted in 70
979assignedspectraataspectrumfalsediscoveryrate(FDR)of
0.07%.Quantitative information for all reporter ionswas
avail-ablein50 947of thesespectraleadingtothequantification of
1788proteinsbasedon6178distinctpeptides(SupportingTable
S6).Themassspectrometryproteomicsdatahavebeendeposited
to the ProteomeXchange Consortium (http://proteomecentral.
proteomexchange.org)viathePRIDEpartnerrepository[30]with
thedatasetidentifierPXD000908 and DOI10.6019/PXD000908.
The data are also available in the pep2pro database at www.
pep2pro.ethz.ch
Allproteomeandtranscriptomeabundancemeasuresforthe
LDexperimentwereintegratedwithintheexistingAGRON-OMICS
database (LeafDB) [18]. A searchable web-interface containing
theseintegrateddatasetsisavailableathttps://www.agronomics.
ethz.ch/
2.4. Statisticalanalysesoftheproteinandtranscriptchanges
Thestatisticalanalyticalmethodswereperformedasdescribed
previously[18]subjectingthelog2-transformedsample/reference
ratios to an analysis of variance (ANOVA) treating stage (S)
and day-time(ND) asmain effects followedby correctionwith
Benjamini-Hochberg[31].TranscriptsandproteinswithapGlobal
(p-value for an overall global change)<0.05 and a maximum
fold-change>log2(1.5) were considered to change significantly
(SupportingTablesS7andS8).Forasignificantdifferencebetween
EODandEONweadditionallyrequiredpND(p-valueforthediurnal
change)<0.05.Thecomparisonoftheproteinandtranscriptlevels
betweentheLD andthetwoshortday(SOWandSWD)
exper-imentsreportedpreviously wasperformedwitha pairedt-test
comparingthevaluesforthe8time-pointsbetweentwo
experi-mentscorrectedwithBenjamini-Hochberg[31]takingintoaccount
allnon-plastidencodedtranscriptswithoutp-valuefiltering.All
statisticalanalyseswereperformedusingR[32].
2.5. GOfunctionalclassification
Assignmentofproteinandtranscriptfunctionalcategorieswas
basedontheTAIRGOcategoriesfromaspectbiologicalprocess
(ATH GO GOSLIM20130731.txt)asdescribedpreviously[18].
3. Resultsanddiscussion
3.1. LDacceleratesArabidopsisgrowthandincreasesindividual
leafareabutdecreasesrosettearea
Whenplottedagainsttimefromleafinitiationtofullexpansion,
leaf6areaincreasedmorerapidlyandreacheditsfinalsize
ear-lierandwas50%largerinLDthaninSD(Fig.1A).Thedynamicsof
cellproductionandexpansionintheupperepidermisofleaf6
indi-catesthatbothcellnumberandcellsizeincreasedmorerapidlyand
reachedtheirfinalvaluesearlierinLDthaninSD(Fig.1B,C).Thus,
photoperiodhasapronouncedeffectonthetimingofleaf
develop-mentbecausecelldivision,cellexpansionandleafexpansionwere
fasterinLDthanSDandceasedearlier.
Similartothefastergrowthofleaf6thewholerosetteleafarea
andleafnumberinitiallyincreasedfasterinLDthanSD(Fig.2A,B).
However,laterindevelopmentanddespitetheincreasedindividual
leafsizeatthefullyexpandedstage(Fig.1A),thewholerosettearea
wassmallerinLDthaninSD.Thiswastheresultofasmallerfinal
numberofrosetteleavesthatwereproduced(Fig.2A).
3.2. Successivecellularstagesofleaf6developmentarea
functionofphotoperiod
Becauseleaf6growthwasacceleratedinthelongphotoperiod
andstages2–4ofleafdevelopmentwerereachedearlierthanin
theshortphotoperiod(Fig.1), biologicalsamplesofleaf6were
harvestedatfourdevelopmentstagescorrespondingtotransitions
associatedwithwell-definedcellularprocesses[18].Thestage1
leafhasmaximum relative areaand thickness expansionrates,
stage2and3leaveshavemaximumanddecreasingabsolutearea
andthicknessexpansionrates,respectively,andinthestage4leaf
expansionends[18].Samplingatdefinedstagesallowsarobust
leafscalecomparisonofphotoperiodeffectsonleafdevelopment
despitedifferentgrowthratesindifferentexperiments.Wefound
thatstage1correspondstothephaseofrapidcelldivisionaround
day7or8afterleafinitiationinbothphotoperiods.Mostofcell
divisionhadceasedatstage2,whichwasaroundday11afterleaf
initiationinLDandday14inSD.Stage3isthephaseofdecreasing
cellexpansionratearound14daysafterleafinitiationinLDandday
Fig.1. KinematicexpansionphenotypesofleavesharvestedforprofilingintheSD (blue)andLD(red)experiments.Changesovertimeinleaf6area(A),meancell numberinleaf6adaxialepidermis(B)andmeancellareainleaf6adaxialepidermis (C).DataaremeanandSDvalues,n=5.Theincreaseofleafarea,cellnumberand cellareaaredescribedbysigmoidcurvesfollowingtheequations:y=A/[1+e (-(X-X0)/B)].Themediandateofthe4harvesttimesarepresentedbyverticallinesfor theSD(bluedotted)andLD(reddot-dashed)experiments.Leaf6initiationoccurred ataroundday12aftersowinginSDandday10inLD.
correspondingtoaroundday21afterleaf6initiationinLDandday
30inSD.
3.3. Photoperiodaffectsindividualleafexpansioninthecontext
ofwholerosettedevelopment
Becausephotoperiodlengthaffectedboth theprogression of
individualleafstagesandwholeplantdevelopment,thefourleaf6
developmentalstagesdidnothavethesamestatuswithregardto
wholerosettedevelopmentinLDandSDplants.Leaf6expansion
inSDwascompletebeforethefinalnumberofrosetteleaveswas
reached,whereasinLDmorethan50%ofleaf6expansionoccurred
afterbolting.Thefloraltransitionattheshootapexoccursseveral
daysbeforebolting,typicallyat10–12daysaftergerminationin
LD[33].Leaf6wasinitiatedat10daysaftersowing,andtherefore
almostallitsgrowthoccurredafterthefloraltransitionattheshoot
apex.
Atstage1inLD,leaf6arearepresentedapproximately5%ofthe
wholerosettearea.Thisproportionincreasedto12–15%during
stages2and3andatstage4declinedtoaround10%.Incontrast,
theproportionofleaf6areacomparedtowholerosetteareaat
Fig.2. Kinematicexpansionphenotypesofwholerosetteleafgrowthofplants har-vestedforleaf6profilingintheSD(blue)andLD(red)experiments.Changesover timeinthenumberofrosetteleaves(A)andwholerosettearea(B).Changesover timeoftheproportionofwholerosetteareacoveredbyleaf6areaispresentedin (C).Theindicatedtrendlinesrepresentpredictionsfromalocalpolynomial regres-sionfitting(loess).The4datesofharvestarepresentedbyverticallinesfortheSD (bluedotted)andLD(reddot-dashed)experiments.
stage4waslessthan5%inSD,confirmingthatleaf6reachesits
smallerfinalsizeinSDbeforewholerosetteexpansionwas
com-plete(Fig.2C).
3.4. Experimentaldesignforassessingmolecularchangesduring
leafdevelopment
Toquantitateproteinandtranscriptlevelsduringthegrowth
ofasingleArabidopsisleafweharvestedleaf6fromplantsgrown
inLDattheendoftheday(EOD)andendofthenight(EON)at
thefoursuccessivestagesofdevelopmentdefinedabove.Proteome
andtranscriptomeprofilingdata,aswellastheamountsofstarch
andsolublesugarswereobtainedfrompooledsamplesofleaf6of
threeindependentbiologicalexperiments.Wethenassessedhow
themolecularprofilesinsingleleavesatprecisestagesof
devel-opmentfromplantsgrowninLDdifferfromleaf6growninSDby
comparingthemtotheSDoptimalwatering(SOW)and40%water
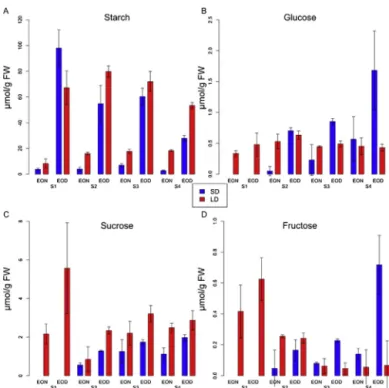
Fig.3.Theamountsof(A)starchandthesolublesugars(B)glucose,(C)sucroseand (D)fructoseing/gFWandtheirstandarddeviationsatEONandEODatthefour leaf6developmentalstagesinSD(blue)andLD(red).
3.5. Photoperiodaffectstheamountanddiurnalturnoverof
carbonreserves
Starchisthemaincarbonreserveforenergyrequirements
dur-ingthenightinArabidopsisandrepresentedabout80–93%ofthe
carbohydratesmeasuredatEODinLDandSD(Fig.3).InLD-grown
plants,theamountofstarchatEODwassimilaratallfour
devel-opmentstages.Althoughstarchalsodecreasedduringthenight
inLDplants,considerablylargeramountsofstarchremainedat
EON,especiallyatstages2,3and4(Fig.3A).InSDadifferent
pat-ternwasfound.ThehighestamountofstarchatEODwasfound
forstage1,withlowerlevelsinstages2,3and,especially,stage
4.Further,inSD,mostofthestarchthataccumulatedatEODwas
consumedduringthenightatalldevelopmentalstages.InLD,the
levelsofglucose,sucroseandfructoseweresimilaratEODandEON
foralldevelopmentalstages,withtheexceptionofstages1and2
forsucrose,wherethelevelswerehigheratEODthanEON.Glucose
levelsinLDweresimilaratalldevelopmentalstages,butfructose
andsucrosewerehighestforstage1.Incontrast,majordifferences
werefoundinSD.First,glucose,fructoseandsucroselevelsinSD
wereconsistentlyhigheratEODthanEON,aspreviouslyreported
forfullrosettes[6].Second,thehighestlevelsofglucose,fructose
andtosomeextentsucroseweredeterminedforstage4atEOD.
Third,sucroselevelsforalldevelopmentalstagesandharvesttimes
wereconsistentlylowerinSDthanLD,aspreviouslyreportedfor
fullrosettes[6](Fig.3B–D).Together,thedatarevealthatin
Ara-bidopsisphotoperiodlengthhasamajorinfluenceonthemetabolic
statusoftheleafduringbothdevelopmentandthediurnalcycle.
3.6. DiurnaltranscriptlevelchangesarelesspronouncedinaLD
photoperiod
Toaccountfortheobservedphenotypicandmetabolic
differ-encesbetweenSDandLDweanalyzedquantitativeproteinand
transcriptdataindetail.WefirstperformedaPrincipal
Compo-nentAnalysis(PCA)toestimatethemainfactorsthatdetermine
changesintranscriptandproteinlevelsinLD.Themain
contribu-tiontothevarianceinthetranscriptdatainthefirsttwoprincipal
componentsisthedifferencebetweenstage1andthelaterstages
2–4,whichaccountedforover60%ofthetotalvariance(Fig.4A).
TheEODandEONsamplesareseparatedonlyinthethird
princi-palcomponent,whichaccountedforabout8%ofthetotalvariance
(Fig.4B).ThisisincontrasttoaPCAofthetranscriptsinSD
condi-tions,wherethetimeofharvestwasthemaincontributiontothe
variationinthedatainthefirstandsecondprincipalcomponents
[18].AssessingthedifferenceintranscriptlevelsbetweenEONand
EONrevealedthatinLDonly21.2%ofalltranscriptsshowed
signifi-cantdiurnaltranscriptlevelfluctuations,incontrastto50.3%inthe
SOWand43.1%intheSWDconditions.Thus,inadditionto
metabo-litechanges,theLDphotoperiodalsohasaconsiderableimpacton
diurnalmRNAexpressionpatterns.Fortheproteindata,the
dif-ferencebetweenthedevelopmentalstagescontributesmosttothe
variationinthedata(Fig.4C),asobservedpreviouslyinSD[18].
3.7. DiurnaltranscriptfluctuationsareshiftedinLDandmost
pronouncedforstressresponse
TranscriptsthatchangedsimilarlybetweenEODandEONboth
inLDandSDincludedthoseencodingthecentralclockproteins
Fig.4.PrincipalComponentAnalysisoftranscriptandproteinprofilesinleaf6growninLD.(A)Firstandsecondprincipalcomponentand(B)firstandthirdprincipal componentinthetranscriptdata,and(C)firstandsecondprincipalcomponentintheproteindata.Thenumbersindicatethegrowthstages1to4andareinblueforthe EONsamplesandinredfortheEODsamples.
Fig.5.(A)ThenumberoftranscriptswithdifferentialdiurnalfluctuationsbetweenSDandLD.(B)ForallthetranscriptswithdifferentialdiurnalfluctuationsbetweenSDand LD,and(C-H)forthetranscriptsinthedifferentsub-categoriesdepictedin(A),thehistogramsrepresentthefrequencyofthenumberoftranscriptswithanexpressionpeak atagivenZTasdeterminedinEdwardsetal.[34].TheZTherecorrespondstothetimeincontinuouslightsincethelastdawnafterplantshadbeenentrainedto12h/12h light/darkcyclesfollowedbyonedayincontinuouslight,andtheexpectedlightanddarkperiodsareindicatedbywhiteandblackbars,respectively.
LATEELONGATEDHYPOCOTYL 1(LHY,AT1G01060),CIRCADIAN
CLOCK ASSOCIATED1 (CCA1, AT2G46830)and TIMING OF CAB
EXPRESSION1(TOC1,AT5G61380).However,asexpectedfromthe
resultsofthePCAanalysis,manymoretranscriptsshoweda
signif-icantchangebetweenEODandEONinSDthaninLD.Wedefined
transcriptstochangeonlyinSDwhentheyhadsignificantly
dif-ferentlevelsbetweenEODandEONinSOWandSWD,butnotin
LD(5238transcripts),andtranscriptstochangeonlyinLDwhen
theyhadsignificantlydifferentlevelsbetweenEODandEONinLD,
butnotinSOWorSWD(835transcripts)(Fig.5A;SupportingTable
S1).Tofurtherexaminethedifferencesinthediurnalfluctuations
betweenSDandLDweusedEONasreferencepointcorresponding
toZeitgeberTime(ZT,hoursafterdawn)–1inbothexperiments.We
thenassessedwhichtranscriptsweresignificantlyhigherorlower
attherespectiveEODcomparedtothereferencepointonlyinSD,
oronlyinLD(Fig.5,SupportingTableS1).Foralltranscriptswith
differentialdiurnalfluctuationsbetweenSDandLDweexamined
whethertheyscoredrhythmicbyCOSOPTinthefree-runningstudy
conductedbyEdwardsetal.[34].Forthosethatwererhythmicwe
plottedtheZeitgeberTime(ZT)peaksdeterminedinEdwardsetal.
[34](Fig.5B–H).TheZTpeaksoftherhythmictranscriptsthatare
loweratEODonlyinSDandhigheratEODonlyinLDpeakinthe
secondhalfofthesubjectivenightaroundZT43-44.Transcriptsthat
arehigheratEODonlyinSDpeakaroundZT33–37corresponding
tothesubjectivedusk,whilethosethatareloweratEODonlyinLD
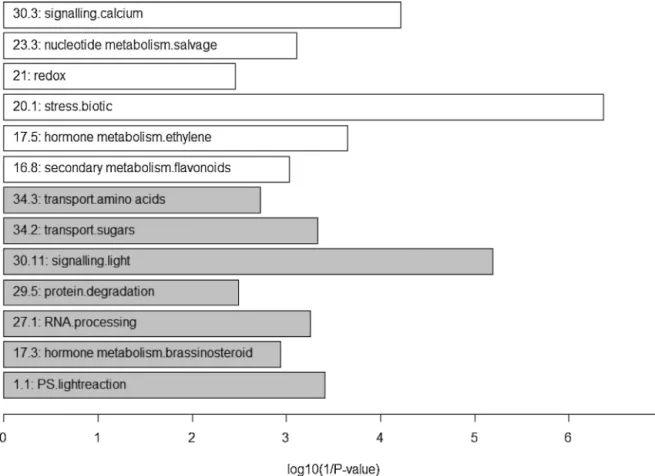
Fig.6.MapMancategoriesthatareover-representedinLD(white)orSD(grey).TheMapManbinswithP-value<0.01areindicatedandthelengthofthebarcorrespondsto thelog-transformedMapMancategoriesthatareover-representedinLD(white)orSD(grey).TheMapManbinswithP-value<0.01areindicatedandthelengthofthebar correspondstothelog-transformedp-value−1.
harvestattherespectiveEODinSDandLDphotoperiodscanaffect
therelativeabundancedifferencebetweenEODandEONfor
tran-scriptspeakingduringthenight,thisisnotthecasefortranscripts
withZTpeaksintheafternoonorearlynight(SupportingFig.S1).
Thedifferentpatternofthesetranscriptsthereforesuggestsashift
intheirdiurnalexpression.Thefunctionalcategorisationagainst
GOBiologicalProcessofthetranscriptshigheratEONonlyinLD
gaveasthetopcategoryresponsetochitin(p-value<1−30).Thelist
of23transcriptsthataccountforthisover-representationcontains
14transcriptionfactorsaccordingtotheAGRISwebsite[35]
(Sup-portingTableS2),andfourofthemarescoredrhythmicwithZT
peaksinthelateafternoon.Together,thissuggeststhatthe
expres-sionpatternsofspecifictranscripts,especiallyfortranscriptslinked
tobioticstressresponse,arechangedinresponsetolightandthe
expectedlengthofthenight.
3.8. Photoperiodandgrowthbehaviourhavespecifictranscript
signatures
Thedifferencesinthediurnaltranscriptaccumulationbetween
SDandLDpromptedustofurtherexaminethetranscriptsthatare
differentiallyexpressedbetweenLDandSD.Weconsideredthose
transcriptstochangeinaphotoperiod-specificmannerthatwere
significantlydifferent(p-value<0.05inapairedt-test,average
fold-change>1.5)intheLDexperimentcomparedtoboththeSOWand
SWDexperiments.Atotalof3469transcriptsfulfilledthesecriteria
with1954beinghigherinLDand1515higherinSD(Supporting
TableS3).
AsplantsgrowfasterinLDthanSOWandSWDconditions,itcan
beexpectedthatsomeofthedifferencesbetweenthetwo
photope-riodswillbeduetotheirdifferentgrowthbehaviours.Comparing
thetwoSDexperimentswehadalreadyfoundthatthetranscript
levelsofproteinsassignedtoGOcategorydefenceresponseto
fun-gusandthosesupportingfastgrowth,suchasproteinsinvolved
inribosomebiogenesisandtranslation, arereduced inleaf6by
waterdeficit[18].Todistinguishbetweeneffectscausedby
dif-ferentgrowthratesandthosespecificforlongdayconditions,we
definedsetsofgrowth-specifictranscriptsbasedonthegradual
increaseingrowthratefromSWDtoSOWandtheLDexperiment.
Wehypothesisedthattranscripts,whichaccumulatetodifferent
levelsbetweenSDandLDandalsoshowasignificantdifferencein
accumulationbetweentheSWDandSOWconditions,arelikelyto
berelatedtogrowth.Applyingthesecriteriawefound134
tran-scriptsthataremosthighlyexpressedinLDand38transcriptsthat
arehighestinSWDconditions(SupportingTableS3).Transcripts
thatarehighestinLDandthereforemightbeassociatedwithfaster
growthareover-representedinvariousresponsepathways,with
responsetochitin,defenceresponsetofungusandresponseto
mechan-icalstimulusasthetopthreecategories.TheGOprocessesthatare
over-representedinthetranscriptshighestintheSWDplantsare
nitrileandprolinebiosyntheticprocess,aswellasphotosynthesis,
con-sistentwithatightenergymanagementinashortphotoperiodand
reducedwatercondition.
3.9. Transcriptsregulatedbyphotoperiodbelongtospecific
functionalcategories
TranscriptsthatweresignificantlyhigherinSDorLD
(Support-ingTableS3) werecategorisedusingMapMan [36]and TAIR10
mapping (AthAGILOCUSTAIR10Aug2012). Over- and
under-representationwasassessedseparatelyforthetranscriptshigherin
Table1
ProteinswithasignificantchangebetweentheLDexperimentandSOW.ProteinsthatwereinadditionsignificantlyincreasedordecreasedintheLDexperimentcompared toSWDareinbold.
Proteinssignificantlyhigherinlongdayconditions
AT1G75040 pathogenesis-relatedgene5,PR5
AT1G75750 GAST1proteinhomolog1
AT2G19730 RibosomalL28eproteinfamily
AT2G21660 cold,circadianrhythm,andrnabinding2,CCR2,GRP7 AT2G29350 senescence-associatedgene13
AT2G45790 phosphomannomutase,PMM
AT3G57260 beta-1,3-glucanase2,ATBG2,ATPR2,BGL2,PR2 AT3G59760 O-acetylserine(thiol)lyaseisoformC
AT4G17830 PeptidaseM20/M25/M40familyprotein AT4G22670 HSP70-interactingprotein1
AT4G32915 FUNCTIONSIN:molecularfunctionunknown;INVOLVEDIN:regulationoftranslationalfidelity AT4G36810 geranylgeranylpyrophosphatesynthase1,GGPS1,GGPPS11
AT5G39570 FUNCTIONSIN:molecular functionunknown;INVOLVEDIN:biologicalprocessunknown;LOCATEDIN:cytososol Proteinssignificantlylowerinlongdayconditions
AT1G54010 GDSL-likeLipase/Acylhydrolasesuperfamilyprotein AT1G76100 plastocyanin1,PETE1
AT2G22230 Thioesterasesuperfamilyprotein AT2G42530 coldregulated15b,COR15B AT2G42540 cold-regulated15a,COR15A AT3G09260 Glycosylhydrolasesuperfamilyprotein AT4G29680 Alkaline-phosphatase-likefamilyprotein AT5G10540 Zincin-likemetalloproteasesfamilyprotein
AT5G15970 stress-responsiveprotein(KIN2)/stress-inducedprotein(KIN2)/cold-responsiveprotein(COR6.6) AT5G51720 2iron,2sulfurclusterbinding
AT5G54160 O-methyltransferase1
ofmeasuredtranscriptswiththenumberthatwouldbeexpected bychance.Fig.6showstheMapManbinswithp-value<0.01and
theAGIsofthegenesinthesecategoriesarelistedinSupporting
TableS4.
3.9.1. RNAprocessingmechanismsdifferdependingon
photoperiodlength
Among the genes for transcripts that have different levels
betweenSD andLDwefoundfewerthanexpectedthatencode
proteinsfortranslation(bin29.2)(p<2.05e−11inaFisher’sexact
test).Thisisinagreementwiththefindingthatribosome
abun-dance does not change between SD and LD grown plants [6].
However,genesinvolvedinRNAprocessingareover-represented
inSD(Fig.6),whilegenesforsmallnucleolarRNAs(snoRNAs)are
over-representedinLD(4.25e−6inaFisher’sexacttest)because
14of45snoRNAsrepresentedonthetilingarrayaresignificantly
more highly expressedin LD. snoRNAsassociate with proteins
toformfunctionalsmallnucleolarribonucleoproteincomplexes
(snoRNPs),whichareinvolvedintheprocessingofprecursorrRNAs
inthenucleolusrequiringexo-andendonucleolyticcleavagesas
wellasmodifications.Thesemodificationsarethoughttoinfluence
ribosomefunction[37].ThedifferentialexpressionofsnoRNAsin
SDandLDconditionsmightreflectaspecificbutcurrentlyunknown
mechanismofadjustingtranslationtotheprevalentphotoperiod
conditions.
3.9.2. FlavonebiosynthesisisenhancedintheLDphotoperiod
Transcriptsthat arehigher inLD areoverrepresented in bin
secondarymetabolism.flavonoids(Fig.6).Flavonoidsareplant
sec-ondary metabolites withbroad physiological functions [38]. Of
thegenesinthiscategory,fiveencodeenzymesintheKEGG[39]
pathway flavonoid biosynthesis, namely TRANSPARENTTESTA 4
(CHS/TT4,AT5G13930),TT5(AT3G55120),F3H/TT6(AT3G51240),
TT7 (AT5G07990) and FLAVONOL SYNTHASE (FLS, AT5G08640)
(SupportingFig.S2).Theseenzymesarerequiredforthe
biosyn-thesis of the three major flavonols quercetin, kaempferol and
myricetin,althoughtheenzymecatalysingthelaststepofmyricetin
productionhasnotyetbeenidentifiedinArabidopsis(Supporting
Fig.S3).Thetranscriptlevelsfortheseenzymesareallincreasedin
LDascomparedtoSDbutgenerallydecreaseduringleaf6
develop-ment(SupportingFig.S4).TT5andTT6/F3Hproteinsweredetected
inLD.TT5proteinlevelsdecreasesignificantlyduringdevelopment
inLDbuttheproteinwasdetectedinallthreeexperimental
condi-tions(SOW,SWDandLD).Transcriptlevelsofflavonoidpathway
geneswerereportedtobeup-regulatedinleavesofsweetpotato
grown inLD that have highconcentrations ofkaempferol [40].
Kaempferolfunctionsasanantioxidantinchloroplasts[41].Higher
transcriptlevelsfortheenzymesintheflavonolbiosynthesis
path-wayinLDthereforecorrelatewellwiththeover-representationof
thebinredoxinLD.Thetranscriptlevelsforenzymesinflavonoid
biosynthesispathwaysinvolvedinresponsetoexcessUVlightor
highlightstress,suchasanthocyaninbiosynthesis,arenothigher
inLDascomparedtoSD.Thisconfirmsthatunderour
experimen-talconditionstheLDphotoperiodisnottriggeringastressresponse
thatwouldrequireenhancedphotoprotection.
3.9.3. LightandhormonesignallingdifferbetweenSDandLD
Plant hormones coordinate developmental processes and
growth through converging pathways [42,43]. We therefore
expectedthatseveralofthegeneswhosetranscriptsaccumulate
todifferentlevelsbetweenSDandLDencodeproteinsinvolvedin
hormonemetabolismandsignalling(SupportingFig.S5).Thebin
hormonemetabolism.ethyleneisover-representedinLDandthelist
ofgenesannotatedtothisbinthathaveincreasedtranscriptlevels
inLDincludes10genesencodingdifferentETHYLENE-RESPONSIVE
ELEMENTBINDINGFACTOR(ERF)proteins.ERFsfunctionindefence
responseandregulatechitinsignalling[44,45].TwooftheseERFs,
DREBANDEARMOTIFPROTEIN1(DEAR1;AT3G50260)andERF6
(AT4G17490),belongtothetranscriptionfactorsthathavehigher
transcriptlevelsatEONonlyinLDandareassignedtoresponseto
chitin(SupportingTableS2).
Ethylenebiosynthesisisrestrictedbythephotoreceptor
phy-tochromeB(PHYB;AT2G18790)[46].PHYBtranscriptlevelsare
decreasedinLDascomparedtoSD,whichcorrelateswithincreased
ethylenebiosynthesisinLD.InadditiontoPHYB,othergenes
encod-ingphytochromessuchasPHYA(AT1G09570)andgenesencoding
proteinsaremorehighlyexpressedinSD,resultinginthe
over-representationofbinsignalling.light(SupportingTableS4).
Photoperiod can be integrated with growth and time to
flowering through regulation of the brassinosteroid hormone
pathway [47]. It was therefore unexpected that bin hormone
metabolism.brassinosteroidwasover-representedinSD,asplants
inSDgrowmoreslowlyandflowerlater.However,themRNAs
withhigherlevelsinSDassignedtothisbinalsoincludethemRNA
forcytochromeP450CYP734A1(AT2G26710).CYP734A1converts
activebrassinosteroidsintotheirinactiveforms[48]andtherefore
actsasanegativeregulatorofbrassinosteroidsignalling.Thus,the
over-representationofthebinhormonemetabolism.brassinosteroid
doesnotimplyincreasedbrassinosteroidsignalling.Infact,theonly
brassinosteroidsignalling-relatedmRNAwithhigherlevelsinLD
encodesBES1/BZR1-LIKEPROTEIN3(BEH3,AT4G18890),whichis
atranscriptionfactorthatishomologoustoBES1/BZR1,apositive
regulatorofbrassinosteroidsignalling[49].
3.9.4. SDincreasestranscriptlevelsforsugartransportand
photosystemproteins
TranscriptsthataresignificantlyhigherinSDthanLDencode
twelvemembersofthemonosaccharidetransporter(MST)(-like)
genefamily[50]andtheSUCROSE-PROTONSYMPORTER9(SUC9,
AT5G06170). Accordingly, the bin sugar.transport is
overrepre-sentedin SD(Fig.6,SupportingTableS4).The membersofthe
MST(-like) gene family are classified into seven distinct
sub-familiesandhaverolesinbothlong-distancesugarpartitioningand
sub-cellularsugar distribution[50].POLYOL/MONOSACCHARIDE
TRANSPORTER2(PMT2,AT2G16130)andSUGARTRANSPORTER
1(STP1, AT1G11260)arelocatedin theplasma membrane and
weresuggestedtoimportmonosaccharidesintoguardcells
dur-ingthenightandfunctioninosmoregulationduringtheday[51].
TheMST(-like)genefamilymembersinvolvedinsub-cellularsugar
distributionincludetheplastid-localisedPLASTIDICGLC
TRANSLO-CATOR(PGLCT,AT5G16150),which contributestotheexportof
themainstarchdegradationproductsmaltose andglucosefrom
chloroplasts[52], and six proteinsencoded by theAtERD6-like
genesub-familythatarelocatedinthevacuolemembrane.AtERD6
homologs are thought to export sugars from the vacuole
dur-ingconditionswhenre-allocationofcarbohydratesisimportant,
includingsenescence,wounding,pathogenattack,C/Nstarvation
anddiurnalchangesintransientstorageofsugarsinthevacuole
[50].Theincreasedtranscriptexpressionofgenesforvarioussugar
transportersinSDisconsistentwiththedifferentamountand
diur-nalturnoverofsugarlevelsinSDascomparedtoLD(Fig.3)and
indicatesthatlong-distanceandsub-cellularsugarpartitioningis
increasedinshorterilluminationperiods.
ThebinPS.lightreactionissignificantlydifferentbetweenSDand
LDandoverrepresentedinSD(Fig.6;SupportingFigs.S5andS6).
MostofthetranscriptsassignedtothisbinthatareincreasedinSD
encodephotosystemIorIIproteins(SupportingTableS4).Someof
theirgenesseemtobelinkedtoreducedgrowth,nevertheless,the
SDcomparedtotheLDphotoperiodapparentlyincreases
photo-systemabundance.Thislikelyincreasestherateofphotosynthesis
tousethelightoftheshorterilluminationperiodmostefficiently.
3.10. ProteinsthatdifferbetweenSDandLDcanmainlybe
attributedtodifferencesingrowth
Wenextexaminedtheproteinsthataredifferentiallyexpressed
intheLDandSOWplants(p-value<0.05inapairedt-test,average
fold-change>1.5).Atotalof24proteinsfulfilledthestrictcut-off
criteriathatwerealsoappliedtothetranscriptdata.Ofthe13
pro-teinsthatwerehigherinLD,5werealsoincreasedinLDcompared
toSWD,andofthe11thatwerelowerinLD,4werealso
signifi-cantlydecreasedinLDcomparedtoSWD.Theseproteinstherefore
showasignificantdifferencebetweenLDandbothSDconditions
(Table1).
ThelistofproteinsthataremoreabundantinLDthaninSD
includesPATHOGENESIS-RELATEDPROTEIN5(PR5,AT1G75040),
PR2(AT3G57260)andribosomalL28efamilyprotein(AT2G19730).
Thisisconsistentwithourpreviousfindingsthatmostofthe
pro-teinsthataccumulatedtohigherlevelsinthefastergrowingSOW
leavesthanintheSWDleavesmainlycomprisedproteinsinvolved
intranslationandthattranscriptswithhigherlevelsintheSOW
leavesareover-represented forGOcategories ribosome
biogene-sis,translation anddefenceresponsetofungus[18].Furthermore,
MapManbinstress.bioticwasover-representedfortranscriptsthat
havehigherlevelsinLD.Thelistofproteinsthataccumulateto
significantlyhigherlevelsinLDalsoincludes
PHOSPHOMANNO-MUTASE(PMM,AT2G45790),whichisinvolvedinthesynthesisof
GDP-mannoseandisthereforerequiredforascorbicacid
biosyn-thesisandN-glycosylation.Interestingly,thepmmmutanthasa
temperature-sensitivephenotypethat was attributedtoa
defi-ciencyinproteinglycosylation[53].Thedifferentabundancelevels
of PMM of in LD and SD might therefore suggest differential
post-translationalmodificationsinLDandSD.GERANYLGERANYL
PYROPHOSPHATESYNTHASE1(GGPPS11,AT4G36810),whichis
requiredforthebiosynthesisofgeranylgeranyldiphosphate(GGPP)
[54],alsoaccumulatestohigherlevelsinleaf6growninLDas
com-paredtoSOWconditions.InArabidopsis,thechloroplast-localized
GGPPS11istheGGPPSisoformwiththehighesttranscriptlevel
inrosetteleavesandmainlyresponsiblefor thebiosynthesis of
GGPP-derived isoprenoidmetabolitesincluding chlorophylland
carotenoids[54].ThehigherproteinlevelofGGPPS11inLDthanin
SDthereforesuggeststheincreasedproductionofthese
metabo-litesinLD.
TheproteinsthataresignificantlymoreabundantinSDthan
in LD are PLASTOCYANIN 1 (PETE1,AT1G76100)and the three
cold response (COR) proteins COR15A (AT2G42540), COR15B
(AT2G42530)andCOR6.6(AT5G15970)(Table1).Although
plasto-cyaninshavebeenimplicatedinphotosyntheticelectrontransport,
theirconcentration is not limiting for electronflow in optimal
growthconditionswith11hlight[55].TheincreasedPETE1
pro-tein levelin SDmight thereforeindicate aspecificrole for this
proteininshortphotoperiods.TheCORproteinsarealso
signifi-cantlymoreabundantinleaf6growninSWDascomparedtoSOW
conditionsandhavebeenimplicatedintheadaptationresponse
tothecontinuous40%waterdeficitcondition[18].However,the
LDdatasuggestthattheaccumulationofthethreeCORproteins
mayalsoberelatedtogrowth.Wedidnotclassifytranscriptsfor
theseproteinsas photoperiod-specificbecausetheyare
signifi-cantlydifferentbetweenSWDandLDbutnotbetweenSOWandLD.
Acrosstalkbetweencoldresponseandfloweringtimeregulation
hasbeenproposedpreviously,withSOC1functioningasanegative
regulatorofCBFsthatbindtotheCORpromoters[56].Here,the
sit-uationisdifferent,becauseSOC1andCBF1(AT4G25490)transcript
levelsarehigherinLDascomparedtoSDandtheCORtranscripts
showadifferentbehaviour.Therefore,thelevelsoftheCOR
pro-teinsseemtoberegulateddifferentlyandrelatedtothegrowth
rateoftheleaves.
3.11. Floweringgeneshavephotoperiod-specifictranscript
signaturesinleaves
LD photoperiods that are characteristic of spring and early
summerinduceflowering in LD plants. Thecore photoperiodic
floweringpathwaycomprisesGIGANTEA(GI,AT1G22770),
FLOW-ERINGLOCUST(FT,AT1G65480)andCONSTANS(CO,AT5G15840)
[57,58].CircadianclockregulationofCOtranscriptlevelandprotein
stabilityiskeytomonitoringchangesinphotoperiodlength,and
[57].ThemRNAlevelsfortheCOtargetFTwerehigherinLD
com-paredtoSDandincreasedduringdevelopment(SupportingFig.S7).
DownstreamofFT,theMADS-boxtranscriptionfactors
AGAMOUS-LIKE20/SUPPRESSOROFCONSTANS1(SOC1,AT2G45660),AGL24
(AT4G24540),FRUITFULL(FUL,AT5G60910)andSHORT
VEGETA-TIVEPHASE(SVP,AT2G22540)functionasfloralintegratorgenes
duringthetransitionoftheshootapicalmeristem(SAM)tothe
flo-ralmeristem[59].Notably,AGL24andFULtranscriptlevelswere
significantlyhigherinLDalsoinleaf6.SOC1transcriptlevelswere
onlyhigherinLDatearlyleaf6developmentalstages,andSVP
tran-scriptlevelswerenotsignificantlydifferentbetweenLDandSD
(SupportingFig.S7).Incontrast,themRNAlevelsforFLOWERING
LOCUSC(FLC,AT5G10140),whichisakeyrepressorofflowering
[60],weresignificantlylowerinLDascomparedtoSD(Supporting
Fig.S7).FLCandSVPformheterodimersduringvegetativegrowth
torepresstranscriptionofFTinleavesandSOC1intheSAM[61].The
reducedlevelsofFLCtranscriptsinLDtogetherwiththeincreased
levelsofFTtranscriptsarethereforeconsistentwithanearlyrelease
offloweringrepressioninLD.
SOC1belongstothegroupofgenesthathaveadiurnal
expres-sionpeakintheafternoon,withSOC1transcriptlevelsbeinghigher
atEODinSD,buthigheratEONinLD(Fig.5).Interestingly,this
patternwasalsofoundfortranscriptlevelsofthepotentialnatural
antisenseRNAgeneAT1G69572,whosegenomicregionoverlaps
withthat of CDF5.Accordingtodata reportedby Bläsinget al.
[62],SOC1transcriptlevelswerehighestintheafternoon(ZT8)ina
12h/12hphotoperiod.Whencomparedtofree-runningconditions
ofcontinuouswhitelight[63],SOC1transcriptlevelswerehighest
atZT8duringthefirstdaybutnosubsequentcircadianoscillation
wasdetectable.SOC1thereforebelongstothegroupgeneswhose
transcriptlevelsarenotregulatedbythecircadianclockbutdirectly
byphotoperiod.
3.12. AtGRP7protein,butnottranscript,ismorehighlyexpressed
inLD
Theglycine-richRNA-bindingproteinAtGRP7(AT2G21660)has
animportantroleinflowering.ExpressionofAtGRP7isdirectly
con-trolledbyCCA1andLHY,anditstranscriptlevelsoscillatewitha
peakintheevening[64].AtGRP7regulatestheamplitudeofthe
circadianoscillationofitsmRNAthroughalternativesplicing.
Ara-bidopsisplantsthatconstitutivelyover-expressAtGRP7producea
short-livedmRNAspliceform,whichdampensAtGRP7transcript
oscillationsandinfluencestheaccumulationofothertranscripts
includingAtGRP8 (AT4G39260)[65].Astheresult, AtGRP7
pro-motesflowering,withamorepronouncedeffectinSDthaninLD
[66].InLDweindeedobservedadampeningofbothAtGRP7and
AtGRP8diurnaltranscriptlevelchangesatallleaf6development
stages,butthetranscriptlevelsofAtGRP7didnotchange
signifi-cantlyduringdevelopment(SupportingFig.S8).Incontrast,AtGRP7
proteinlevelsweresignificantlyhigherintheLDexperimentas
comparedtoSOW(Table1),didnotdisplaydiurnallevelchanges,
anddecreasedduringdevelopmentbothinSDandLD(Supporting
Fig.S8).ThehigherAtGRP7proteinlevelsinLDascomparedtoSD
provideanexplanationforearlierobservationsthattheeffectof
AtGRP7overexpressionontimetofloweringisstrongerinSDthan
inLD.
4. Conclusions
Inadditiontophotoperiod,whichmayactatmultiplepoints
inthecircadianclock[67–69],therhythmic,diurnalendogenous
sugarsignalscanentraincircadianrhythmsinArabidopsis[70].
Fur-thermore,inan18hphotoperiodconsiderableamountsofstarch
remainatEONwhiletherateofphotosynthesisisdecreased
com-paredtoa4-,6-,8-,and12-hphotoperiod.Consequently,inlong
photoperiodsgrowthis notlongerlimitedbytheavailabilityof
carbonandthecarbonconversionefficiencydecreases[6].By
sys-tematicallyinvestigatingthemolecularchangesinasingleleafthat
areinvolvedintheadaptationtodifferentphotoperiodsinhighly
controlledconditionswedemonstratedthatfewertranscripts
dis-playsignificantchangesbetweenEODandEONinLDthaninSD.We
previouslydiscussedthatdifferentmRNAlevelsatspecifictimes
duringthediurnalcyclemightberequiredforthetime-dependent
regulationofthecellularenergystatusinprevailingenvironmental
conditions[18].Ifdiurnaltranscriptlevelfluctuationsareindeed
requiredforefficientresourceallocation,thismightexplainwhy
plantsgrowninlongdaysdonotdependonastrictdiurnal
regula-tionoftranscriptiontotightlyeconomisetheirenergybudget.We
alsoestablishedthattranscriptsregulatedbyphotoperiodbelong
to specificfunctional categories that areimportant for
adapta-tiontotheprevailingphotoperiodcondition.Incontrast,identified
proteinsthatdiffersignificantlybetweenphotoperiodsaremainly
relatedtothedifferentgrowthratesofleaf6.Together,changesin
thecomplexmolecularpatternunderlyingleafgrowthindifferent
photoperiodsaretightlylinkedtotheavailableenergy.
Conflictsofinterest
none.
Acknowledgements
WethanktheFunctionalGenomicsCenterZurich(FGCZ)for
pro-vidinginfrastructureandtechnicalsupport,PascalSchläpferand
JohannesFütterer(ETHZurich)forhelpfuldiscussionsandcritical
readingofthemanuscript.We thankNicoleKrohnandBeatrice
Encke(MPIMP)formetaboliteanalyses.Thisworkwassupported
bytheAGRON-OMICSintegratedprojectfundedintheEuropean
FrameworkProgramme6(LSHG-CT-2006-037704).TheUMREEF
issupportedbytheFrenchNationalResearchAgencythroughthe
LaboratoryofExcellenceARBRE(ANR-12-LABXARBRE-01).
AppendixA. Supplementarydata
Supplementarydataassociatedwiththisarticlecanbefound,in
theonlineversion,athttp://dx.doi.org/10.1016/j.cpb.2015.07.001
References
[1]A.N.Dodd,N.Salathia,A.Hall,E.Kévei,R.Tóth,F.Nagy,etal.,Plantcircadian clocksincreasephotosynthesis,growth,survival,andcompetitiveadvantage, Science309(2005)630–633,http://dx.doi.org/10.1126/science.1115581
[2]C.Troein,J.C.W.Locke,M.S.Turner,A.J.Millar,Weatherandseasonstogether demandcomplexbiologicalclocks,Curr.Biol.19(2009)1961–1964,http://dx. doi.org/10.1016/j.cub.2009.09.024
[3]E.M.Farré,S.E.Weise,Theinteractionsbetweenthecircadianclockand primarymetabolism,Curr.Opin.PlantBiol.15(2012)293–300,http://dx.doi. org/10.1016/j.pbi.2012.01.013
[4]B.Y.Chow,S.A.Kay,Globalapproachesfortellingtime:Omicsandthe Arabidopsiscircadianclock,Semin.CellDev.Biol.24(2013)383–392,http:// dx.doi.org/10.1016/j.semcdb.2013.02.005
[5]A.Graf,A.M.Smith,Starchandtheclock:thedarksideofplantproductivity, TrendsPlantSci.16(2011)169–175,http://dx.doi.org/10.1016/j.tplants.2010. 12.003
[6]R.Sulpice,A.Flis,A.aIvakov,F.Apelt,N.Krohn,B.Encke,etal.,Arabidopsis coordinatesthediurnalregulationofcarbonallocationandgrowthacrossa widerangeofphotoperiods,Mol.Plant7(2014)137–155,http://dx.doi.org/ 10.1093/mp/sst127
[7]R.Hayama,G.Coupland,Sheddinglightonthecircadianclockandthe photoperiodiccontrolofflowering,Curr.Opin.PlantBiol.6(2003)13–19,
http://dx.doi.org/10.1016/S1369-5266
[8]F.Andrés,G.Coupland,Thegeneticbasisoffloweringresponsestoseasonal cues,Nat.Rev.Genet.13(2012)627–639,http://dx.doi.org/10.1038/nrg3291
[9]H.A.Kinmonth-Schultz,G.S.Golembeski,T.Imaizumi,Circadian
CellDev.Biol.24(2013)407–413,http://dx.doi.org/10.1016/j.semcdb.2013. 02.006
[10]A.Srikanth,M.Schmid,Regulationoffloweringtime:allroadsleadtoRome, Cell.Mol.LifeSci.68(2011)2013–2037, http://dx.doi.org/10.1007/s00018-011-0673-y
[11]K.S.Sandhu,K.Hagely,M.M.Neff,Geneticinteractionsbetween
brassinosteroid-inactivatingP450sandphotomorphogenicphotoreceptorsin Arabidopsisthaliana,G32(2012)1585–1593,http://dx.doi.org/10.1534/g3. 112.004580
[12]S.J.Cookson,K.Chenu,C.Granier,Daylengthaffectsthedynamicsofleaf expansionandcellulardevelopmentinArabidopsisthalianapartiallythrough floraltransitiontiming,Ann.Bot.99(2007)703–711,http://dx.doi.org/10. 1093/aob/mcm005
[13]T.Usami,G.Horiguchi,S.Yano,H.Tsukaya,Themoreandsmallercells mutantsofArabidopsisthalianaidentifynovelrolesforSQUAMOSA PROMOTERBINDINGPROTEIN-LIKEgenesinthecontrolofheteroblasty, Development136(2009)955–964,http://dx.doi.org/10.1242/dev.028613
[14]R.S.Poethig,Thepast,present,andfutureofvegetativephasechange,Plant Physiol.154(2010)541–544,http://dx.doi.org/10.1104/pp.110.161620
[15]M.R.Willmann,R.S.Poethig,TheeffectofthefloralrepressorFLConthe timingandprogressionofvegetativephasechangeinArabidopsis, Development138(2011)677–685,http://dx.doi.org/10.1242/dev.057448
[16]N.Wuyts,C.Massonnet,M.Dauzat,C.Granier,Structuralassessmentofthe impactofenvironmentalconstraintsonArabidopsisleafgrowth:a3D approach,PlantCellEnv.35(2012)1631–1646,http://dx.doi.org/10.1111/j.
1365-3040.2012.02514.x
[17]A.Telfer,K.M.Bollman,R.S.Poethig,Phasechangeandtheregulationof trichomedistributioninArabidopsisthaliana,Development124(1997) 645–654.
[18]K.Baerenfaller,C.Massonnet,S.Walsh,S.Baginsky,P.Bühlmann,L.Hennig, etal.,Systems-basedanalysisofArabidopsisleafgrowthrevealsadaptationto waterdeficit,Mol.Syst.Biol.8(2012),http://dx.doi.org/10.1038/msb.2012.39
[19]C.Granier,L.Aguirrezabal,K.Chenu,S.J.Cookson,M.Dauzat,P.Hamard,etal., PHENOPSIS,anautomatedplatformforreproduciblephenotypingofplant responsestosoilwaterdeficitinArabidopsisthalianapermittedthe identificationofanaccessionwithlowsensitivitytosoilwaterdeficit,New Phytol.169(2006)623–635,http://dx.doi.org/10.1111/j.1469-8137.2005. 01609.x
[20]C.Massonnet,D.Vile,J.Fabre,M.A.Hannah,C.Caldana,J.Lisec,etal.,Probing thereproducibilityofleafgrowthandmolecularphenotypes:acomparisonof threeArabidopsisaccessionscultivatedintenlaboratories,PlantPhysiol.152 (2010)2142–2157,http://dx.doi.org/10.1104/pp.109.148338
[21]J.M.Cross,M.vonKorff,T.Altmann,L.Bartzetko,R.Sulpice,Y.Gibon,etal., Variationofenzymeactivitiesandmetabolitelevelsin24Arabidopsis accessionsgrowingincarbon-limitedconditions,PlantPhysiol.142(2006) 1574–1588,http://dx.doi.org/10.1104/pp.106.086629
[22]Y.Gibon,O.E.Blaesing,J.Hannemann,P.Carillo,M.Höhne,J.H.M.Hendriks, etal.,ARobot-basedplatformtomeasuremultipleenzymeactivitiesin Arabidopsisusingasetofcyclingassays:comparisonofchangesofenzyme activitiesandtranscriptlevelsduringdiurnalcyclesandinprolonged darkness,PlantCell16(2004)3304–3325,http://dx.doi.org/10.1105/tpc.104. 025973
[23]H.Rehrauer,C.Aquino,W.Gruissem,S.R.Henz,P.Hilson,S.Laubinger,etal., AGRONOMICS1:anewresourceforArabidopsistranscriptomeprofiling,Plant Physiol.152(2010)487–499,http://dx.doi.org/10.1104/pp.109.150185
[24]M.Müller,A.Patrignani,H.Rehrauer,W.Gruissem,L.Hennig,Evaluationof alternativeRNAlabelingprotocolsfortranscriptprofilingwithArabidopsis AGRONOMICS1tilingarrays,PlantMet.8(2012)18,http://dx.doi.org/10. 1186/1746-4811-8-18
[25]P.L.Ross,Y.N.Huang,J.N.Marchese,B.Williamson,K.Parker,S.Hattan,etal., MultiplexedproteinquantitationinSaccharomycescerevisiaeusing amine-reactiveisobarictaggingreagents,Mol.CellProteomics3(2004) 1154–1169.
[26]A.Pierce,R.D.Unwin,C.A.Evans,S.Griffiths,L.Carney,L.Zhang,etal., Eight-channeliTRAQenablescomparisonoftheactivityofsixleukemogenic tyrosinekinases,Mol.CellProteomics7(2008)853–863.
[27]P.Lamesch,T.Z.Berardini,D.Li,D.Swarbreck,C.Wilks,R.Sasidharan,etal., TheArabidopsisInformationResource(TAIR):improvedgeneannotationand newtools,NucleicAcidsRes.40(2012)D1202–D1210,http://dx.doi.org/10. 1093/nar/gkr1090
[28]K.Baerenfaller,M.Hirsch-Hoffmann,J.Svozil,R.Hull,D.Russenberger,S. Bischof,etal.,pep2pro:anewtoolforcomprehensiveproteomedataanalysis torevealinformationaboutorgan-specificproteomesinArabidopsisthaliana, Integr.Biol.(Camb)3(2011)225–237,http://dx.doi.org/10.1039/c0ib00078g
[29]M.Hirsch-Hoffmann,W.Gruissem,K.Baerenfaller,pep2pro:the
high-throughputproteomicsdataprocessing,analysis,andvisualizationtool, Front.PlantSci.3(2012),http://dx.doi.org/10.3389/fpls.2012.00123
[30]J.A.Vizcaíno,R.G.Côté,A.Csordas,J.A.Dianes,A.Fabregat,J.M.Foster,etal., ThePRoteomicsIDEntifications(PRIDE)databaseandassociatedtools:status in2013,NucleicAcidsRes.41(2013)D1063–D1069,http://dx.doi.org/10. 1093/nar/gks1262
[31]Y.Benjamini,Y.Hochberg,Controllingthefalsediscoveryrate:apracticaland powerfulapproachtomultipletesting,J.R.Stat.Soc.Ser.B57(1995)289–300.
[32]RCoreTeam,R.A.languageandenvironmentforstatisticalcomputing,2012.
http://www.r-project.org
[33]V.Wahl,J.Ponnu,A.Schlereth,S.Arrivault,T.Langenecker,A.Franke,etal., Regulationoffloweringbytrehalose-6-phosphatesignalinginArabidopsis thaliana,Science339(2013)704–707,http://dx.doi.org/10.1126/science. 1230406
[34]K.D.Edwards,P.E.Anderson,A.Hall,N.S.Salathia,J.C.W.Locke,J.R.Lynn,etal., FLOWERINGLOCUSCmediatesnaturalvariationinthehigh-temperature responseoftheArabidopsiscircadianclock,PlantCell18(2006)639–650,
http://dx.doi.org/10.1105/tpc.105.038315
[35]A.Yilmaz,M.K.Mejia-Guerra,K.Kurz,X.Liang,L.Welch,E.Grotewold,AGRIS: theArabidopsisGeneregulatoryinformationserver,anupdate,NucleicAcids Res.39(2010)D1118–D1122,http://dx.doi.org/10.1093/nar/gkq1120
[36]O.Thimm,O.Bläsing,Y.Gibon,A.Nagel,S.Meyer,P.Krüger,etal.,MAPMAN: auser-driventooltodisplaygenomicsdatasetsontodiagramsofmetabolic pathwaysandotherbiologicalprocesses,PlantJ.37(2004)914–939.
[37]W.A.Decatur,M.J.Fournier,rRNAmodificationsandribosomefunction, TrendsBiochem.Sci.27(2002)344–351.
[38]J.B.Harborne,C.A.Williams,Advancesinflavonoidresearchsince1992, Phytochemistry55(2000)481–504.
[39]M.Kanehisa,S.Goto,Y.Sato,M.Furumichi,M.Tanabe,KEGGforintegration andinterpretationoflarge-scalemoleculardatasets,NucleicAcidsRes.40 (2012)D109–D114,http://dx.doi.org/10.1093/nar/gkr988
[40]I.S.Carvalho,T.Cavaco,L.M.Carvalho,P.Duque,Effectofphotoperiodon flavonoidpathwayactivityinsweetpotato(Iopmeabatatas(L.)Lam.)leaves, FoodChem.118(2010)384–390.
[41]U.Takahama,RedoxReactionsbetweenkaempferolandilluminated chloroplasts,PlantPhysiol.71(1983)598–601.
[42]G.Krouk,S.Ruffel,R.A.Gutiérrez,A.Gojon,N.M.Crawford,G.M.Coruzzi,etal., Aframeworkintegratingplantgrowthwithhormonesandnutrients,Trends PlantSci.16(2011)178–182,http://dx.doi.org/10.1016/j.tplants.2011.02.004
[43]M.Vanstraelen,E.Benková,Hormonalinteractionsintheregulationofplant development,AnnuRevCellDevBiol.28(2012)463–487,http://dx.doi.org/ 10.1146/annurev-cellbio-101011-155741
[44]K.C.McGrath,B.Dombrecht,J.M.Manners,P.M.Schenk,C.I.Edgar,D.J. Maclean,etal.,Repressor-andactivator-typeethyleneresponsefactors functioninginjasmonatesignalinganddiseaseresistanceidentifiedviaa genome-widescreenofArabidopsistranscriptionfactorgeneexpression, PlantPhysiol.139(2005)949–959,http://dx.doi.org/10.1104/pp.105.068544
[45]G.H.Son,J.Wan,H.J.Kim,X.C.Nguyen,W.S.Chung,J.C.Hong,etal., Ethylene-responsiveelement-bindingfactor5,ERF5,isinvolvedin chitin-inducedinnateimmunityresponse,Mol.PlantMicrobe.Interact25 (2012)48–60,http://dx.doi.org/10.1094/-06-11-0165
[46]R.Bours,M.vanZanten,R.Pierik,H.Bouwmeester,A.vanderKrol,Antiphase lightandtemperaturecyclesaffectPHYTOCHROMEB-controlledethylene sensitivityandbiosynthesis,limitingleafmovementandgrowthof Arabidopsis,PlantPhysiol.163(2013)882–895,http://dx.doi.org/10.1104/pp. 113.221648
[47]E.M.Turk,S.Fujioka,H.Seto,Y.Shimada,S.Takatsuto,S.Yoshida,etal.,BAS1 andSOB7actredundantlytomodulateArabidopsisphotomorphogenesisvia uniquebrassinosteroidinactivationmechanisms,PlantJ.42(2005)23–34, 10.1111/j.1365-313X.2012.05093.x.
[48]E.M.Turk,S.Fujioka,H.Seto,Y.Shimada,S.Takatsuto,S.Yoshida,etal., CYP72B1inactivatesbrassinosteroidhormones:anintersectionbetween photomorphogenesisandplantsteroidsignaltransduction,PlantPhysiol.133 (2003)1643–1653,http://dx.doi.org/10.1104/pp.103.030882
[49]Y.Yin,D.Vafeados,Y.Tao,S.Yoshida,T.Asami,J.Chory,Anewclassof transcriptionfactorsmediatesbrassinosteroid-regulatedgeneexpressionin Arabidopsis,Cell120(2005)249–259,http://dx.doi.org/10.1016/j.cell.2004. 11.044
[50]M.Büttner,Themonosaccharidetransporter(-like)genefamilyin Arabidopsis,FEBSLett.581(2007)2318–2324,http://dx.doi.org/10.1016/j. febslet.2007.03.016
[51]R.Stadler,M.Büttner,P.Ache,R.Hedrich,N.Ivashikina,M.Melzer,etal., Diurnalandlight-regulatedexpressionofAtSTP1inguardcellsofArabidopsis, PlantPhysiol.133(2003)528–537,http://dx.doi.org/10.1104/pp.103.024240
[52]M.H.Cho,H.Lim,D.H.Shin,J.S.Jeon,S.H.Bhoo,Y.IlPark,etal.,Roleofthe plastidicglucosetranslocatorintheexportofstarchdegradationproducts fromthechloroplastsinArabidopsisthaliana,NewPhytol.190(2011) 101–112,http://dx.doi.org/10.1111/j.1469-8137.2010.03580.x
[53]F.A.Hoeberichts,E.Vaeck,G.Kiddle,E.Coppens,B.vandeCotte,A. Adamantidis,etal.,ATemperature-sensitivemutationintheArabidopsis thalianaphosphomannomutasegenedisruptsproteinglycosylationand triggerscelldeath,J.Biol.Chem.283(2008)5708–5718,http://dx.doi.org/10. 1074/jbc.M704991200
[54]G.Beck,D.Coman,E.Herren,M.A.Ruiz-Sola,M.Rodríguez-Concepción,W. Gruissem,etal.,CharacterizationoftheGGPPsynthasegenefamilyin Arabidopsisthaliana,PlantMol.Biol.82(2013)393–416,http://dx.doi.org/10. 1007/s11103-013-0070-z
[55]P.Pesaresi,M.Scharfenberg,M.Weigel,I.Granlund,W.P.Schröder,G.Finazzi, etal.,Mutants,overexpressors,andinteractorsofArabidopsisplastocyanin isoforms:revisedrolesofplastocyanininphotosyntheticelectronflowand thylakoidredoxstate,Mol.Plant2(2009)236–248,http://dx.doi.org/10. 1093/mp/ssn041
[56]E.Seo,H.Lee,J.Jeon,H.Park,J.Kim,Y.-S.Noh,etal.,Crosstalkbetweencold responseandfloweringinArabidopsisismediatedthroughthe
flowering-timegeneSOC1anditsupstreamnegativeregulatorFLC,PlantCell 21(2009)3185–3197,http://dx.doi.org/10.1105/tpc.108.063883
[57]Y.Kobayashi,D.Weigel,Moveonup,it’stimeforchange-mobilesignals controllingphotoperiod-dependentflowering,GenesDev.21(2007) 2371–2384,http://dx.doi.org/10.1101/gad.1589007
[58]F.Turck,F.Fornara,G.Coupland,Regulationandidentityofflorigen: FLOWERINGLOCUSTmovescenterstage,Annu.Rev.PlantBiol.59(2008) 573–594,http://dx.doi.org/10.1146/annurev.arplant.59.032607.092755
[59]S.Torti,F.Fornara,AGL24actsinconcertwithSOC1andFULduring Arabidopsisfloraltransition,PlantSignalBehav.7(2012)1251–1254,http:// dx.doi.org/10.4161/psb.21552
[60]S.D.Michaels,R.M.Amasino,FLOWERINGLOCUSCencodesanovelMADS domainproteinthatactsasarepressorofflowering,PlantCell11(1999) 949–956.
[61]D.Li,C.Liu,L.Shen,Y.Wu,H.Chen,M.Robertson,etal.,ARepressorComplex GovernstheIntegrationofFloweringSignalsinArabidopsis,Dev.Cell15 (2008)110–120,http://dx.doi.org/10.1016/j.devcel.2008.05.002
[62]O.E.Bläsing,Y.Gibon,M.Günther,M.Höhne,R.Morcuende,D.Osuna,etal., Sugarsandcircadianregulationmakemajorcontributionstotheglobal regulationofdiurnalgeneexpressioninArabidopsis,PlantCell17(2005) 3257–3281,http://dx.doi.org/10.1105/tpc.105.035261
[63]M.F.Covington,J.N.Maloof,M.Straume,S.A.Kay,S.L.Harmer,Global transcriptomeanalysisrevealscircadianregulationofkeypathwaysinplant growthanddevelopment,GenomeBiol.9(2008)R130,http://dx.doi.org/10. 1186/gb-2008-9-8-r130
[64]C.Heintzen,M.Nater,K.Apel,D.Staiger,AtGRP7,anuclearRNA-binding proteinasacomponentofacircadian-regulatednegativefeedbackloopin Arabidopsisthaliana,Proc.Natl.Acad.Sci.94(1997)8515–8520,USA.
[65]D.Staiger,L.Zecca,D.A.WieczorekKirk,K.Apel,L.Eckstein,Thecircadian clockregulatedRNA-bindingproteinAtGRP7autoregulatesitsexpressionby influencingalternativesplicingofitsownpre-mRNA,PlantJ.33(2003) 361–371.
[66]C.Streitner,S.Danisman,F.Wehrle,J.C.Schöning,J.R.Alfano,D.Staiger,The smallglycine-richRNAbindingproteinAtGRP7promotesfloraltransitionin Arabidopsisthaliana,PlantJ.56(2008)239–250,http://dx.doi.org/10.1111/j. 1365-313X,2008.03591.x.
[67]A.J.Millar,Inputsignalstotheplantcircadianclock,J.Exp.Bot.55(2004) 277–283,http://dx.doi.org/10.1093/jxb/erh034
[68]A.Pokhilko,A.P.Fernández,K.D.Edwards,M.M.Southern,K.J.Halliday,A.J. Millar,TheclockgenecircuitinArabidopsisincludesarepressilatorwith additionalfeedbackloops,Mol.Syst.Biol.8(2012)574,http://dx.doi.org/10. 1038/msb.2012.6
[69]M.L.Rugnone,A.Faig,S.E.Sanchez,R.G.Schlaen,C.E.Hernando,K.Danelle, etal.,LNKgenesintegratelightandclocksignalingnetworksatthecoreofthe Arabidopsisoscillator,Proc.Natl.Acad.Sci.U.S.A110(2013)12120–12125,
http://dx.doi.org/10.1073/pnas.1302170110
[70]M.J.Haydon,O.Mielczarek,F.C.Robertson,K.E.Hubbard,A.A.R.Webb, PhotosyntheticentrainmentoftheArabidopsisthalianacircadianclock, Nature502(2013)689–692,http://dx.doi.org/10.1038/nature12603.