HAL Id: hal-03086548
https://hal.inrae.fr/hal-03086548
Submitted on 22 Dec 2020
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Distributed under a Creative Commons Attribution - NonCommercial - NoDerivatives| 4.0
International License
Bartonella gabonensis sp. nov., a new bartonella species
from savannah rodent Lophuromys sp. in Franceville,
Gabon
J.B. Mangombi, N. N’Dilimabaka, H. Medkour, O.L. Banga, M.L. Tall, M.
Ben Khedher, J. Terras, S. Abdi, Mathieu Bourgarel, E. Leroy, et al.
To cite this version:
J.B. Mangombi, N. N’Dilimabaka, H. Medkour, O.L. Banga, M.L. Tall, et al..
Bartonella
gabonensis sp.
nov., a new bartonella species from savannah rodent Lophuromys sp.
in
Franceville, Gabon. New Microbes and New Infections, Wiley Online Library 2020, 38, pp.100796.
�10.1016/j.nmni.2020.100796�. �hal-03086548�
Characterization and antibiotic resistance pattern of diffusely adherent
Escherichia coli (DAEC), isolated from paediatric diarrhoea in Shiraz,
southern Iran
K. Javadi1), S. Mohebi1), M. Motamedifar1),2)and N. Hadi1),3)
1) Department of Bacteriology and Virology, School of Medicine, 2) Shiraz HIV/AIDS Research Centre, Institute of Health and 3) Bioinformatics and Computational Biology Research Centre, Shiraz University of Medical Sciences, Shiraz, Iran
Abstract
Diarrhoea is a major health concern, especially in developing countries. Research has implicated diffusely adherent Escherichia coli (DAEC) strains as a cause of diarrhoea. In this study, we investigated the prevalence, adherence assay, virulence gene profiles and antimicrobial resistance of DAEC at a hospital in southern Iran. In this cross-sectional study, 309 infants and children under the age of 13 years with diarrhoea who had been referred to Shahid Dastgheib Hospital, Shiraz between October 2018 and May 2019 were recruited. Microbiological methods, PCR, HEp-2 adherence assay and antimicrobial susceptibility test were used. Of the 309 stool samples, 207 (66.9%) were found to contain E. coli by biochemical tests and culture. Molecular analysis of Afa/Dr and AIDA-I adhesin-encoding genes showed that 14 (6.7%) out of 207 E. coli isolates were DAEC. All DAEC isolates in HEp-2 cells showed a diffusely adherent pattern. The virulence genes sat, pet, sigA, pic, astA andfimH were found in 50%, 0%, 14.2%, 14.2%, 21.4% and 100% of DAEC isolates, respectively. The most effective antibiotic against the DAEC isolates was imipenem (92.8%) and the least effective was ampicillin (0%). Ourfindings expand the knowledge on DAEC prevalence and its characteristics in Iran. It also explains the role of virulence genes in DAEC pathogenesis. The results showed that although the prevalence of DAEC is low, these strains exhibit a high rate of antimicrobial resistance as well as high frequency for carrying virulence genes.
© 2020 Published by Elsevier Ltd.
Keywords: Adherence, antibiotic resistance, diarrhoea, diffusely adherent Escherichia coli, virulence genes Original Submission: 19 August 2020; Revised Submission: 25 September 2020; Accepted: 5 October 2020 Article published online: 10 October 2020
Corresponding author: N. Hadi, Department of Bacteriology and Virology, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran.
E-mails:hadina@sums.ac.ir,nahalhadi@gmail.com
Introduction
Diarrhoea is a major health problem, especially in developing countries, and one of the most important causes of mortality among children under the age of 5 [1]. The aetiological agents of diarrhoea include a wide range of viruses, bacteria and parasites. Among the bacterial pathogens, diarrhoeagenic Escherichia coli is an important cause of endemic and epidemic diarrhoea worldwide [2].
Diarrhoeagenic E. coli strains are classified into six main pathotypes based on their specific virulence characteristics, association with some serotypes and epidemiological charac-teristics: enteropathogenic E. coli, enterohaemorrhagic E. coli, enterotoxigenic E. coli, enteroaggregative E. coli, enteroinvasive E. coli and diffusely adherent E. coli (DAEC) [2].
DAEC strains are defined based on the presence of a diffuse adherence pattern (DA) on HeLa and HEp-2 epithelial cells. In the DA pattern, bacteria uniformly cover the cell surface [3].
According to the adhesin expression, two groups of DAEC strains have been identified, Afa/Dr DAEC and AIDA-I DAEC. Furthermore, these adhesins are also responsible for the DA phenotype [4]. Afa/Dr DAEC strains are associated with acute diarrhoea in children, especially in those 6 months and older, with persistent diarrhoea. Therefore, the DAEC pathogroup contains Afa/Dr adhesin-encoding genes and can cause
New Microbe and New Infect 2020; 38: 100780 © 2020 Published by Elsevier Ltd This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
diarrhoea in patients [5]. The Afa/Dr family includesfimbrial and afimbrial adhesins, afimbrial adhesins Afa-I, Afa-II, Afa-III, Afa-V, Afa-VII, Afa-VIII, plus Dr-2 as well as Dr and F1845 fimbrial adhesins. Many of these adhesins have been identified in E. coli strains isolated from human urinary tract infections or diarrhoea, except Afa-VII, which was only found in E. coli iso-lated from bovine faeces [6]. F1845 adhesin wasfirst identified in an E. coli strain (C1845) isolated from a child with chronic diarrhoea [6]. Until now, only the genes encoding for I, Afa-II, Afa-III and Afa-V have been identified in E. coli strains isolated from individuals with diarrhoea [7].
Afa-I and Afa-V adhesins only bind to the decay-accelerating factor, whereas AfaE- III, Dr and F1845 can each also bind to carcinoembryonic antigen-related cell adhesin molecules, and the Dr adhesin can also bind to type IV collagen [8–10].
Enterobacteriaceae have autotransporters called Serine pro-tease autotransporters of Enterobacteriaceae (SPATEs). SPATEs are classified into two classes. Pet (plasmid-encoded toxin), Sat (secreted autotransporter toxin) and SigA are members of Class I SPATEs, which are cytotoxic to epithelial cells; Pic (protease involved in intestinal colonization) are members of Class II SPATEs, and are non-cytotoxic. The distribution of SPATEs among the diarrhoeagenic E. coli has been shown in many studies [11].
Type I pili are a type of composite surfacefibre present in DAEC and are encoded by thefim gene cluster. FimH is the pilus adhesin [12]. Some DAEC strains through activation of Src and the mitogen-activated protein kinase, elicit a secondary interleukin-8 production by polymorphonuclear lymphocytes in a type-1 pili-dependent manner [13].
Other virulence factor genes are recorded in DAEC strains, including astA, the gene for enteroaggregative heat-stable toxin 1 (EAST1), which wasfirst identified in the enteroaggregative E. coli. EAST1 similarly stimulates the guanylate cyclase receptor to both enterotoxigenic E. coli St toxin and guanylin [14].
Enteropathogenic E. coli strains might show a DA pattern on HEp-2 and HeLa cells; therefore, we cannot use cell adhesins assay alone to detect Afa/Dr DAEC [15]. PCR is a simpler and faster method to identify Afa/Dr adhesins; consequently, PCR methods are more suitable to describe Afa/Dr DAEC strains [16,17]. Overall, these studies have clearly shown that it is important to determine the prevalence of DAEC selecting Afa or Daa target genes [18].
Although recent epidemiological studies indicated a high prevalence of DAEC strains isolated from diarrhoeal faeces, its pathogenicity has not been identified in adults [19]. It was found that the relative risk of diarrhoea associated with DAEC in-creases with the child’s age from 18 months to 5 years [20]. The reason for such an age-related phenomenon and the mode of DAEC acquisition are yet to be determined [21].
The objectives of this study were to determine the preva-lence of DAEC by PCR and HEp-2 cell adhesin assays among bacterial enteropathogens recovered from children with diar-rhoea in Shahid Dastgheib Hospital, Shiraz, Iran, as well as to determine the presence of virulence genes in DAEC strains. We also determined the antimicrobial resistance profiles of the DAEC strains.
Materials and methods
Sample collection
This cross-sectional study included infants and children under the age of 13 years with diarrhoea who were referred to Shahid Dastgheib Hospital from October 2018 to May 2019. Patient information, such as age, gender, occult blood, pus cells and red blood cells, were obtained. Our exclusion criteria were age >13 years, no diarrhoea, insufficient data, and dry or suspected contaminated sample.
Laboratory processing
The stool samples were immediately cultured on xylose lysine deoxycholate and MacConkey agar media in the laboratory of Shahid Dastgheib Hospital and then transferred to the labora-tory of Bacteriology and Virology at Shiraz University of Med-ical Sciences for definitive diagnosis and identification. After incubation for 24 h at 37°C, lactose-positive colonies were re-cultured with standard biochemical methods to detect E. coli.
The tests included Gram staining, oxidase test, catalase test, motility test, triple-sugar iron fermentation, the citrate test, methyl red staining, the Voges–Proskauer test, indole test, ortho-nitrophenylgalactoside test, and acid production from carbohydrates.
Escherichia coli isolates were stored at–70°C in tryptic soy broth containing 20% glycerol (Merck Co., Darmstadt, Ger-many) for further characterization.
DNA extraction
DNA was extracted using the boiling method. In this method, a single colony was grown on eosin methylene blue agar plates, mixed with 300μL distilled water and boiled at 100°C for 10 minutes. After centrifugation at 13 000 g for 15 min, the su-pernatant containing the extracted DNA was transferred into a new sterile tube and stored at – 20°C.
Identification of DAEC isolates
All E. coli isolates were tested by PCR to detect six adherence genes: afaE-1, afaE-2, afaE-3, afaE-5, daaE and aida/aah for identification of DAEC isolates. The primers that were used to amplify these genes are listed inTable 1.
2 New Microbes and New Infections, Volume 38 Number C,---2020
NMNI
© 2020 Published by Elsevier Ltd, NMNI, 38, 100780
The PCR assay was performed in a total volume of 25 mL containing 0.5 mL of each primer (10 pM), 12.5 mL of DNA Polymerase Master Mix RED (Ampliqon Co., Inc., Odense, Denmark), 1 mL of DNA and 10.5 mL of water (DNase- and RNase-free water) in a T100™ thermal cycler (Bio-Rad, Her-cules, CA, USA) under the following conditions: initial dena-turation at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 1 min, annealing at 52°C–61°C for 1 min (Table 1), and extension at 72°C for 1 min, and a singlefinal extension at 72°C for 5 min. PCR products underwent elec-trophoresis in 1.5% agarose gels in the 0.5 Tris/EDTA/boric acid buffer and were visualized using ultraviolet light after staining with safe stain load dye (CinnaGen Co., Tehran, Iran). HEp-2 adherence assay
DAEC isolates were examined for HEp-2 adherence as described by Scaletsky et al. [22], with slight modifications. Briefly,
mono-layers of 105 HEp-2 cells were grown in Dulbecco’s modified
Eagle’s medium (DMEM) with 10% fetal bovine serum in Leighton tubes containing a cover slip and incubated for 48 h at 37°C. DAEC strains were grown in 3 mL of trypticase soy broth for 16–18 h at 37°C. The tubes were then washed with phosphate-buffered saline and 1 mL of DMEM with 10% fetal bovine serum was added to each tube. The monolayers were infected with 40 μL of bacterial cultures added to 1 mL of DMEM and incubated for 3 h at 37°C. The infected monolayers were washed with sterile phosphate-buffered saline, fixed with methanol, stained with Giemsa stain and examined for DA pattern under a light microscope using a × 100 objective.
Standard strains of E. coli were used as the control to detect daaE and aida/aah, which were C1845 and 2787, respectively [18,23].
Detection of virulence genes
All DAEC isolates were examined by PCR to detect six viru-lence genes: sat, pet, sigA, pic, astA andfimH. The primers used to amplify these genes are listed inTable 1. The same condi-tions as for the adherence genes were employed for the detection and amplification of virulence genes expect that the annealing temperature was 56°C–58°C.
Antimicrobial susceptibility testing
Antibiotic susceptibility testing of the DAEC isolates to ampicillin, ampicillin-sulbactam, cefotaxime, ceftriaxone, ceftazidime, imipe-nem, gentamicin, nalidixic acid, trimethoprim-sulfamethoxazole, moxifloxacin and chloramphenicol (Mast Group Ltd, Bootle, UK) was carried out on Müller–Hinton agar (Merck Co.) by disc diffusion method according to the Clinical and Laboratory Stan-dards Institute. Escherichia coli ATCC25922 was used as the quality control strain.
DNA sequence analysis
To verify the accuracy of amplified genes of afaE-1, afaE-2, afaE-3, afaE-5, the amplicons were submitted for sequencing (Bioneer Co., Munpyeongseoro, Daedeok-gu, Daejeon, South Korea) and the resulting sequences were analysed using online BLAST software (https://blast.ncbi.nlm.nih.gov/ Blast.cgi).
TABLE 1.Primers used inPCR analysis
Gene Primer sequences
Annealing temperature (°C) Amplicon size (bp) Reference afaE-1 5ʹ-CGAAAACGGCACTGACAAG-3ʹ 58°C 230 pb [26] 5ʹ-AGGCTTCCGTGAATACAACC-3ʹ afaE-2 5ʹ-TTAGACCGTACTGTTGTGTTACC-3ʹ 60°C 375 pb [26] 5ʹ-TTTCCCAGTAGACTGGAATGAAGC-3ʹ afaE-3 5ʹ-TTAGACCGTACTGTTGTGTACC-3ʹ 58°C 408 pb [26] 5ʹ-ACCATTGTCGGTCGTCCAGGC-3ʹ afaE-5 5ʹ-TTAGACCGTACTGTTGTGTTACC-3ʹ 61°C 429 pb [26] 5ʹ-AGCATCGGCGCGGTATACGGT-3ʹ daaE 5ʹ-TGACTGTGACCGAAGAGTGC-3ʹ 56°C 380 pb [26] 5ʹ-TTAGTTCGTCCAGTAACCCCC-3ʹ aida/aah 5ʹ-TCGATACCGAAACGCATACGCAGA-3ʹ 52°C 204 pb [29] 5ʹ-ACGCCGATCGGTGATGATGAAGAT-3ʹ sat 5ʹ-TCAGAAGCTCAGCGAATCATTG-3ʹ 58°C 930 pb [35] 5ʹ-CCATTATCACCAGTAAAACGCACC-3ʹ
pet 5ʹ-GGCACAGAAT AAAGGGGTGTTT-3ʹ 58°C 302 pb [35] 5ʹ-CCTCTTGTTTCCACGACATAC-3ʹ sigA 5ʹ-CCGACTTCTCACTTTCTCCCG-3ʹ 58°C 430 pb [35] 5ʹ-CCATCCAGCTGCATAGTGTTTG-3ʹ pic 5ʹ-ACTGGATCTTAAGGCTCAGGAT-3ʹ 58°C 572 pb [35] 5ʹ-GACTTAATGTCACTGTTCAGCG-3ʹ astA 5ʹ-ATGCCATCAACACAGTATAT-3ʹ 58°C 110 pb [38] 5ʹ-GCGAGTGACGGCTTTGTAGT-3ʹ fimH 5ʹ-TGCAGAACGGATAAGCCGTGG-3ʹ 61°C 508 pb [39] 5ʹ-GCAGTCACCTGCCCTCCGGTA-3ʹ
Statistical analysis
SPSS software, version 26.0 (IBM Corp., Armonk, NJ, USA) was used for statistical analysis. The chi-square test and Fisher’s exact test were performed for analysis of the data and p values < 0.05 were considered to be statistically significant.
Results
Subject
A total of 309 stool samples from children under 13 years old with diarrhoea were included in our study. The samples were divided into three groups, 131 (42.3%) samples from children younger than 3 years; 98 (31.7%) from children aged 3–6 years and 80 (25.8%) from children aged 7–12 years. Male to female ratio was 52.7% to 47.3% (163 males versus 146 female). Prevalence of DAEC strains
Escherichia coli isolates were identified in 207 (66.9%) samples by biochemical tests and culture. Strains of E. coli harbouring the Afa/Dr and AIDA-I adhesin-encoding genes were found in the stool samples of 14 (6.7%) children with diarrhoea (Table 2); positive samples were from six boys and eight girls. Seven (50%) children with DAEC were under 3 years old,five (35.7%) and two (14.3%) were aged 3–6 years and 7–12 years, respectively (Table 3). Most DAEC samples contained occult blood, red blood cells and pus cells (Table 4).
Adherence pattern analysis of DAEC strains
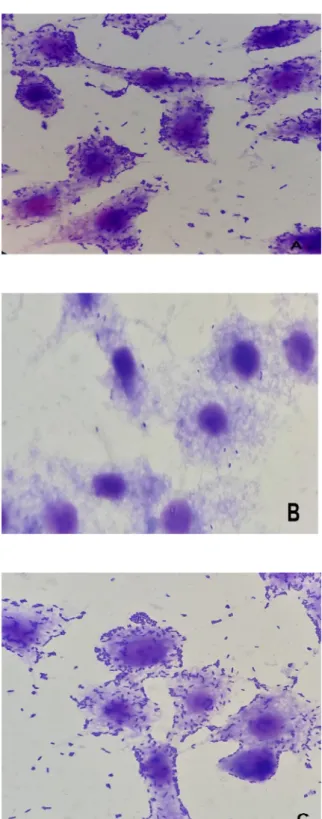
The analysis of the adherence patterns of the 14 isolates studied in HEp-2 cells showed that 100% (14 isolates) expressed the DA pattern (Fig. 1).
Presence of virulence genes
All DAEC isolates were tested by PCR to detect the genes of the proposed DAEC virulence factors, including sat, pet, sigA, pic, astA and fimH. Seven DAEC strains were positive for sat (50%). The astA gene was identified in three (21.4%) DAEC strains and the pic and sigA genes were found in two (14.2%) of the 14 DAEC strains. All DAEC isolates were negative for the pet gene and all DAEC isolates were positive for thefimH gene (Table 5). Several different combinations of the virulence markers were found among the DAEC isolates (Table 6).
Antimicrobial resistance among DAEC isolates The results of antimicrobial susceptibility testing of the diar-rhoeagenic DAEC isolates with 11 antibiotics by disc diffusion method are shown in Table 7. The most effective antibiotic against DAEC was imipenem and the least effective was ampicillin.
Discussion
The DAEC strains are a heterogeneous group of E. coli strains that generate a diffuse adherence pattern on HeLa and HEp-2 cells [20]. Food or water polluted with human or animal faeces is the main transmission route for the DAEC pathotype [24]. DAEC strains destroy the intestinal epithe-lium by binding to proteins that accelerate degradation. They account for a large proportion of diarrhoeal cases observed in inpatients who have no other identified enteropathogen [19]. Recent epidemiological studies have implicated DAEC strains as a cause of diarrhoea in children in developing countries [25].
TABLE 3.Age and gender of 207 Escherichia coli isolates and
positive diffusely adherent E. coli (DAEC) in different age groups Age (years) No. of samples Males, n (%) Females, n (%) DAEC, n (%) <3 113 58 (53.7) 55 (55.5) 7 (6.2) 3−6 56 29 (26.8) 27 (27.3) 5 (8.9) 7−12 38 21 (19.4) 17 (17.2) 2 (5.2) Total 207 108 (52.7) 99 (47.3) 14 (6.7)
TABLE 4.Stool analysisfindings of Escherichia coli isolates and
positive diffusely adherent E. coli (DAEC) isolates from children with diarrhoea
Stool characteristic All E. coli (n[ 207), n (%) DAEC (n[ 14), n (%) Occult blood 149 (72) 10 (71.4) Red blood cells 118 (57) 7 (50) Pus cells 145 (70) 8 (57.1)
TABLE 2. Characterization of the adhesin of diffusely
adherent Escherichia coli (DAEC) isolates from children with diarrhoea
Isolate afaE-1 afaE-2 afaE-3 afaE-5 daaE aida /aah HEp-2 adherence DAEC1 + — — + — — DA DAEC2 + — + — — — DA DAEC3 + — — — + — DA DAEC4 + — — — — — DA DAEC5 + — — — — — DA DAEC6 — + + — — — DA DAEC7 + — — — + — DA DAEC8 + — — — — — DA DAEC9 — — + — — — DA DAEC10 — + — — — — DA DAEC11 — — — + — — DA DAEC12 + — — — — — DA DAEC13 + — — + — — DA DAEC14 — — + — + — DA n (%) 9 (64.3) 2 (14.2) 4 (28.6) 3 (21.5) 3 (21.5) 0 (0)
DA, diffuse adherence.
4 New Microbes and New Infections, Volume 38 Number C,---2020
NMNI
© 2020 Published by Elsevier Ltd, NMNI, 38, 100780
The proportion of DAEC among E. coli in our study was 6.7%, which is comparable to previous reports in Shiraz (7.9%) [25]. DAEC occurrence in our study was higher than in northwest Mexico (6.2%) [26] and in South American coun-tries, such as Peru (4%) and Colombia (1.2%) [27,28]. In contrast, the identified rate of DAEC was 8.18% in China [29], 15.7% in Brazil [30] and 35% in Mexico [14], which is higher than the rate found in this study.
We investigated all sample data tofind any association be-tween these data and DAEC positivity. Accordingly, we classi-fied our data based on age, gender and stool analysis findings, such as occult blood, red blood cells and pus cells. There was no significant association between these data and DAEC, which shows that all diarrhoeal stools should be examined for DAEC isolates.
The Afa/Dr adhesin-encoding genes afaE-1, afaE-2, afaE-3, afaE-5 and daaE were identified in 64.3%, 14.2%, 28.6%, 21.5%
FIG. 1.Microscopic appearance of adherence patterns on HEp2 cells of
diffusely adherent Escherichia coli (DAEC) isolates from children with diarrhoea: (a) DAEC C1845, (b) E. coli K12, (c) DAEC isolated from children with diarrhoea.
TABLE 5. Incidence of virulence genes among diffusely
adherent Escherichia coli (DAEC) isolates from children with diarrhoea
Sat pet sigA pic astA fimH DAEC 7 (50%) 0 (0) 2 (14.2%) 2 (14.2%) 3 (21.4%) 14 (100%)
TABLE 6.The prevalence of different virulence genes among
diffusely adherent Escherichia coli (DAEC) isolates from children with diarrhoea
Genetic profile No. (%) of DAEC
fimH 6 (42.8)
fimH, sat 2 (14.2)
fimH, sigA 1 (7.1)
fimH, sat, pic 1 (7.1) fimH, sat, astA 3 (21.5) fimH, sat, sigA, pic 1 (7.1)
TABLE 7. Antimicrobial susceptibility of diffusely adherent
Escherichia coli isolates from children with diarrhoea
Antimicrobial agent Resistant, n (%) Susceptible, n (%) Intermediate, n (%) Ampicillin 14 (100) 0 (0) 0 (0) Ampicillin-sulbactam 10 (71.4) 3 (21.5) 1 (7.1) Cefotaxime 11 (78.6) 2 (14.3) 1 (7.1) Ceftriaxone 7 (50) 4 (28.6) 3 (21.5) Ceftazidime 8 (57.1) 5 (35.7) 1 (7.1) Imipenem 0 (0) 13 (92.8) 1 (7.1) Gentamicin 2 (14.3) 12 (85.7) 0 (0) Nalidixic acid 7 (50) 4 (28.6) 3 (21.5) Trimethoprim-Sulfamethoxazole 11 (78.6) 2 (14.3) 1 (7.1) Moxifloxacin 5 (35.7) 6 (42.9) 3 (21.5) Chloramphenicol 5 (35.7) 7 (50) 2 (14.3)
and 21.5% of DAEC isolates, respectively (Table 4). Mansan-Almeida et al. detected afaE-1, afaE-2, afaE-3, afaE-5 and daaE in, respectively, 44%, 10%, 2%, 2% and 6% of DAEC isolated from children with diarrhoea [26]. Therefore, the most frequent gene among Afa/Dr DAEC strains was afaE-1 in our study.
AIDA-I is the E. coli adhesin involved in diffuse adherence [31]. In the current study, aida/aah was not detected in any DAEC isolates, which is in line with the Abbasi et al. study [24]. These results indicate that the prevalence of aida/aah was low in DAEC isolates.
The PCR results for the Afa/Dr genes showed significant relation with the HEp-2 cell adhesin assay, as 100% of Afa/Dr-positive isolates were confirmed as DAEC. This is the first report on the adhesin assay of DAEC in Iran.
The sat gene was detected in seven (50%) DAEC isolates in this study. Guignot et al. showed that sat might play a role in Afa/Dr DAEC intestinal pathogenesis by inducing lesions at the junctional barrier in Caco-2/TC7 monolayers [32]. Spano et al. found sat in 7.1% of DAEC isolated from children [33], Li et al. reported that 44.44% of DAEC was positive for sat [27] and Mansan-Almeida et al. found sat in 46% of DAEC isolated from children [26]. The rate of sat genes in our study was higher than those reported by Li et al. [27] and Mansan-Almeida et al. [26], and much higher than the percentage reported by Spano et al. [33]. Therefore, it seems that sat is common in the pathogen-esis of DAEC.
In the present study, pet was not identified in any DAEC isolates. Pet generates diarrhoeagenic effects by changes in the actin cytoskeleton [34]. Spano et al. reported that 14.2% of DAEC isolated from children was positive for pet [33] but other studies did not detect the pet gene in any DAEC isolates [14,27,35].
A total of 14.2% of the DAEC isolates harboured sigA and pic genes in this study. SigA is a cytotoxin that was found to have significant cytotoxic effects on HEp-2 cells [36]. Pic is the protein involved in intestinal colonization in children with diarrhoea, and unlike sigA it does not have a cytotoxic effect on HEp-2 cells [37]. Spano et al. identified pic in 2.3% of DAEC
isolated from children [33]. Boisen et al. detected pic in 10% of DAEC strains, whereas sigA was not detected in any DAEC [38]. Our results showed that the existence of pic and sigA in DAEC isolates was higher than in other studies.
The astA gene encodes the EAST-1 toxin with a positive rate of 21.4% in DAEC isolates in this study. EAST-1 toxin is thought to be a supplementary determinant in the pathogenesis of E. coli diarrhoea [39]. Spano et al. detected astA in 9.5% of DAEC isolated from children [33]. Lopes et al. reported that 15.2% of DAEC was positive for astA [35], and Patzi-Vargas et al. iden-tified astA in 3.1% of DAEC [14]. Thesefindings are contrary to
our findings, which showed the occurrence of astA in DAEC isolates to be much higher.
In this study, the type 1 fimbriae-encoding gene fimH was identified in all DAEC isolates. Li et al. found fimH in 100% of DAEC strains [27], and Lopes et al. detected thefimH gene in 48.2% of DAEC strains [35]. Johnson and Stell identified this adhesin in nearly all E. coli strains [40].
In the present study, we observed high percentages of resistance to ampicillin, cefotaxime and cotrimoxazole; only imipenem and gentamicin were effective against 92.8% and 85.7% of DAEC isolates, respectively. Moreover, resistance to more than one antibiotic was found in 100% of DAEC strains. Different studies have reported that most DAEC strains were resistant to several antibiotics, such as sulfonamide, doxycy-cline, tetracydoxycy-cline, ampicilin, cefotaxime and cotrimoxazole, whereas the isolates were sensitive to antibiotics such as imi-penem, amikacin, gentamicin and nitrofurantoin [25,27,28,33].
Conclusions
Ourfindings expand our knowledge of DAEC prevalence and characteristics in Iran, and explain the role of virulence genes in DAEC pathogenesis. Although the prevalence of DAEC was low, these strains exhibited high rates of antimicrobial resis-tance and high frequency for carrying virulence genes. Furthermore, preventing infections caused by this bacterium among children is essential and further researches are war-ranted, which should include other cities in Iran, using larger number of DAEC isolates.
Con
flict of interest
The authors declare that there is no conflict of interest in relation to this article.
Authors
’ contribution
All authors participated in the research design and contributed to different parts of the research.
Funding
This paper was part of the MSc thesis of K. Javadi supervised by N. Hadi and was supported by Shiraz University of Medical Sciences grant No. 97–18400.
6 New Microbes and New Infections, Volume 38 Number C,---2020
NMNI
© 2020 Published by Elsevier Ltd, NMNI, 38, 100780
Ethical approval
The study design was approved by the Ethics Committee of Shiraz University of Medical Sciences IR.SUMS.REC.1398.613 and followed the statements of the Declaration of Helsinki.
Acknowledgements
The authors wish to thank the staff of the microbiology labo-ratories of the hospitals for their kind collaboration.
References
[1] Bern C, Martines J, De Zoysa I, Glass R. The magnitude of the global problem of diarrhoeal disease: a ten-year update. Bull World Health Organ 1992;70:705.
[2] Mansour Amin MS, Javaherizadeh H, Motamedifar M, Saki M, Veisi H, Ebrahimi S, et al. Antibiotic resistance pattern and molecular charac-terization of extended-spectrum β-lactamase producing enter-oaggregative Escherichia coli isolates in children from southwest Iran. Infect Drug Resist 2018;11:1097.
[3] Scaletsky IC, Fabbricotti SH, Carvalho RL, Nunes CR, Maranhao HS, Morais MB, et al. Diffusely adherent Escherichia coli as a cause of acute diarrhea in young children in Northeast Brazil: a case-control study. J Clin Microbiol 2002;40:645–8.
[4] Scaletsky IC, Fabbricotti SH, Silva SO, Morais MB, Fagundes-Neto U. HEp-2–adherent Escherichia coli strains associated with acute infantile diarrhea, São Paulo, Brazil. Emerg Infect Dis 2002;8:855.
[5] Lozer DM, Souza TB, Monfardini MV, Vicentini F, Kitagawa SS, Scaletsky IC, et al. Genotypic and phenotypic analysis of diar-rheagenic Escherichia coli strains isolated from Brazilian children living in low socioeconomic level communities. BMC Infect Dis 2013;13:418.
[6] Lalioui L, Jouve M, Gounon P, Le Bouguenec C. Molecular cloning and characterization of the afa-7and afa-8 gene clusters encoding afimbrial adhesins inEscherichia coli strains associated with diarrhea or septi-cemia in calves. Infect Immun 1999;67:5048–59.
[7] Servin AL. Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): current insights and future challenges. Clin Microbiol Rev 2014;27:823–69.
[8] Guignot J, Peiffer I, Bernet-Camard M-F, Lublin DM, Carnoy C, Moseley SL, et al. Recruitment of CD55 and CD66e brush border-associated glycosylphosphatidylinositol-anchored proteins by mem-bers of the Afa/Dr diffusely adhering family of Escherichia coli that infect the human polarized intestinal Caco-2/TC7 cells. Infect Immun 2000;68:3554–63.
[9] Berger CN, Billker O, Meyer TF, Servin AL, Kansau I. Differential recognition of members of the carcinoembryonic antigen family by Afa/ Dr adhesins of diffusely adhering Escherichia coli (Afa/Dr DAEC). Mol Microbiol 2004;52:963–83.
[10] Nowicki B, Moulds J, Hull R, Hull S. A hemagglutinin of uropathogenic Escherichia coli recognizes the Dr blood group antigen. Infect Immun 1988;56:1057–60.
[11] Tapader R, Chatterjee S, Singh A, Dayma P, Haldar S, Pal A, et al. The high prevalence of serine protease autotransporters of
Enterobacteriaceae (SPATEs) in Escherichia coli causing neonatal septicemia. Eur J Clin Microbiol Infect Dis 2014;33:2015–24. [12] Pizarro-Cerdá J, Cossart P. Bacterial adhesin and entry into host cells.
Cell 2006;124:715–27.
[13] Sémiramoth N, Gleizes A, Turbica I, Sandré C, Gorges R, Kansau I, et al. Escherichia coli type 1 pili trigger late IL-8 production by neutrophil-like differentiated PLB-985 cells through a Src family kinase-and MAPK-dependent mechanism. J Leukoc Biol 2009;85:310–21. [14] Patzi-Vargas S, Zaidi MB, Perez-Martinez I, León–Cen M,
Michel-Ayala A, Chaussabel D, et al. Diarrheagenic Escherichia coli carrying supplementary virulence genes are an important cause of moderate to severe diarrhoeal disease in Mexico. PLoS Negl Trop Dis 2015;9: e0003510.
[15] Labigne-Roussel AF, Lark D, Schoolnik G, Falkow S. Cloning and expression of an afimbrial adhesin (AFA-I) responsible for P blood group-independent, mannose-resistant hemagglutination from a py-elonephritic Escherichia coli strain. Infect Immun 1984;46:251–9. [16] Le Bouguenec C, Archambaud M, Labigne A. Rapid and specific
detection of the pap, afa, and sfa adhesin-encoding operons in uro-pathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol 1992;30:1189–93.
[17] Le Bouguénec C, Lalioui L, du Merle L, Jouve M, Courcoux P, Bouzari S, et al. Characterization of AfaE adhesins produced by extraintestinal and intestinal human Escherichia coliIsolates: PCR as-says for detection of afa adhesins that do or do not recognize Dr blood group Antigens. J Clin Microbiol 2001;35:1738–45.
[18] Bilge S, Clausen C, Lau W, Moseley S. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol 1989;171: 4281–9.
[19] Jallat C, Livrelli V, Darfeuille-Michaud A, Rich C, Joly B. Escherichia coli strains involved in diarrhea in France: high prevalence and heteroge-neity of diffusely adhering strains. J Clin Microbiol 1993;31:2031–7. [20] Servin AL. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli.
Clin Microbiol Rev 2005;18:264–92.
[21] Nataro JP, Kaper JB. Diarrheagenic escherichia coli. Clin Microbiol Rev 1998;11:142–201.
[22] Scaletsky I, Silva M, Trabulsi LR. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun 1984;45: 534–6.
[23] Benz I, Schmidt MA. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect Immun 1989;57:1506–11.
[24] Abbasi P, Kargar M, Doosti A, Mardaneh J, Ghorbani-Dalini S, Dehyadegari MA. Molecular detection of diffusely adherent Escherichia coli strains associated with diarrhea in Shiraz, Iran. Arch Pediatr Infect Dis 2016;5.
[25] Levine MM, Ferreccio C, Prado V, Cayazzo M, Abrego P, Martinez J, et al. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am J Epidemiol 1993;138:849–69.
[26] Canizalez-Roman A, Flores-Villaseñor HM, Gonzalez-Nuñez E, Velaz-quez-Roman J, Vidal JE, Muro-Amador S, et al. Surveillance of diar-rheagenic Escherichia coli strains isolated from diarrhea cases from children, adults and elderly at Northwest of Mexico. Front Microbiol 2016;7:1924.
[27] Gómez-Duarte OG, Arzuza O, Urbina D, Bai J, Guerra J, Montes O, et al. Detection of Escherichia coli enteropathogens by multiplex po-lymerase chain reaction from children’s diarrheal stools in two Caribbean–Colombian cities. Foodborne Pathog Dis 2010;7:199–206. [28] Ochoa TJ, Ruiz J, Molina M, Del Valle LJ, Vargas M, Gil AI, et al. High frequency of antimicrobial drug resistance of diarrheagenic Escherichia coli in infants in Peru. Am J Trop Med Hyg 2009;81:296–301.
[29] Li D, Shen M, Xu Y, Liu C, Wang W, Wu J, et al. Virulence gene profiles and molecular genetic characteristics of diarrheagenic Escherichia coli from a hospital in western China. Gut Pathog 2018;10:35.
[30] Mansan-Almeida R, Pereira AL, Giugliano LG. Diffusely adherent Escherichia coli strains isolated from children and adults constitute two different populations. BMC Microbiol 2013;13:22.
[31] Charbonneau M-È, Berthiaume F, Mourez M. Proteolytic processing is not essential for multiple functions of the Escherichia coli auto-transporter adhesin involved in diffuse adherence (AIDA-I). J Bacteriol 2006;188:8504–12.
[32] Guignot J, Chaplais C, Coconnier-Polter MH, Servin AL. The secreted autotransporter toxin, Sat, functions as a virulence factor in Afa/Dr diffusely adhering Escherichia coli by promoting lesions in tight junc-tion of polarized epithelial cells. Cell Microbiol 2007;9:204–21. [33] Spano LC, da Cunha KF, Monfardini MV, Fonseca RdCB, Scaletsky ICA.
High prevalence of diarrheagenic Escherichia coli carrying toxin-encoding genes isolated from children and adults in southeastern Brazil. BMC Infect Dis 2017;17:773.
[34] Navarro-Garcia F, Sonnested M, Teter K. Host-toxin interactions involving EspC and Pet, two serine protease autotransporters of the Enterobacteriaceae. Toxins (Basel) 2010;2:1134–47.
[35] Lopes LM, Fabbricotti SH, Ferreira AJ, Kato MA, Michalski J, Scaletsky IC. Heterogeneity among strains of diffusely adherent Escherichia coli isolated in Brazil. J Clin Microbiol 2005;43:1968–72. [36] Ruiz-Perez F, Nataro JP. Bacterial serine proteases secreted by the
autotransporter pathway: classification, specificity, and role in viru-lence. Cell Mol Life Sci 2014;71:745–70.
[37] Navarro-Garcia F, Gutierrez-Jimenez J, Garcia-Tovar C, Castro LA, Salazar-Gonzalez H, Cordova V. Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect Immun 2010;78:4101–9. [38] Boisen N, Ruiz-Perez F, Scheutz F, Krogfelt KA, Nataro JP. High
prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg 2009;80:294–301.
[39] Yuste M, De La Fuente R, Ruiz-Santa-Quiteria J, Cid D, Orden J. Detection of the astA (EAST1) gene in attaching and effacing Escher-ichia coli from ruminants. J Vet Med B Infect Dis Vet Public Health 2006;53:75–7.
[40] Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000;181:261–72.
8 New Microbes and New Infections, Volume 38 Number C,---2020
NMNI
© 2020 Published by Elsevier Ltd, NMNI, 38, 100780