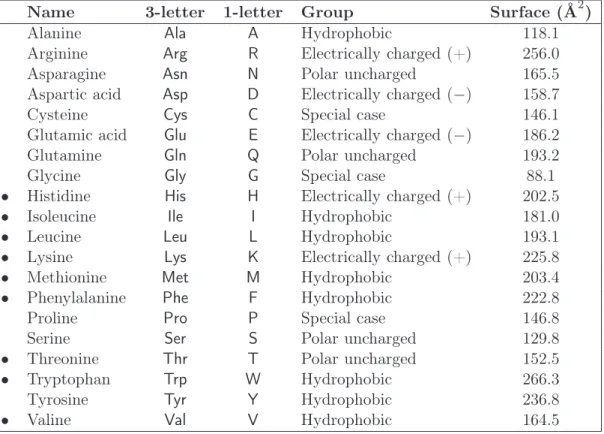
Protein Structural Annotation: Multi-Task Learning and Feature Selection
Texte intégral
Figure




Documents relatifs
Abstract: First-principles calculations are performed to study the structural, electronic, and thermalproperties of the AlAs and BAs bulk materials and Al 1 -x B x As ternary
The heat transfer fluid (humid air) licks the salt, water vapor diffuses within, allowing the chemical reaction to happen, and heat to be stored or released.. The salt thickness
Differential expression analysis on the complete transcriptome including both reference and novel transcripts with cuffdiff (step IV) identified 4,111 DE genes accounting for 4,735
of Geology, Quaternary Sciences, Lund Univ., S¨olvegatan 12, 22362 Lund, Sweden 2 Geology and Geophysics Department, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA..
Since activation of tyrosine kinases may be involved in the pathogen- esis of scleroderma, we have decided to test the effect of imatinib mesylate, a tyrosine kinases inhibitor,
To test whether the protein can mediate a more specific recombination reaction, the invasion of a duplex DNA by a ssDNA D-loop formation, hPOMp100 was incubated with the
The original analysis by TIGR provided 625 function predictions (Bult et al., 1996) (37% of the chromosomal genome), with the update increasing them to 809 assignments (Kyrpides et
Prediction of functional features: predicted aspects include ConSurf annotations and visualizations of functionally im- portant sites (26,27), protein mutability landscape