HAL Id: hal-02627204
https://hal.inrae.fr/hal-02627204
Submitted on 26 May 2020
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Distributed under a Creative Commons Attribution| 4.0 International License
Improved drought stress tolerance in Arabidopsis by
CRISPR/dCas9 fusion with a Histone AcetylTransferase
Joaquim Felipe Paixao Roca, François-Xavier Gillet, Thuanne Pires Ribeiro,
Caroline Bournaud, Isabela Tristan Lourenco-Tessutti, Daniel D. Noriega,
Bruno Paes de Melo, Janice de Almeida-Engler, Maria Fatima Grossi-De-Sa
To cite this version:
Joaquim Felipe Paixao Roca, François-Xavier Gillet, Thuanne Pires Ribeiro, Caroline Bournaud,
Isabela Tristan Lourenco-Tessutti, et al..
Improved drought stress tolerance in Arabidopsis by
CRISPR/dCas9 fusion with a Histone AcetylTransferase. Scientific Reports, Nature Publishing Group,
2019, 9, �10.1038/s41598-019-44571-y�. �hal-02627204�
www.nature.com/scientificreports
Improved drought stress tolerance
in Arabidopsis by CRIspR/
dCas9 fusion with a Histone
AcetylTransferase
Joaquin Felipe Roca paixão
1,2, François-Xavier Gillet
1, Thuanne Pires Ribeiro
1,
Caroline Bournaud
1, Isabela Tristan Lourenço-Tessutti
1, Daniel D. Noriega
1,
Bruno paes de Melo
1, Janice de Almeida-Engler
2& Maria Fatima Grossi-de-Sa
1,3Drought episodes decrease plant growth and productivity, which in turn cause high economic losses. Plants naturally sense and respond to water stress by activating specific signalling pathways leading to physiological and developmental adaptations. Genetically engineering genes that belong to these pathways might improve the drought tolerance of plants. The abscisic acid (ABA)-responsive element binding protein 1/ABRE binding factor (AREB1/ABF2) is a key positive regulator of the drought stress response. We investigated whether the CRISPR activation (CRISPRa) system that targets AREB1 might contribute to improve drought stress tolerance in Arabidopsis. Arabidopsis histone acetyltransferase 1 (AtHAT1) promotes gene expression activation by switching chromatin to a relaxed state. Stable transgenic plants expressing chimeric dCas9HAT were first generated. Then, we showed that the
CRISPRa dCas9HAT mechanism increased the promoter activity controlling the β-glucuronidase (GUS)
reporter gene. To activate the endogenous promoter of AREB1, the CRISPRa dCas9HAT system was set
up, and resultant plants showed a dwarf phenotype. Our qRT-PCR experiments indicated that both
AREB1 and RD29A, a gene positively regulated by AREB1, exhibited higher gene expression than the
control plants. The plants generated here showed higher chlorophyll content and faster stomatal aperture under water deficit, in addition to a better survival rate after drought stress. Altogether, we report that CRISPRa dCas9HAT is a valuable biotechnological tool to improve drought stress tolerance
through the positive regulation of AREB1.
Improving agronomic traits that give plants resistance to biotic and abiotic stresses to increase their economic value is a recurrent concern worldwide. The awareness of climate change and global warming emphasizes the need to implement some efficient and sustainable solutions. In several countries, a current important issue is the maintenance of crop production during drought1–3. Drought varies spatially and temporally as well as in strength. Accordingly, plants have diversified their responses and have evolved to exhibit multiple morphological and phys-iological behaviours4,5. These behaviors consist of different degrees of drought escape, avoidance and tolerance. Exploiting genetic traits that enhance the drought stress response while maintaining high yields remains of criti-cal interest for crop management. Conventional breeding and transgenic approaches have been shown to improve drought stress tolerance in plants such as maize, soybean, rice and wheat6,7.
Comprehensive molecular analyses have deciphered the cellular pathways that orchestrate the drought stress response8,9. Abscisic acid (ABA) is a phytohormone that plays a role as a key regulator of the drought stress response in plants by regulating gene expression and controlling stomatal closure to prevent water losses by transpiration10. The basic leucin zipper (bZIP) transcription factors, termed ABA-responsive element binding proteins/ABRE binding factors (AREB/ABFs), are important determinants in ABA signalling8. Over-expression 1Embrapa Genetic Resources and Biotechnology, Brasília, DF, Brazil. 2INRA, Université Côte d’Azur, CNRS, ISA, Sophia Antipolis, France. 3Catholic University of Brasilia - Post-Graduation Program in Genomic Sciences and Biotechnology, Brasília, DF, Brazil. Joaquin Felipe Roca Paixão and François-Xavier Gillet contributed equally. Correspondence and requests for materials should be addressed to J.F.R.P. (email: joaquinfrp@gmail.com) or M.F.G.-d.-S. (email: fatima.grossi@gmail.com)
Received: 11 September 2018 Accepted: 9 May 2019 Published: xx xx xxxx
www.nature.com/scientificreports
www.nature.com/scientificreports/
of AREB1 (also named ABF2) exhibited enhanced drought stress tolerance in Arabidopsis, rice and soybean, while AREB1 loss of function causes drought stress sensitivity11–15. Indeed, AREB1 regulates a large set of genes downstream of the ABA signalling pathway in response to drought stress13 and participates in osmotic stress protection, ABA biosynthesis and antioxidant signalling12,16. Thus, AREB1 represents an attractive candidate gene for improving the drought stress response.
Targeted plant genome editing using CRISPR nucleases has become a promising approach to create new plant varieties17–19. Beyond genome editing, the CRISPR mechanism has been remodeled to accomplish CRISPR acti-vation (CRISPRa) and CRISPR interference (CRISPRi)20. The catalytically inactive form of Cas9 (dead Cas9, abbreviated dCas9) has been fused with transcription activators and inhibitors to modify transcription through specific gene promoters and with chromatin modulator domains to facilitate targeted epigenome editing21–23. Histone acetyltransferase (HAT) catalyzes the acetylation of core histones through the addition of an acetyl group to the lysine residue on the terminal tail of the histones24. Histone acetylation triggers DNA relaxation and leads to exposure of DNA to the transcriptional machinery25. Thus, HAT activity is correlated with gene expression activation. In this way, the use of dCas9 in fusion with HAT (dCas9HAT), combined with the directed targeting of sgRNAs, appears promising for positively regulating the activity of a targeted promoter26,27. CRISPR/dCas9 epigenome editing of specific drought stress response genes therefore emerges as an encouraging strategy for improving stress tolerance in plants.
Herein, we asked whether an engineered dCas9HAT could efficiently enhance AREB1 gene expression in
Arabidopsis thaliana in response to drought stress. We first generated stable Arabidopsis transgenic lines
express-ing dCas9 fused with the core catalytic domain of an Arabidopsis HAT. Next, we validated our CRISPR system using sgRNA targeting a β-glucuronidase (GUS) reporter system. Finally, we transformed dCas9HAT lines with a construct containing sgRNAs targeting an AtAREB1 promoter region and observed, by molecular and physiolog-ical approaches, an enhanced response to drought stress in these transgenic plants.
Results and Discussion
Generation of Arabidopsis transgenic lines expressing dCas9
HAT.
We first designed the dCas9HAT construct to assay the transcriptional regulation of the gene of interest. The catalytic core from the Arabidopsis Histone AcetylTransferase 1 gene (AtHAC1, AT1G79000), was fused to the N-terminal part of dCas9 and cloned into a modified version of the plant binary vector pGreenII (Fig. 1A, Table S1). The T-DNA was designed to perform two rounds of plant transgenic selection based on antibiotic (kanamycin) resistance and the level of a fluorescence reporter gene (a nuclear mOrange fluorescent protein, mOFP). Once positive transformants were selected, fluorescence intensity was assessed by confocal microscopy (Fig. 1B). The fluorescence correspondingFigure 1. Molecular characterization of transgenic A. thaliana dCas9HAT lines. (A) Schematic representation of the construct allowing the selection of A. thaliana dCas9HAT lines. KanR: the kanamycin resistance gene; mOFP: monomeric orange fluorescent protein; NLS: nuclear localization signal. The white arrows indicate the cauliflower mosaic virus (CaMV) 35 S promoter; the white squares indicate the CaMV poly(A) signal (terminator); the black arrows indicate qPCR oligos (B) Fluorescence microscopy imaging of A. thaliana leaves, primordia and roots expressing the mOFP in the nucleus in three dCas9HAT lines compared with Col-0 plants. All confocal images were acquired under identical parameters (excitation: 549 nm/emission: 565 nm). Scale bars in the left inferior corner = 50 μm (C) Assessment of dCas9HAT expression. RT-qPCR was performed in Col-0 plants and in three lines of dCas9HAT-transformed plants. Transcript levels were normalized against the geometric mean of the transcript levels of the housekeeping genes (GAPDH and Actin2). The mean and standard deviation (SD) were calculated from three independent biological replicates. The calibrator was chosen as the sample with the lowest expression of the transgene (excluding Col-0 plants). Asterisks indicate significant differences between Col-0 plants and the different lines (Wilcoxon test, *P < 0.05).
www.nature.com/scientificreports
www.nature.com/scientificreports/
to mOFP could be seen all over the roots and leaves and was present at the highest intensity in the nuclei. Some transgenic lines displayed stronger fluorescence, suggesting that the cassette was inserted in an actively expressed region of the genome. Among the lines, the occurrence of the dCas9 construct was checked by PCR (Fig. S1), and gene expression was analyzed by qPCR (Fig. 1C). The transgenic line dCas9HAT number 2 had the highest mOFP and dCas9 expression and was retained for this study (hereafter named dCas9HAT).
Challenge of the dCas9 constructs in a GUS reporter system.
To evaluate the dCas9HAT construct, we set up a surrogate reporter system based on regulation of GUS reporter gene expression. We used the 170-bp minimal truncated version of the Glycine max ubiquitin promoter, herein designated GmUcesMin28,29. We selected two sgRNAs near the transcription start site (TSS) of GmUcesMin (Fig. 2A, Table S2). The efficiency of dCas9HAT in activating the GUS reporter system was quantified by its enzymatic activity. Seedlings of stably transformed Arabidopsis lines expressing dCas9HAT were incubated with Agrobacterium carrying Ti plasmids to perform transient ectopic expression of GmUcesMin-GUS in combination with the expression of one or two sgRNAs. Significantly elevated enzymatic activity was observed for sgRNA1 (~2.4-fold increase) and sgRNA2 (~2-fold increase), while enzymatic activity was elevated ~1.4-fold for the combination of the two sgRNAs. This result indicates that the expression of dCas9HAT enhanced the expression of the GUS gene in trans when targeted to GmUcesMin promoter. Remarkably, some substantial differences were noted depending on the location of the sgRNA and/or the sgRNA combination. Previous studies have suggested that the distance of the sgRNA from the TSS might influence the transcriptional regulation of the gene of interest. While some studies have reported that a specific sgRNA binding distance from the TSS (−50 bp to +300 bp) corresponds to higher target gene expression30,31, others have pointed out that dCas9 might generate steric hindrance and thus interfere with tran-scriptional machinery activities21,32. Regarding the construct GmUcesMin, our two sgRNAs are separated only by 118 bp. Considering the 3D conformation of the DNA, the 30 bp length occupied by the dCas9 on DNA33 and the space taken by the HAT domain, a steric hindrance effect could be responsible for destabilizing locally the protein complexes standing on DNA. Compared to the use of only one sgRNA, this collateral effect might result to lower gene expression rather than improve it.We determined the expression profile of GmUcesMin-GUS by GUS staining (Fig. 2C). Since dCas9HAT and the sgRNA are supposed to be broadly co-expressed in the whole Arabidopsis plant, we expected to observe GUS activity in every tissue. However, GUS activity was restricted to the roots and was higher in transgenic plants than in controls. We hypothesized that some root-specific transcription activators might trigger GUS expression
Figure 2. Challenge of dCas9HAT in a GUS reporter system. (A) Schematic representation of the GmUcesMin promoter with the sgRNA positions (black and white squares). The TATA box is represented by a black square. The black and white curved arrows represent the TSS and ATG codon, respectively. (B) Arabidopsis seedlings from stably transformed dCas9HAT fusions were transiently transformed with GmUcesMin and different combinations of sgRNAs. The results are presented as the mean and SD of 6 independent experiments (n = 20 pooled seedlings per experiment). Asterisks indicate significant difference between the GUS activity for each dCas9 fusion guided by one sgRNA or a combination of two sgRNAs compared to mock controls (Student’s t-test, *P < 0.05). Bars indicate the standard error. (C) GUS staining was performed for the GmUcesMin promoter in the same transiently transformed seedlings.
www.nature.com/scientificreports
www.nature.com/scientificreports/
specifically in roots. However, the mapping of different regulatory boxes in GmUcesMin identified several reg-ulatory elements but did not discriminate whether they are specific to the root (Fig. S2). The transgenic plants differed from the controls only in intensity. These results suggest that dCas9HAT in combination with sgRNA enhances gene expression in the roots but does not activate gene expression in other tissues. Notably, Zhang et al.29 previously reported that the full-length GmUcesMin (called GmScreamM2, length: 1391 bp) triggers gene expression principally in soybean seeds. It is tempting to postulate that the truncated form of GmUcesMin and/or our expression conditions might favor the expression of the target gene in roots.
Molecular and phenotypic characterization of dCas9
HAT-sgA.
We next inquired whether AREB1 gene expression could be regulated by dCas9HAT. We designed two sgRNAs to target the endogenous promoter of AtAREB1 (Fig. 3A). One sgRNA is located at 3′ from the TSS (−479 bp), and the second is in the 5′ UTR (+356 bp). The two sgRNAs (sg-pAREB1.1 and sg-pAREB1.2) were cloned in tandem within a single T-DNA and transformed into the Arabidopsis dCas9HAT transgenic lines to generate dCas9HAT-sgA. We verified AREB1 gene expression in three transgenic lines by real-time qPCR (Fig. 3B). In each experiment, the control line was the parental line, dCas9HAT. We observed a slight but significant 1.7-fold increase in AREB1 expression in the dCas9HAT-sgA1 line and a 2-fold increase in the dCas9HAT-sgA2 line compared to that in the control line, sug-gesting that targeting dCas9HAT to the AREB1 gene could trigger its transcription. Three weeks after germination, the rosette diameter in the dCas9HAT-sgA2 and dCas9HAT-sgA1 plants was ~3-fold smaller than in the controls (Figs 3C, S3A,B). The leaf length was also smaller for both lines, respectively, than in the control line (Fig. S3C). These results suggest that the mutant caused a dwarf phenotype under normal plant growth conditions. They also corroborate phenotypic traits related to drought stress shown by Fujita et al.12, in which the over-expression of the constitutive form of AREB1, AREB1ΔQT, presented smaller phenotypes and the areb1 mutants had larger rosettes. Interestingly, we observed that without water deficit, AREB1 is slightly positively regulated, indicating that dCas9HAT activates AREB1 apart of the context of drought.The activation of promoters via histone H3K27 acetylation through the use of dCas9p300Core domains has been evaluated in animal cells, and the dCas9p300Core domains have been shown to be more potent than activation domains21,34,35. Further studies have also reported that the over-expression of histone deacetylases (HDACs) plays a role in the ABA response and that the use of an HDAC inhibitor induces hyperacetylation and increases AREB1
Figure 3. Challenge of dCas9HAT constructs in the transcriptional regulation of AREB1 by targeting pAREB1. (A) Schematic representation of pAREB1 with the two sgRNAs designated. The TATA box is represented by a black square. (B) Relative expression of the AREB1 and (C) Phenotypic analysis of dCas9HAT-sgA. Rosette diameter of three-week-old plants. The results represent the mean of n = 13. dCas9HAT control plants are represented by a – symbol, and dCas9HAT-sgA2 plants are represented by a + symbol. Asterisks indicate significant difference between dCas9-sgA lines and dCas9 control lines (Wilcoxon test, *P < 0.05). Bars indicate standard error.
www.nature.com/scientificreports
www.nature.com/scientificreports/
gene expression in peanut36,37. Altogether, our findings suggest that histone acetylation by dCas9HAT at AREB1 loci is a determinant parameter in AREB1 expression and allows local chromatin rearrangement.
dCas9
HAT-sgA plants have enhanced drought stress tolerance.
We next analyzed the efficiency of the dCas9HAT activator at a molecular level by measuring the gene expression of AREB1 and RD29A, a gene posi-tively regulated by AREB1 and a reporter gene for AREB1 activity under drought stress12,38. The expression levels of AREB1 and RD29A were significantly increased by ~2-fold and ~3-fold respectively in dCas9HAT-sgA2 plants (Fig. 4A,B), and ~2-fold and ~2.6-fold in dCas9HAT-sgA1 plants (Fig. S4). These data suggest that the drought stress response is enhanced in the dCas9HAT-sgA lines. We next investigated the physiological traits of transgenic Arabidopsis seedlings during drought. Only the dCas9HAT-sgA2 was selected for further physiological analyses. We performed this investigation under two drought conditions. Severe drought stress (SDS) was produced by removing plants from the soil and keeping them at 20% humidity on plates on the laboratory bench. In anotherFigure 4. Molecular and physiological analyses of drought stress responses in dCas9HAT-sgA. Transcript levels of (A) AREB1 and (B) RD29A in dCas9HAT and dCas9HAT-sgA2 plants during drought stress. Expression levels were normalized against the geometric mean of the expression of the housekeeping genes (GAPDH and Actin2). The mean and SD were obtained from three biological replicates. Asterisks indicate significant differences between the control and transformed plants (Wilcoxon test, *P < 0.05). For each gene, the expression level in the dCas9HAT control was defined as the calibrator (1.0). (C) Total chlorophyll content in non-stressed plants 4 h after SDS and after MSDS. The results represent the mean of n = 6. (D) Stomatal aperture measurements after 2 h and 4 h of severe stress and 20 days of drought stress. The results are presented as the mean of n = 30. Asterisks indicate significant difference between co-transformed plants and control lines (Student’s t-test, *P < 0.05). The bars indicate standard error. (E) Survival rates after 6 h of SDS and 48 h and 96 h of rehydration. The results represent the percentage of surviving plants (n = 20). An asterisk indicates a significant difference between the control dCas9HAT and dCas9HAT-sgA2 plants (chi-square test, *P < 0.05). dCas9HAT control plants are represented by a – symbol, and dCas9HATsgA2 plants are represented by a + symbol.
www.nature.com/scientificreports
www.nature.com/scientificreports/
experiment, mild-severe drought stress (MSDS) was produced through water withdrawal for up to 20 days. The photosynthesis is affected by drought stress, and a decrease of chlorophyll content is an indicator of photosyn-thesis activity39,40. We solvent-extracted the chlorophyll from leaves and subsequently measured the chlorophyll fluorescence by UV-visible spectroscopy41. When plants were normally irrigated, the content of chlorophyll was comparable between the dCas9HAT-sgA2 line and the control line (Fig. 4C). In contrast, 4 h after SDS, the content of total chlorophyll was 1.7-fold higher in dCas9HAT-sgA2 plants. After twenty days of MSDS, dCas9HAT-sgA2 plants had significantly higher content of chlorophyll (1.3-fold) than the control plants. Our findings likely indi-cate that the higher chlorophyll content found in the dCas9HAT-sgA2 line is in agreement with an improved drought stress response at a physiological level.
Another characteristic of the drought stress response is a decrease in stomatal aperture to limit water loss42. We measured stomatal width and length by confocal imaging and calculated the stomatal aperture (width/length) in non-stressed versus stressed plants after SDS (1 h, 2 h) and MSDS. In dCas9HAT-sgA2 plants without drought stress, the stomatal aperture was comparable to that of the control plants. However, 1 h after drought stress, the stomatal aperture was 1.6-fold lower in dCas9HAT-sgA2 plants than in control plants (Fig. 4D). After 2 h, the stomatal aperture was still significantly lower in dCas9HAT-sgA2 plants, even though control plants also partially closed their stomata between 1 h and 2 h of stress. However, our data revealed that stomatal closure was triggered more rapidly in dCas9HAT-sgA plants. After 20 days of stress, the stomatal aperture of the dCas9HAT-sgA plants was comparable to that of the control plants, suggesting that dCas9HAT-sgA2 does not regulate stomatal aperture differently during long-term drought stress. Our results indicate that the expression of dCas9HAT-sgA2 leads to faster stomatal closure after severe drought stress and corroborate a previous study stating that AREB1 might be partially associated with stomatal closure13.
Finally, we performed a survival assay to analyze the ability of transformed plants to recover after SDS and MSDS (Figs 4E; S5). After SDS followed by 48 h of rehydration, we observed that 85% of the dCas9HAT-sgA2 plants showed significant total recovery, whereas only 50% of the control plants were still alive. However, after 96 h of rehydration, pictures showed that transgenic plants present a recovered vigor compared to control plants (Fig. 4E). When we subjected the plants to MSDS and then rehydration for 48 h, we observed that all the trans-genic plants survived, whereas all the controls died (Fig. S5). Altogether, our data indicate that dCas9HAT directed to AREB1 loci improves the Arabidopsis drought response.
Conclusion
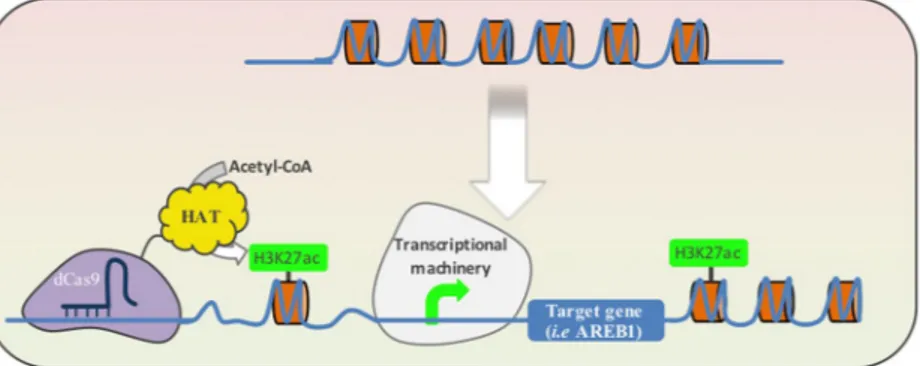
In addition to the use of modulators to aid the recruitment of RNA polymerase (Pol II) transcription machinery, the use of domains that modify chromatin folding is another interesting way to fine-tune gene expression. The expression of dCas9HAT allows acetylation of lysine 27 of Histone 3 (H3K27ac) favoring the unwind of chromatin and enhancing the interaction with transcriptional enhancers as the assembly of the transcriptional machin-ery (Fig. 5). This approach has been demonstrated in animals. In the present study, our main finding was that dCas9HAT positively regulates AREB1 and produces an enhanced drought stress response. It is noteworthy that dCas9HAT activity depends on the cellular context. The enhancer effect of dCas9HAT is stronger when the drought stress response is activated. This finding suggests that the chromatin folding at the AREB1 locus constitutes a reg-ulatory mechanism for AREB1 gene expression. We also report that GUS expression varies based on number of sgRNAs used and their respective positions. Having a better understanding of the chromatin context of a specific locus will help in the rational design of CRISPRi/a strategies.
experimental procedures
Plasmid construction and sgRNA design.
The sequence encoding the A. thaliana acetyltransferase domain was gathered from TAIR from the protein HAC1 (AT1G79000, between amino acids (AA) 1119 and 1408), synthesized (EPOCH) and cloned into a vector containing the sequence encoding dCas9 (pAWG-dCas9-VPR, which was a gift from George Church; plasmid #63802; http://n2t.net/addgene:63802; RRID:Figure 5. Schematic illustration of a model for dCas9HAT function in transcriptional activation of a target gene. Upper: the histone compaction (in orange) induces DNA condensation and limits transcription. Below: the dCas9HAT in complex with a single guide RNA (in dark blue) binds DNA on a target locus. The histone acetyl-transferase (HAT) triggers histone acetylation on the lysine 27 (H3K27ac) and induces local DNA relaxation. The DNA relaxation strengthens the interaction of the transcriptional machinery and /or transcriptional enhancers with the target locus.
www.nature.com/scientificreports
www.nature.com/scientificreports/
Addgene_63802)43. The cassette containing dCAS9HAT was cloned into a pGreen KII (pGKII_dCas9-VPR) vector containing a NeoR/KanR selection region and a sequence coding for the orange fluorescent protein (OFP). The sgRNAs were designed and screened using CHOPCHOP (http://chopchop.cbu.uib.no). The in-silico analysis of cis-acting elementos in UcesMin promoter was also considered for the design of sgRNAs 1, 2 and 3 (Fig. S2). The promoter sequence of AREB1 (AT1G45249.3) was extracted from the SeqViewer tool on the TAIR website, and sgRNAs targeting the AREB1 promoter (sg-pAREB1.1 and sg-pAREB1.2) were designed at −479 bp and +356 bp from the predicted TSS (approximately 25 bp downstream from the TATA box). A second vector was designed containing a sgRNA scaffold and two cassettes for sgRNA expression, with sg-pAREB1.1 and sg-pAREB1.2 con-trolled by the U6 promoter. Box sequences called Gibson boxes44 were designed at the edges and the middle of each sgRNA template to facilitate amplification and cloning. sgRNAs targeting the GmUcesMin promoter were chosen to have two different locations around the TSS. The sgRNA templates were amplified in a three-step PCR with one pair of primers in the Gibson boxes (extremities) and a second pair spanning the new 20-nt sgRNA and 20 nt in the template (Table S3). Each sgRNA template was sub-cloned into SmaI-linearized pGKII0229.
Plant material, growth conditions and stable transformation.
Col-0 A. thaliana seeds were surface-sterilized and germinated in vitro in Murashige and Skoog (MS) medium including vitamins (with or without selection agents). After stratification (2 days at 4 °C), the seeds were germinated and grown under a 12-h photoperiod in a growth chamber at 21 °C. Alternatively, plants were sown on a soil/sand mixture, strat-ified and grown under a 16 h photoperiod in a growth chamber at 21 °C. The constructs were transferred intoAgrobacterium tumefaciens GV3101 via heat-shock (30 minutes at 0 °C, 5 minutes at 37 °C and 2 h at 28 °C in
Luria-Bertani (LB) medium)45. The floral dip method was used to transform the A. thaliana plants46.
transient expression in A. thaliana seedlings.
Transient expression in Arabidopsis seedlings was adapted from the FAST and AGROBEST protocols47,48. Briefly, 20 seedlings were stratified and germi-nated in ½ MS in 96-well plates. After 4 days in a growth chamber (12 h photoperiod, 21 °C), seedlings were co-incubated with the A. tumefaciens strain GV3101 (OD600 = 0.2) carrying constructs to allow the expression of GmUcesMin-GUS, sg-UcesMin.1 and/or sg-UcesMin.2. The co-culture medium consisted of a combination of 50% ½ MS, 0.25% sucrose and 50% ABMES salt medium (17.2 mM K2HPO4, 8.3 mM NaH2PO4, 18.7 mM NH4Cl, 2 mM KCl, 1.25 mM MgSO4, 100 µM CaCl2, 10 µM FeSO4, 50 mM MES, 2% glucose, and 200 μM acetosyrin-gone). To control the agrobacterium population, we used an identical ratio o A. tumefasciens with each construct corresponding to 1/3 of the total bacterial population. If only one sgRNA was used, the rest 1/3 was composed of A. tumefasciens expressing an empty vector. After two days of co-cultivation, a fraction of the seedlings (5 of 20) were washed in distilled water for GUS staining. The remaining seedlings were put on MS with carbenicillin (100 µg/mL) for 1 day and then collected for GUS activity assays.Promoter activity assays.
β-Glucuronidase (GUS) histochemical staining was performed by adding X-Gluc solution (2 mM X-Gluc ((5-bromo-4-chloro-3-indolyl) β-D-glucuronic acid) in 50 mM Na2HPO4 (pH 7.0) and 0.1% Triton X-100) to A. thaliana seedlings for 10 h at 37 °C. Seedlings were washed in 70% ethanol overnight and observed using bright field optics with a digital camera (Olympus MVX10). Promoter activity was monitored by a fluorometric GUS enzymatic assay49. The total soluble protein was extracted with a GUS extraction buffer (50 mM sodium phosphate buffer, 10 mM EDTA, 0.1% sarkosyl, 0.1% Triton X-100 and 10 mM β-mercaptoethanol) and quantified using the Bradford method (Bradford, 1976). The assay was carried out by adding 20 µg oftotal soluble protein to 1 mM 4-MUG (4-methyl-umbelliferyl-glucuronide) fluorogenic substrate (excitation: 356 nm; emis-sion: 455 nm) and sampling 200 µLinto a stop buffer (0.2 M sodium carbonate) at three different time points (0, 15 and 30 minutes). A standard dilution curve was created with different concentrations of 4-MU (4-methyl umbel-liferone). The fluorescence was measured using a SpectraMax M3 fluorometer (Molecular Devices).In silico analysis of the GmUcesMin promoter.
The presence of cis-acting elements in the GmUcesMin promoter was examined using the bioinformatics tool MatInspector version 8.0 (Genomatix®
). The term “plants” was used as the matrix group, “0.85” was used as the value for the similarity of the main bases that constitute each cis-acting element (core similarity), and “Optimized +1” was used as the value for the matrix similarity50.Gene expression analysis by qPCR.
RNA extraction from Arabidopsis thaliana leaves was performed with a Concert™
Plant RNA Reagent kit (Invitrogen) following the manufacturer’s protocol. For cDNA synthesis, the following reagents were added to a 0.2-mL tube: 1 µg of total RNA, 1 µL of 10 µM NV-dT30 (2 µM), 1 µL of 10 mM dNTP (2 mM), and Milli-Q water to a total volume of 12 µL. The reaction was then incubated at 65 °C for 5 minutes. Then, 4 µL of 5 × First-Strand Buffer with 0.1 M DTT and 1 µL of RNase Out were added, and the tion was incubated at a 37 °C for 2 minutes. Finally, 1 µL of the MMLV enzyme was added to the reaction; the reac-tion was incubated for 50 minutes at 37 °C and then inactivated for 15 minutes at 70 °C. The cDNA obtained in the previous step was diluted 1:20 and analyzed in biological and experimental triplicates for the differential expres-sion of the AREB1, RD29A and dCAS9 genes by real-time PCR. The qPCR reaction was performed as follows: 2 µL of cDNA (1:20), 5 µL of 2 × GoTaq qPCR Master Mix SYBR Green, 0.5 µL of each primer and 2 µL of water to a final volume of 10 µL. Amplification was performed in a CFX96 machine (BioRad). GAPDH (AT1G16300) and Actin2 (AT3G18780) were used as reference genes for relative quantification with the 2^(ΔΔCt) method51,52.Drought stress and morphological and physiological analyses.
Transgenic A. thaliana seeds were sown in pots containing similar proportions of soil and sand and were regularly watered. After one week, plants were transferred to pots containing 3 transgenic plants and 3 control plants. For mild to severe drought stress (MSDS), after approximately 3–4 weeks, the soil was water saturated, and the plants were then kept without water for 25 days. Subsequently, plants were rehydrated for survival rate measurements. Rosette radius measurementswww.nature.com/scientificreports
www.nature.com/scientificreports/
were made and leaf morphology was examined in three-week-old plants53 with the aid of the LeafJ plugin for ImageJ54. Severe drought stress (SDS) conditions were created by removing whole plants from pots or by cutting the rosettes and placing them on the bench for 1 h to 6 h. Whole-rosette dehydration during SDS treatment was determined by cutting rosettes from three-week-old plants and weighing them hourly. To determine survival rates, whole seedlings were removed from the soil without harming the roots, and SDS was applied for 6 h on the bench. Both transformed and control plants that presented root damage were eliminated. The plants that had survived after rehydration for 48 h were recorded. Stomatal aperture was assessed from stressed leaves collected after 2 h, 4 h and 20 days. Confocal images of stomata were acquired under identical settings and processed with a Zeiss LSM 880 confocal fluorescence microscope using the software package LSM 510 version 3.2. The width and length of the stomatal apertures were captured by confocal microscopy, and the width-to-length ratios were calculated. After 24 h of treatment, chlorophyll was extracted from leaves with 80% acetone, and the chlorophyll content was estimated. After centrifugation at 12.000 × g for 5 minutes, absorbance was measured with a spectro-photometer at wavelengths of 645 and 663 nm. The chlorophyll concentration was estimated following Arnon’s equations: (Chlorophyll a (μg/mL) = 12.7 (A663) − 2.69 (A645); Chlorophyll b (μg/mL) = 22.9 (A645) − 4.68 (A663); Total chlorophyll (μg/mL) = 20.2 (A645) + 8.02 (A663))55.
Statistical analysis.
At least three independent replicates were conducted for each determination. For gene expression, Wilcoxon test was used to compare Ct values. For Gus activity of Physiological tests, Student’s t-test was used to compare the means. A difference was statistically significant when P < 0.05. Error bars in figures represent standard deviation (SD) of the means.Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
1. Lobell, D. B. & Gourdji, S. M. The Influence of Climate Change on Global Crop Productivity. Plant Physiology 160, 1686–1697 (2012). 2. Zhang, X. & Cai, X. Climate change impacts on global agricultural water deficit: more water needed for agriculture? Geophysical
Research Letters 40, 1111–1117 (2013).
3. Alizadeh, V., Shokri, V., Soltani, A. & Yousefi, M. A. Effects of Climate Change and Drought-Stress on Plant Physiology (2014). 4. Fahad, S. et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Frontiers in Plant
Science 8 (2017).
5. Tiwari, S., Lata, C., Chauhan, P. S., Prasad, V. & Prasad, M. A Functional Genomic Perspective on Drought Signalling and its Crosstalk with Phytohormone-mediated Signalling Pathways in Plants. Current Genomics 18 (2017).
6. Ashraf, M. Inducing drought tolerance in plants: Recent advances. Biotechnology Advances 28, 169–183 (2010).
7. Wani, S. H., Dutta, T., Neelapu, N. R. R. & Surekha, C. Transgenic approaches to enhance salt and drought tolerance in plants. Plant
Gene 11, 219–231 (2017).
8. Nakashima, K., Yamaguchi-Shinozaki, K. & Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Frontiers in Plant Science 5 (2014).
9. Singh, D. & Laxmi, A. Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Frontiers in
Plant Science 6 (2015).
10. Osakabe, Y., Osakabe, K., Shinozaki, K. & Tran, L.-S. P. Response of plants to water stress. Frontiers in Plant Science 5 (2014). 11. Oh, S.-J. Arabidopsis CBF3/DREB1A and ABF3 in Transgenic Rice Increased Tolerance to Abiotic Stress without Stunting Growth.
Plant Physiology 138, 341–351 (2005).
12. Fujita, Y. AREB1 Is a Transcription Activator of Novel ABRE-Dependent ABA Signaling That Enhances Drought Stress Tolerance in Arabidopsis. The Plant Cell Online 17, 3470–3488 (2005).
13. Yoshida, T. et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant Journal 61, 672–685 (2010).
14. Barbosa, E. G. G. et al. Overexpression of the ABA-Dependent AREB1 Transcription Factor from Arabidopsis thaliana Improves Soybean Tolerance to Water Deficit. Plant Molecular Biology Reporter 31, 719–730 (2013).
15. Todaka, D., Shinozaki, K. & Yamaguchi-Shinozaki, K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Frontiers in Plant Science 6 (2015).
16. Li, X.-Y. et al. Overexpression of Arachis hypogaea AREB1 Gene Enhances Drought Tolerance by Modulating ROS Scavenging and Maintaining Endogenous ABA Content. International Journal of Molecular Sciences 14, 12827–12842 (2013).
17. Kumar, V. & Jain, M. The CRISPR–Cas system for plant genome editing: advances and opportunities. Journal of Experimental Botany
66, 47–57 (2015).
18. Abdallah, N. A., Prakash, C. S. & McHughen, A. G. Genome editing for crop improvement: Challenges and opportunities. GM Crops
& Food 6, 183–205 (2015).
19. Malzahn, A., Lowder, L. & Qi, Y. Plant genome editing with TALEN and CRISPR. Cell & Bioscience 7 (2017).
20. Brocken, D. J. W., Tark-Dame, M. & Dame, R. T. dCas9: A Versatile Tool for Epigenome Editing. Current Issues in Molecular Biology 15–32, https://doi.org/10.21775/cimb.026.015 (2018).
21. Hilton, I. B. et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers.
Nature Biotechnology 33, 510–517 (2015).
22. Liu, X. S. et al. Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–247.e17 (2016).
23. Dominguez, A. A., Lim, W. A. & Qi, L. S. Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nature Reviews Molecular Cell Biology 17, 5–15 (2016).
24. Schneider, A. et al. Acetyltransferases (HATs) as Targets for Neurological Therapeutics. Neurotherapeutics 10, 568–588 (2013). 25. Eberharter, A. Histone acetylation: a switch between repressive and permissive chromatin: Second in review series on chromatin
dynamics. EMBO Reports 3, 224–229 (2002).
26. Bordoli, L. Plant orthologs of p300/CBP: conservation of a core domain in metazoan p300/CBP acetyltransferase-related proteins.
Nucleic Acids Research 29, 589–597 (2001).
27. Deng, W. et al. Involvement of the Histone Acetyltransferase AtHAC1 in the Regulation of Flowering Time via Repression of FLOWERING LOCUS C in Arabidopsis. Plant Physiology 143, 1660–1668 (2007).
28. Grossi de Sa, M. F. et al. Compositions and methods for modifying gene expression using the promoter of Ubiquitin Conjugating Protein Coding Gene of soybean plants. Patent: US9012720B2 (2015).
29. Zhang, N., McHale, L. K. & Finer, J. J. Isolation and characterization of “GmScream” promoters that regulate highly expressing soybean (Glycine max Merr.) genes. Plant Science 241, 189–198 (2015).
www.nature.com/scientificreports
www.nature.com/scientificreports/
30. Gilbert, L. A. et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661 (2014). 31. Radzisheuskaya, A., Shlyueva, D., Müller, I. & Helin, K. Optimizing sgRNA position markedly improves the efficiency of CRISPR/
dCas9-mediated transcriptional repression. Nucleic Acids Research 44, e141–e141 (2016).
32. Larson, M. H. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nature Protocols 8, 2180–2196 (2013).
33. Jinek, M. et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 343, 1247997–1247997 (2014).
34. de Groote, M. L., Verschure, P. J. & Rots, M. G. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Research 40, 10596–10613 (2012).
35. Cheng, A. W. et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell
Research 23, 1163–1171 (2013).
36. Sridha, S. & Wu, K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. The Plant Journal 46, 124–133 (2006).
37. Su, L.-C. et al. Isolation and characterization of an osmotic stress and ABA induced histone deacetylase in Arachis hygogaea.
Frontiers in Plant Science 6 (2015).
38. Msanne, J., Lin, J., Stone, J. M. & Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107 (2011).
39. Chen, Y.-E. et al. Different response of photosystem II to short and long-term drought stress in Arabidopsis thaliana. Physiologia
Plantarum 158, 225–235 (2016).
40. Baker, N. R. Chlorophyll Fluorescence: A Probe of Photosynthesis In Vivo. Annual Review of Plant Biology 59, 89–113 (2008). 41. Liang, Y. et al. A nondestructive method to estimate the chlorophyll content of Arabidopsis seedlings. Plant Methods 13 (2017). 42. Daszkowska-Golec, A. & Szarejko, I. Open or Close the Gate – Stomata Action Under the Control of Phytohormones in Drought
Stress Conditions. Frontiers in Plant Science 4 (2013).
43. Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods. 2015 Mar 2, 10.1038/nmeth.3312.
https://doi.org/10.1038/nmeth.3312 PubMed 25730490.
44. Gibson, D. G. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Research 37, 6984–6990 (2009).
45. Holsters, M. et al. Transfection and transformation of Agrobacterium tumefaciens. Molecular and General Genetics MGG 163, 181–187 (1978).
46. Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation ofArabidopsis thaliana: Floral dip transformation of Arabidopsis. The Plant Journal 16, 735–743 (1998).
47. Li, J. F. & Nebenfuhr, A. FAST Technique for Agrobacterium-Mediated Transient Gene Expression in Seedlings of Arabidopsis and Other Plant Species. Cold Spring Harbor Protocols 2010, pdb.prot5428-pdb.prot5428 (2010).
48. Wu, H.-Y. et al. AGROBEST: an efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods 10, 19 (2014).
49. Jefferson, R. A. The GUS reporter gene system. Nature 342, 837–838 (1989).
50. Cartharius, K. et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 (2005).
51. Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, 45e–45 (2001). 52. Bakó, A. et al. Monitoring transgene expression levels in different genotypes of field grown maize (Zea mays L.). South African
Journal of Botany 84, 6–10 (2013).
53. Maloof, J. N., Nozue, K., Mumbach, M. R. & Palmer, C. M. LeafJ: An ImageJ Plugin for Semi-automated Leaf Shape Measurement.
Journal of Visualized Experiments, https://doi.org/10.3791/50028 (2013).
54. Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675 (2012).
55. Arnon, D. I. Copper enzymes in isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant physiology 24, 1–15 (1949).
Acknowledgements
We thank Professor Denis Henrique Pinheiro for his technical support in the statistical analysis. We also thank CAPES-COFEUB Sv.622/18.
Author Contributions
J.F.R.P., F.X.G. and M.F.G.S. conceived the presented idea. J.F.R.P., F.X.G. developed the theory and designed the strategy. J.F.R.P. and T.P.R. drafted and performed the experiments with the support of F.X.G. and C.B., I.T.L., D.D.N. and B.P.M. performed the gene expression experiments. J.F.R.P., F.X.G. interpreted the results and wrote the manuscript with the support from C.B., M.F.G.S. and J.d.A.E. All authors discussed the results and contributed to the final manuscript.
Additional Information
Supplementary information accompanies this paper at https://doi.org/10.1038/s41598-019-44571-y.
Competing Interests: The authors declare no competing interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and
institutional affiliations.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International
License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Cre-ative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not per-mitted by statutory regulation or exceeds the perper-mitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.