Integration of two essential virulence modulating
signals at the Erwinia chrysanthemi pel gene
promoters: a role for Fis in the growth-phase
regulation
Thomas Lautier,1Nicolas Blot,2
Georgi Muskhelishvili3and William Nasser1* 1Université de Lyon, F-69003, Université Lyon 1, F-69622; INSA-Lyon, Villeurbanne, F-69621, CNRS, UMR 5240, Unité Microbiologie Adaptation et Pathogénie, F-69622, France.
2Station Biologique de Roscoff; CNRS, UMR7144; Roscoff, F-29680, France.
3Jacobs University, Bremen, Campus Ring1, D-28759 Bremen, Germany.
Summary
Production of the essential virulence factors, called pectate lyases (Pels), in the phytopathogenic bacte-rium Erwinia chrysanthemi is controlled by a complex regulation system and responds to various stimuli, such as the presence of pectin or plant extracts, growth phase, temperature and iron concentration. The presence of pectin and growth phase are the most important signals identified. Eight regulators modulating the expression of the pel genes (encoding Pels) have been characterized. These regulators are organized in a network allowing a sequential function-ing of the regulators durfunction-ing infection. Although many studies have been carried out, the mechanisms of control of Pel production by growth phase have not yet been elucidated. Here we report that a fis mutant of E. chrysanthemi showed a strong increase in tran-scription of the pel genes during exponential growth whereas induction of expression in the parental strain occurred at the end of exponential growth. This reveals that Fis acts to prevent an efficient transcrip-tion of pel genes at the beginning of exponential growth and also provides evidence of the involve-ment of Fis in the growth-phase regulation of the pel genes. By using in vitro DNA–protein interactions and transcription experiments, we find that Fis directly represses the pel gene expression at the transcription
initiation step. In addition, we show that Fis acts in concert with KdgR, the main repressor responding to the presence of pectin compounds, to shut down the pel gene transcription. Finally, we find that active Fis is required for the efficient translocation of the Pels in growth medium. Together, these data indicate that Fis tightly controls the availability of Pels during patho-genesis by acting on both their production and their translocation in the external medium.
Introduction
Bacteria maintain intricate signalling networks that sense any variation in the environmental conditions and adjust cellular physiology accordingly. These controls are exerted primarily at the level of transcription initiation (Browning and Busby, 2004). An attractive model for learning how bacteria integrate various regulatory mechanisms to control transcription initiation is the pel genes in Erwinia chrysanthemi. These genes encode pectate lyases (Pels), essential virulence factors, which are secreted by a type II system encoded by the out genes (He et al., 1991; Condemine et al., 1992; Login and Shevchik, 2006). Expression of the pel genes is under the control of a complex regulation system and it varies greatly in response to growth of the bacteria under different environmental or physiological condi-tions. These include growth phase, catabolic repression, pectin (the main component of plant cell walls) or plant extract, temperature, osmolarity, pH and iron (Hugouvieux-Cotte-Pattat et al., 1996; Expert, 1999; Sepulchre et al., 2007). Among these conditions, the effects of pectin and growth phase are predominant (Hugouvieux-Cotte-Pattat et al., 1996; Sepulchre et al., 2007). Several regulators (KdgR, Pir, PecS, PecT, Fur, CRP, H-NS) modulating the expression of pel genes in E. chrysanthemi have been previously characterized (Reverchon et al., 1991; 1994; 1997; 1998; Praillet et al., 1996; 1997; Surgey et al., 1996; Castillo and Reverchon, 1997; Nomura et al., 1998; Franza et al., 1999; Nasser and Reverchon, 2002; Rouanet et al., 2004). The induc-tion of pel gene expression by pectic compounds is mediated by KdgR (Reverchon et al., 1991; Nasser
Accepted 13 October, 2007. *For correspondence. E-mail William.nasser@insa-lyon.fr; Tel. (+33) 4 72 43 26 95; Fax (+33) 4 72 43 15 84.
First published online 19 November 2007
et al., 1992; 1994). The induction by plant extracts and iron starvation is mediated by Pir (Nomura et al., 1998) and Fur (Franza et al., 1999) respectively. ExpR is a quorum-sensing regulator (Nasser et al., 1998; Castang et al., 2006) that moderately participates in the modula-tion of pel gene expression in response to N-acyl-homoserine lactones (acyl-HSLs) generated by ExpI synthase (Nasser et al., 1998). Catabolic repression is mediated by the cAMP–CRP complex, which acts as the main activator of the pel gene expression (Reverchon et al., 1997; Nasser et al., 1998; Rouanet et al., 1999). Finally, the signals to which PecS, PecT and H-NS respond have not yet been elucidated. However, H-NS is thought to respond to changes in environmental con-ditions, and in particular to nutritional stress and varia-tion in temperature (Nasser et al., 2001a; Sepulchre et al., 2007), whereas PecS is thought to respond to phenolic compounds or reactive oxygen species pro-duced by the plant defence reactions. Most of these regulators act by binding to the regulatory regions of pel genes (Nasser et al., 1994; Praillet et al., 1996; Nomura et al., 1999; Rouanet et al., 1999; Robert-Baudouy et al., 2000; Franza et al., 2002; Nasser and Reverchon, 2002). Their mode of action has been studied and several interactions between these regulators have been characterized, supporting the existence of a regulatory network (Reverchon et al., 1998; Nasser and Rever-chon, 2002; Rodionov et al., 2004). Mathematical and computational studies of the regulatory network strongly suggest that the regulators CRP and H-NS correspond to nodes allowing a sequential functioning of the network (Sepulchre et al., 2007).
However, the mode of action of some of these stimuli, particularly growth phase, has not yet been elucidated. In the accompanying paper, we show that the E. chry-santhemi nucleoid-associated protein (NAP) Fis is highly produced during the early exponential growth and that Fis co-ordinates the production of the main virulence factors, including Pels. In a fis mutant, the induction of the Pel activity is delayed and the synthesis increases during the stationary growth phase. This observation suggests a growth-phase regulation of the pel gene expression by Fis via a complex mechanism because the pattern of regulation does not fit with the transient nature of Fis production. We investigate here the mechanisms by which Fis controls the production of Pels and demonstrate that Fis directly represses the expression of the pel genes by preventing transcription initiation. Moreover, we reveal that Fis and KdgR act in concert to shut down the pel gene expression. Finally we show that Fis is required for the efficient transloca-tion of the Pels in the growth medium. The relevance to pathogenesis of these multiple controls on Pel availabil-ity by Fis is discussed.
Results
The negative control of Fis on the transcription of pel genes is growth phase-dependent
In the accompanying paper we have reported that in a fis mutant the induction of the Pel activity is often delayed but that the synthesis is always increased during the station-ary growth phase. As the E. chrysanthemi fis mutant has a reduced growth rate, we express the Pel activity both in terms of the bacterial number (OD600) and the length of time after inoculation (Fig. 1). The new bacterial-number-dependent representation (Fig. 1C) confirmed that the absence of Fis modifies the growth-phase distribution of Pels in the culture supernatants. These results are con-sistent with the quantification of four of the five major pectate lyases (PelB, C, D and E) in the supernatant of Fig. 1. Time-course induction of pectate lyase activity in the culture
supernatants of Erwinia chrysanthemi strains A350 (ps) and A4374 (fis).
A. Growth curves.
B. Time-dependent expression of pectate lyase (Pel) specific activity (SA).
C. Bacterial number-dependent expression of Pel SA. Bacteria were grown at 30°C in liquid LB medium containing
polygalacturonate (4 g l-1) and 1 ml samples were taken every hour.
Pectate lyase-specific activity is expressed as nmol of unsaturated product liberated per min per mg of bacterial dry weight. Each value represents the mean of six experiments. Bars indicate the standard deviation.
the fis cultures. In contrast, the amount of PelA varied differently from the four other major enzymes because it was decreased in the fis mutant throughout growth.
We further analysed the expression of individual pel genes in a fis background by using the following chromo-somal gene fusions: pelA::uidA, pelB::uidA, pelD::uidA and pelE::uidA. The level of pelB, pelD and pelE transcrip-tion was in general higher in the fis mutant than in the parental strain (Fig. 2 and Fig. S1). However, the increase in the expression of these three genes, in the fis back-ground, starts at the beginning of the exponential growth phase and is particularly pronounced at lower cell concentration. This large increase was evident throughout the exponential phase, and yet there was little difference from the parental strain during stationary phase. These results, contrary to what is observed by Pel activity quan-tification in the culture supernatants, correlate better with the growth-phase cellular content of Fis and therefore support the idea that Fis is acting to repress the pelB, pelD and pelE transcription. Moreover, the induction rate of pelB and pelD gene expression by polygalacturonate
(PGA) is higher in the fis mutant than in the parental strain. Thus it seems that Fis and KdgR (the main regu-lator mediating the induction of pel gene expression by pectic compounds) might synergistically repress the expression of both pel genes. The results obtained on pelE in the presence of PGA appear to be more difficult to interpret because the expression of this gene fluctuates along the growth curve in the fis background. For pelA, significant expression was only observed in the presence of PGA and the results obtained generally correspond with previous observation in isoelectrofocusing experi-ments (Fig. 2B and Fig. S1B). The results obtained on pelD and pelE genes were further investigated by using a quantitative polymerase chain reaction (qPCR) approach. As the data in Fig. 3 show, the increase transcription of both genes in the fis background was more pronounced at the beginning of the exponential phase of growth than in the early stationary phase. The results obtained from the pelE transcript quantification, in the presence of PGA, were not in accordance with those determined using the pelE::uidA transcriptional fusion because no strong
differ-Fig. 2. Bacterial number-dependent
expression of pelA::uidA, pelB::uidA,
pelD::uidA and pelE::uidA in Erwinia chrysanthemi strains A350 (ps) and A4374
(fis). Bacteria were grown at 30°C in liquid LB medium (A) or in LB plus polygalacturonate (4 g l-1) (B) and 1 ml samples were taken
every hour.b-Glucuronidase-specific activity is expressed as nmol of p-nitrophenol produced per min per mg of bacterial dry weight.
ence in the fusion expression was observed in the paren-tal strain and in the fis mutant (Fig. 2B and Fig. S1B). However, as a similar pattern of pelE transcript accumu-lation was observed in primer extension experiments (data not shown), we decided to concentrate on the tran-script quantification results rather than those obtained with pelE::uidA transcriptional fusion. Thus, we conclude that Fis represses the expression of pelB, pelD and pelE genes and that this action is more pronounced at the beginning of the exponential phase of growth.
Finally, as recent data (Lenz and Bassler, 2007) revealed an involvement of Fis in the Vibrio cholerae quorum-sensing circuit, we looked for the existence of a similar mechanism in E. chrysanthemi. However, no effect of the Fis absence was observed either on pheromone synthesis or on the expression of the quorum-sensing system genes expI and expR (data not shown). Thus, Fis does not seem to be involved in the E. chrysanthemi quorum-sensing circuit.
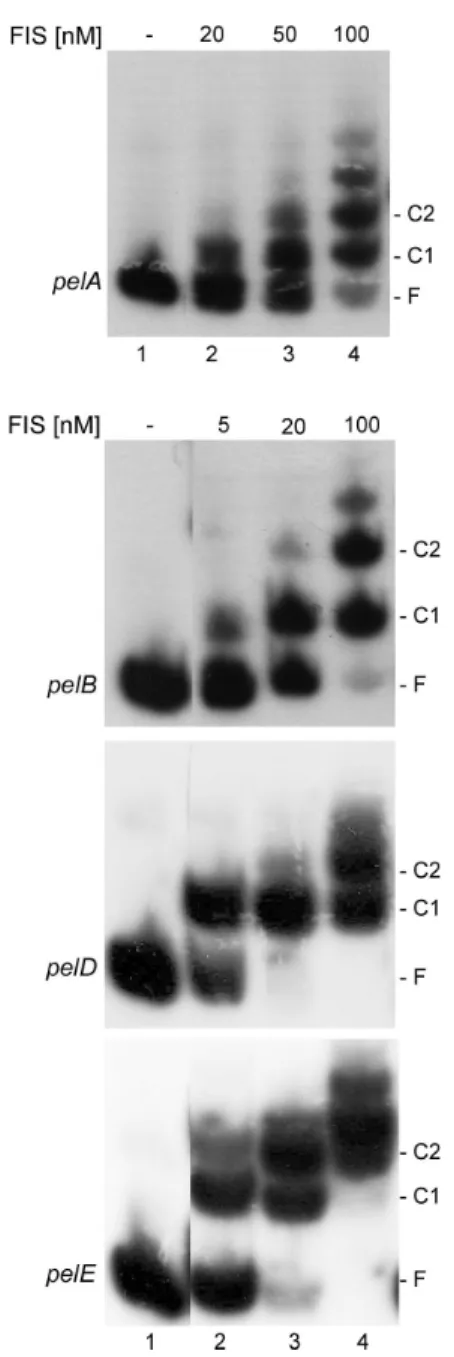
Fis binds the pelA, pelB, pelD and pelE promoters To test whether transcriptional regulation of the pel genes is achieved by a direct binding of Fis to the promoter region, in vitro DNA–protein interactions were performed. Purified Fis was found to bind the regulatory regions of these four genes, as shown by a shift in the migration of the DNA probes (Fig. 4). In the case of the regulatory region of pelB, pelD and pelE genes, a shift was seen at the lowest concentration (5 nM) of Fis used, though it was more pronounced with pelD, followed by pelE, and then
pelB. A higher concentration (20 nM) was needed to detect a complex between Fis and the regulatory region of pelA. These data show that Fis interacts directly with pelA, pelB, pelD and pelE genes, albeit with different affinities. Furthermore, with increasing Fis concentrations, highly retarded complexes appeared at the expense of the lower complexes. This suggests the existence, in the four tested operators, of several Fis binding sites. A particular situa-tion was observed for the pelE operator on which a
Fig. 3. Quantification of the increase in pelD and pelE gene
transcript accumulation in the fis background using real-time PCR analysis.
Fig. 4. Band-shift assay for Fis–DNA binding. Lane 1, no protein;
lanes 2–4, DNA with increasing concentration of Fis, indicated on the top. The position of free DNA (F) and the main Fis–DNA complexes (C) are indicated, C1 corresponds to the complex with the high-affinity binding site, whereas C2 corresponds to the complex obtained by binding to the high-affinity sites and the additional lower site.
second highly retarded complex was observed at a low Fis concentration (5 nM), though it was less pronounced than the lower retarded band. Thus it is reasonable to conclude that pelE contains two relatively high-affinity binding sites for Fis.
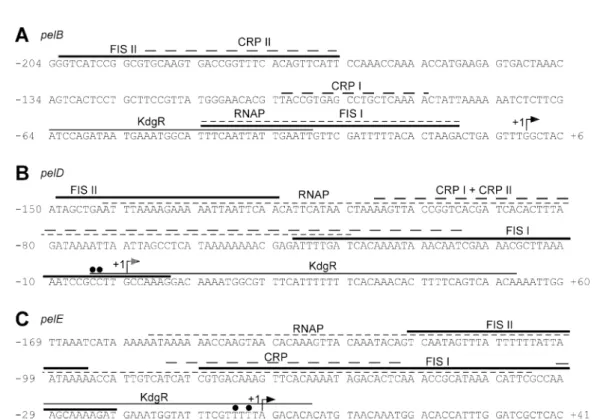
DNase I footprinting experiments were conducted next to establish the precise location of the Fis binding site(s) on the regulatory regions of pelB, pelD and pelE. These experiments were not performed for pelA because the organization of its regulatory region has not yet been established. Moreover, the effect of Fis on pelA expres-sion was relatively low compared with that observed for the pelB, pelD and pelE genes. At low Fis concentrations (10–20 nM), a single region that was clearly protected was observed on the three operators (Fig. 5). This pro-tected area (Fis I) extends from nucleotides-44 to -9, -47 to+7 and -79 to -20 with respect to the transcription start site+1 of pelB, pelD and pelE respectively. Increasing the Fis concentration up to 100 nM resulted in the protection of a second region (Fis II) located in the upstream regions of the three genes. Fis binding sites II extend from nucle-otides-202 to -165, -149 to -120 and -120 to -94 with respect to the transcription start site of pelB, pelD and pelE respectively. In addition to these protected sites, Fis binding induces the appearance of several DNase I-hypersensitive sites (Fig. 5). Thus, it appears that these three pel genes contain two Fis binding sites and that the high-affinity binding sites, Fis I, overlap one or both RNA polymerase binding sites. Because the increase in Fis concentration beyond the saturation point does not reveal any additional protected region, it seems that the stability of some highly retarded complexes observed in band-shift experiments is not sufficient for their identification by DNase I footprinting experiments.
Fis is able to form a nucleoprotein complex with RNAP, CRP and KdgR at the pel promoter/operator region Fis protects DNA regions located both upstream from, and overlapping, the pelB, pelD and pelE promoters. These DNA sequences also bind in vitro RNAP, the CRP activator and the KdgR repressor (Reverchon et al., 1991; Nasser et al., 1997). Indeed, previous footprinting by KdgR revealed a single protected region on the promoter of the pelB, pelE and pelD genes (Nasser et al., 1994). Similar experiments performed in the presence of CRP (Nasser et al., 1997; Rouanet et al., 1999) showed a single pro-tected region on pelE, whereas two CRP binding sites were revealed on pelB and pelD. Further analysis revealed that CRP and RNAP synergistically bind to the pelD promoter and that KdgR prevents binding of RNAP at the region around the-10 promoter sequence (Rouanet et al., 1999). Hence, we performed DNase I digestions on pelB and pelE promoters in the presence of CRP, RNAP and KdgR. We
firstly investigated if CRP and RNAP synergistically bind at the pelB and pelE promoter regions. In the presence of RNAP alone, a weakly protected region spanning from-44 to-9 for pelB and from -156 to -34 for pelE was observed. In addition to the protected regions, typical DNase I-hypersensitive sites induced by RNAP were observed around positions-35 for pelB and -130 and -80 for pelE. In the presence of both CRP and RNAP, the regions protected by each of the two proteins at the pelB and pelE promoters became more pronounced (Fig. 5A, compare lanes 4 and 6 with 9; Fig. 5C, compare lanes 9 and 13 with 16). Thus, as previously reported for the pelD promoter, CRP and RNAP synergistically bind at the pelB and pelE promoters. More notably, a new DNase I-hypersensitive site was observed around positions-60 for pelB and -20 for pelE, whereas the hypersensitive site induced by RNAP binding at the position around-80 on the pelE promoter disappeared. The added presence of KdgR showed that this protein prevents the binding of RNAP to the full (pelB) or the downstream part (pelE) of the core promoters, without affecting the interaction of the RNAP–CRP complex with the upstream region of both genes (Fig. 5A, compare lane 9 with 14; Fig. 5C, compare lane 16 with 21). Thus, KdgR might repress the pel gene expression by inhibiting tran-scription initiation. As the Fis high-affinity binding site (Fis I) overlaps both the KdgR and CRP binding sites on the pelD and pelE promoter regions (Fig. 6), we next analysed whether Fis and CRP or Fis and KdgR can interact in a co-operative, independent or antagonistic manner on these two operators. Examples of the results obtained are shown in Fig. 5B and C and suggest that Fis binds the pelD and pelE operators independently of KdgR and CRP.
We investigated further the effects that Fis exerts on the binding of RNAP and of the CRP–RNAP complex on the pel genes. In the presence of low concentrations of Fis (10–20 M) and high concentrations of RNAP (200 nM), the protection pattern obtained globally corresponded to that obtained with Fis alone at the pelB promoter. Similar experiments conducted at the pelD promoter revealed that the binding of Fis is preferential in the region which con-tains the Fis high-affinity site (-47 to +7), whereas binding of RNAP is preferential and increases in the upstream region of the promoter (-142 to -40). Similarly, at a high concentration of both Fis (100 nM) and RNAP (200 nM), the pattern observed at the pelB promoter corresponded to that observed in the presence of Fis alone. In the same conditions, binding of Fis is preferential in the region con-taining its high-affinity sites I at the pelD and pelE promot-ers, whereas in the upstream regions of both promoters a footprint that combines both the Fis and RNAP protected regions, with the typical DNase I-hypersensitive sites induced by each protein, was observed. This indicates that Fis and RNAP can simultaneously bind to the upstream region of pelD and pelE promoters. Overall, it appeared
that Fis could displace RNAP from the core promoter regions of the pelB, pelD and pelE genes. The relevance of the increase in RNAP binding by low Fis concentrations in the upstream region of pelD was not further investigated within the framework of these studies. Footprinting of the pelB operator in the presence of CRP, RNAP and Fis gave a digestion pattern similar to that obtained with Fis alone in the region containing the Fis high-affinity binding site, without displacing the RNAP–CRP complex in the upstream region of the promoter. However, the inhibition of RNAP–CRP complex binding around the pelB core pro-moter by Fis appeared to be more pronounced at a high Fis concentration (100 nM), as judged by the disappear-ance of the DNase I-hypersensitive site around position -60, attributable to the CRP–RNAP complex binding. In similar DNase I digestion experiments performed on pelE and pelD promoters, at low concentrations of Fis, a foot-print that combines both the Fis and RNAP–CRP binding pattern was observed. However, at higher Fis concentra-tions the pattern obtained around the core promoter, which contains the Fis high-affinity binding site I, became similar
to that obtained in the presence of Fis alone. This was particularly evident at the pelE promoter in which the DNase I-hypersensitive site at a position around -20, attributable to the CRP–RNAP complex binding, com-pletely disappeared in the presence of the three proteins. Thus, it appears that Fis prevents positioning of the RNAP on the pelB, pelD and pelE transcription initiation regions rather than inhibiting binding of the CRP–RNAP complex and that this action is dependent on the Fis concentration. Finally, footprinting of the three operators in the pres-ence of CRP, RNAP, Fis and KdgR revealed that Fis and KdgR occupy the downstream regions of the operators, which encompass either all (pelB) or the downstream part (pelD and pelE) of the core promoters, the KdgR and Fis high-affinity binding sites. In contrast, the digestion pattern of the upstream regions of the promoters was close to that observed with Fis and the CRP–RNAP complex. These data revealed that RNAP, CRP, Fis and KdgR form a nucleoprotein complex at the pel gene pro-moters and suggest that Fis and KdgR act in concert to repress the pelB, pelD and pelE gene expression. Fig. 5. DNase I footprinting of Fis, CRP, KdgR and RNAP binding at the pelB (A), pelD (B) and pelE (C) promoters. The protein
concentrations used are indicated. The regions binding Fis, KdgR, CRP, RNAP and CRP–RNAP are indicted by thick, thin, standard dashed, close dashed, and bold dashed lines respectively. The black, grey and open arrowheads indicate hypersensitivities induced by binding of Fis, CRP and RNAP respectively. The open circles indicate hypersensitivities induced by Fis and RNAP. The stars indicate hypersensitivities induced by RNAP and CRP.
Fig. 6. Sequence of the pelB, pelD and pelE promoters. The binding sites for the proteins are indicated as in Fig. 5. The KMnO4-sensitive
bases are indicated by closed circles. © 2007 The Authors
Fis and KdgR act in concert to inhibit transcription initiation at the pelD and pelE promoters
The effect of Fis upon CRP-dependent transcription was firstly investigated by using potassium permanganate (KMnO4) footprinting on supercoiled plasmid-containing RRpelD (pTL4) and RRpelE (pTL5). Following the addi-tion of RNAP, we observed that two bases, located between the +1 transcription initiation position and the -10 RNAP binding site of both genes are sensitive to KMnO4 (Figs 6 and 7A, lane 4 and Fig. 7B, lane 11) (-3 and -4 for pelD, -2 and -4 for pelE). The addition of CRP substantially increased the KMnO4reactivity of the two bases at both the pelD and pelE promoters. Thus, as predicted by the DNase I results, CRP enhanced open complex formation by RNAP at the pelD and pelE promoters. The presence of Fis or KdgR decreased the open complex formation. When Fis and KdgR were added in combination, the base reactivity to KMnO4was strongly reduced, suggesting a cooperative effect (Fig. 7A, compare lanes 5, 7 and 10 with lane 13 for pelD; Fig. 7B, compare lanes 12, 14 and 16 with lane 18 for pelE).
We next used in vitro transcription to directly follow the effect of Fis and KdgR on the RNAP–CRP complex activity. For this purpose, we monitored pelD and pelE transcription using pTL4 and pTL5 DNAs with RNAP, CRP, Fis and KdgR, added either alone or in combination. The results demonstrate a similar dependence of both pel promoter activities on CRP, Fis and KdgR concentrations (Fig. 7A, compare lanes 5, 7 and 10 with lane 13; Fig. 7B, compare lanes 2, 3 and 6 with lane 9). Under the same conditions, the transcription of the reference bla promoter located on the same plasmid was not noticeably affected. We thus infer that CRP directly activates the pel gene expression and that Fis and KdgR cooperate to repress the pel gene promoters in vitro. The cellular concentration of CRP, and particularly that of Fis, is subject to a strong fluctuation and the active form of the repressor KdgR is supposed to vary in relation to the cellular content of pectin degradation products. We therefore monitored pelD transcription in the presence of various concentra-tions of the three regulators CRP, KdgR and Fis. Impor-tantly, these experiments revealed that Fis and KdgR are able to repress pelD expression when used at a lower concentration (10 nM) than that of the activator CRP Fig. 7. Fis and KdgR prevent transcription initiation at the pelD (A) and pelE (B) promoters. The KMnO4reactivity and transcription
experiments were performed on supercoiled templates. The protein concentrations used are indicated.
(20 nM) (Fig. 7A, right hand section; compare lane 16 with lanes 18 and 20).
To clarify the mechanism of cooperation between Fis and KdgR, in vivo quantification of pel-uidA transcriptional fusion expression in the parental strain and in fis, kdgR and fis–kdgR mutants was undertaken. The data obtained on pelB, pelD and pelE (Fig. 8 and Fig. S2) revealed that the derepression observed in the two single mutants, particularly at the beginning of the exponential growth phase, was lower than that obtained in the double fis–
kdgR mutant. We thus infer that Fis and KdgR cooperate to repress the pel gene expression in vivo. Similar results were obtained in the in vitro experiments by using Fis from Escherichia coli or from E. chrysanthemi, indicating that both proteins have the same effect on the activity of the E. colis70RNA polymerase used in the this work.
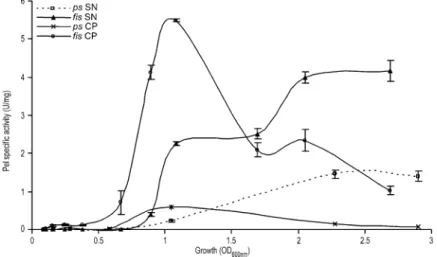
Fis is required for full translocation of the Pels in the external medium
Our data show that Fis mostly represses the pel gene transcription at the beginning of the exponential phase of growth, whereas quantification on supernatants from cul-tures performed in the presence of PGA revealed that the induction of the Pel activity is increased during the sta-tionary growth phase in a fis mutant. This discrepancy led us to question whether the translocation of Pels is modi-fied or not in the fis mutant. To clarify this issue, we quantified the Pel activity present both in the supernatants and in the cell pellets, throughout growth (Fig. 9 and Fig. S3). In the parental strain and its fis derivatives, most of the activity was observed in the cells at the beginning of the growth period. However, the activity content of fis cell was increased more than 10-fold compared with the parental strain. The activity content of cells subsequently decreased, concomitant with an increase in the activity of the supernatants. Importantly, although the parental cells content activity completely disappeared in the stationary phase of growth, significant activity remained in the fis mutant cells. Moreover, the intracellular activity in the fis background was much higher than that obtained in the parental strain throughout growth. We can thus conclude that active Fis is required for an efficient translocation of the Pels.
Discussion
The late exponential phase induction of Pels in E. chrysan-themi is a dramatic effect, whether observed by enzyme activity quantification or in a number of studies using pel::uidA fusions (Hugouvieux-Cotte-Pattat et al., 1992; Nasser et al., 1998). Contrary to observations made on the taxonomically related bacteria Erwinia carotovora (Jones et al., 1993; Pirhonen et al., 1993; Burr et al., 2006), this control does not seem to be directed by a quorum-sensing mechanism (Nasser et al., 1998; S. Reverchon and S. Castang, unpubl. data). In this work, we document a role for the NAP, Fis, as a negative regulatory element for pel genes, acting directly at the transcription level. This model is intuitive because Fis abundance varies inversely with Pels. Synthesis of Fis is under transcriptional control and Fis abundance varies dramatically from being undetect-able in the stationary phase to 40 000 dimers per cell upon dilution into fresh medium (Ball et al., 1992; Lautier and Fig. 8. Bacterial number-dependent expression of pelB::uidA,
pelD::uidA and pelE::uidA in Erwinia chrysanthemi parental strain
and its fis, kdgR and kdgR-fis derivatives. Bacteria were grown at 30°C in liquid LB medium and 1 ml samples were taken every hour. b-Glucuronidase-specific activity is expressed as nmol of
p-nitrophenol produced per min per mg of bacterial dry weight. For pelD, two different graphs were made: – upper graph shows the
expression obtained in the parental strain and its fis derivative, – lower graph shows the expression obtained in kdgR and
kdgR-fis backgrounds.
Nasser, 2007). Consequently, Fis is responsible for orga-nizing the DNA during logarithmic growth (Schneider et al., 1999; 2001; Dorman and Deighan, 2003) and for adjusting cells to the onset of rapid growth by directly interacting with the promoters of numerous genes in E. coli (Nasser et al. 2001b; 2002; Dorman and Deighan, 2003). Recently Fis was also shown to regulate virulence in various animal pathogenic bacteria including Shigella flexneri, enteroinva-sive E. coli, Salmonella typhimurium and V. cholerae (Falconi et al., 2001; Dorman and Deighan, 2003; Kelly et al., 2004; Lenz and Bassler, 2007). However, apart from the control on vir genes in S. flexneri and enteroinvasive E. coli, the action of Fis on the other virulence genes was not fully characterized.
Using a gene fusion approach, we have shown that Fis regulates the expression of pelA, pelB, pelD and pelE genes. Fis slightly activates pelA expression throughout the growth period, whereas it strongly represses the expression of pelB, pelD and pelE genes, particularly at the beginning of the exponential phase of growth. This repression by Fis was further confirmed by pel gene tran-script quantification and therefore in general Fis prevents enzyme production at the beginning of exponential growth.
Gel shift assays demonstrated that purified Fis specifi-cally binds to the regulatory region of the pelA, pelB, pelD and pelE genes. DNase I footprinting experiments further revealed that Fis interacts with the promoter region of the pelB, pelD and pelE genes via two binding sites, the highest affinity site either partly (pelE) or fully (pelB and pelD) overlapping the RNA polymerase binding sites (core promoter) on these genes. Consistently, Fis was shown to be able to displace RNAP from the core promoter regions of the pelB, pelD and pelE genes. As full expression of the pel genes requires the presence of the CRP activator (Nasser et al., 1997; Reverchon et al., 1997), we next investigated the effect of Fis on the activity of the CRP– RNAP complex. Our DNase I footprinting digestions show
that Fis, RNAP and CRP simultaneously bind on the pelB, pelD and pelE regulatory regions to form a nucleoprotein complex. However, the binding of Fis was shown to be preferential and occurs at the expense of RNAP in the regions of the core promoters of these genes, where the binding sites of both proteins are superimposed (Figs 5 and 6). The involvement of the upstream low affinity Fis binding site II in the regulation of pel gene expression is unclear, but we suppose that binding of Fis at this site, which overlaps the upstream binding site of RNAP on pelD and pelE or the CRP binding site II on pelB, may contribute, in association with the high-affinity site I, to driving the promoters into an inhibitory state. This asser-tion is particularly relevant in condiasser-tions of a high cellular content of Fis. Finally, KMnO4reactivity and in vitro tran-scription experiments revealed that Fis prevents access of the CRP–RNAP complex to the pelD and pelE gene promoters, thereby directly repressing transcription initiation. Furthermore, transcription assays revealed that Fis is able to repress pel gene transcription even at a low concentration and in the conditions that are necessary for efficient activity of the CRP–RNAP complex (Fig. 7). Overall these data provide a good correlation between the pattern of the control on pel genes and the growth-phase cellular content of Fis: a strong repression at the begin-ning of the exponential phase of growth, when the Fis concentration is high, followed by a decreased effect at the advanced stages of growth correlated with a reduction in the cellular content of Fis. These findings support the idea that Fis is acting to directly repress, in vivo, the pel gene transcription in a concentration-dependent manner and they provide a significant step forward in elucidating the growth-phase regulation of pel gene expression in E. chrysanthemi. Future investigations should clarify the role of each of the two Fis binding sites involved in the regulation of pel gene expression.
Having clarified the action of Fis in the growth-phase regulation of pel gene transcription, we turned our
atten-Fig. 9. Bacterial number-dependent
expression of the Pel SA present both in the supernatants and in the cell pellets in Erwinia
chrysanthemi parental strain A350 and its fis
derivative A4374. Bacteria were grown at 30°C in liquid LB medium containing polygalacturonate (4 g l-1) and 1 ml samples
were taken every hour. Pel-specific activity is expressed as nmol of unsaturated product liberated per min per mg of bacterial dry weight. Each value represents the mean of two experiments. Bars indicate the standard deviation. SN corresponds to culture supernatant and CP corresponds to cell pellet.
tion to the elucidation of the mechanisms underlying the increased derepression ratio of pel gene expression between the parental strain and its fis derivative in the presence of PGA (Figs 2 and 8, Figs S1 and S2). We postulated that this may involve the KdgR repressor, the activity of which is modulated by the presence of pectic compounds. In vivo gene fusion studies and in vitro KMnO4 reactivity and transcription experiments clearly revealed that the effect of each separate protein on the pel gene expression is lower than that observed in the presence of both proteins. Thus, it appeared that Fis and KdgR cooperate to shut off pel gene expression (Fig. 7). As these two repressors respond to different signals, this organization could allow for a gradual, but co-ordinated derepression of the pel gene expression during pathogenesis. Whether Fis indeed cooperates or exerts an antagonistic action with other proteins of the pel gene regulatory network is currently under investigation.
Finally, quantification of the Pel activity present both in the supernatants and in the cell pellets throughout growth has revealed that the efficiency of Pels translocation is reduced in the fis background, compared with that of the parental strain (Fig. 9 and Fig. S3). This observation sug-gests that Fis might activate the production of the con-stituents of the type II secretion system Out, which direct the translocation of the Pels across the outer membrane. Moreover, Fis might also activate the synthesis of some constituents of the Sec machinery, used for Pels translo-cation across the inner membrane, as previously revealed for SecD and SecF in E. coli (Slany and Kersten, 1992). Finally, we could not rule out an involvement of Fis in the regulation of the production of components necessary to maintain the integrity of the E. chrysanthemi outer mem-brane, which might in turn modify the efficiency of Pel translocation in the external medium. Further investiga-tions should elucidate the mechanism of Fis action on the Pels translocation machinery.
Together, these results suggest a key role for Fis in the availability of Pels during pathogenesis, by acting on both their production and their translocation. By integrating the results described here with the data accumulated over recent years, we are able to propose a model to describe how KdgR and Fis might regulate the availability of Pels for appropriate infection of the plant host by E. chrysanthemi. Moreover, this model integrates the effect of the two events (growth-phase regulation and presence of pectin compounds) having the strongest effect on the pel gene expression identified so far. During the first steps of infection, the bacterium faces nutritional starvation with regard to metabolites in the apoplast. Moreover, it is well documented that production of Pels at this step is detrimental to successful pathogenesis (Nasser et al., 2005; Burr et al., 2006) because of the low cell number and the fact that a strong degradation of the
plant cell wall would result in a premature stimulation of the plant defence reactions. Thus, in the initial steps of infection, pel genes are mostly repressed by Fis and KdgR, the strong repression by KdgR being indicated by the relatively low amount of pectin-degradation products present in the plant intercellular spaces. During this period, Fis stimulates in parallel the production of the components of the machinery used for the translocation of Pels in the external medium. After a certain time, the basal production of Pels leads to the initiation of pectin degra-dation, which in turn results in a more favourable environ-ment for bacteria multiplication. The increase in pectin degradation products and in cell number leads to the decrease of the cellular content of Fis and of the active form of KdgR. These events result in a strong production of Pels, which are suddenly released in the external medium by the translocation machinery, which is thought to be particularly efficient at this stage. This sudden release of Pels allows for a strong attack on the host plant and the consequent development of soft rot symptoms.
Experimental procedures
Bacterial strains, plasmids, culture conditions and DNA manipulation techniques
Bacterial strains and plasmids used in this study are described in Table 1. E. chrysanthemi and E. coli were grown at 30°C and 37°C respectively. Luria broth (LB) medium (Miller, 1972), supplemented by PGA at 0.4% (w/v) when required, was used. Media were solidified by the addition of 1.5% agar. When required, the antibiotics were as follows:
ampicillin (Ap), 100mg ml-1
; kanamycin (Km) and
chloram-phenicol (Cm) 50mg ml-1. DNA manipulations were
per-formed using standard methods (Sambrook et al., 1989). The plasmids pTL4 and pTL5 were generated by cloning the pelD and pelE regulatory regions (-202 to +126 and -186 to+32 relative to the transcription initiation +1 site of pelD and pelE respectively) in the vector pBN4 (Bardonnet and Blanco, 1992). The pelD and pelE regulatory regions were obtained from plasmids pWN2481 and pSR1175 (Nasser et al., 1994; Rouanet et al., 1999).
Genetic techniques
Transduction with phage phiEC2 was performed as
described by Résibois et al. (1984).
Protein and enzyme assays
Pectate lyase activity was determined by the degradation of PGA to unsaturated products that absorb at 235 nm (Moran et al., 1968). Experiments were usually performed on super-natants of the bacteria cultures except in the cases of comparison of the cell content activity with that of the supernatants. In these cases, supernatant from 1 ml of the bacteria cultures was collected by a 3 min centrifugation at
12 000 r.p.m., then the pellets were concentrated fourfold (in order to detect any low level of pectate lyase activity) in the culture medium (LB) and, finally, toluenized. Assays were performed on both supernatants and permeabilized
cells. Specific activity is expressed asmmol of unsaturated
products liberated min-1mg-1 (dry weight) of bacteria.
b-Glucuronidase assay was performed on cell extracts treated with toluene by monitoring the degradation of p-nitrophenyl-b-D-glucuronide into p-nitrophenol that absorbs at 405 nm (Bardonnet and Blanco, 1992). Specific activity for the enzyme is expressed as nmol of product liberated
min-1 mg-1
(dry weight) of bacteria. For growth in synthetic medium, bacterial concentration was estimated by measuring turbidity at 600 nm given that an optical density (OD) of 1.0 at
600 nm corresponds to 109bacteria per ml and to 0.47 mg of
bacteria (dry weight) per ml.
RNA isolation, primer extension and qPCR analysis
Total RNA was extracted from E. chrysanthemi by the frozen-phenol method described by Maes and Messens (1992) or by
Table 1. Bacterial strains, plasmids, phages and oligonucleotides used in this work.
Strain Relevant characteristics and usea Reference or source
Escherichia coli
NM522 D(lac-proAB) thi hsd-5 supE (F⬘ proAB + lacIq lacZDM15) Stratagene DH5a F⬘ j80 dLacZ D(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rk-1, mk+) phoA
supE44l-thi-1 gyrA96 relA1/F⬘ proAB+lacIqZDM15 Tn10-Tc
Life Technology
Erwinia chrysanthemi
A350 lmrT(con) lacZ2 Hugouvieux-Cotte-Pattat and
Robert-Baudouy (1985) A4374 lmrT(con) lacZ2 fis::Cm Lautier and Nasser (2007) A4392 lmrT(con) lacZ2 pelA::uidA Km Bourson et al. (1993) A4393 lmrT(con) lacZ2 fis::Cm, pelA::uidA Km This work
A1787 lmrT(con) lacZ2 pelB::uidA Km Hugouvieux-Cotte-Pattat et al. (1992) A4395 lmrT(con) lacZ2 fis::Cm, pelB::uidA Km This work
A1798 lmrT(con) lacZ2 pelD::uidA Km Hugouvieux-Cotte-Pattat et al. (1992) A4390 lmrT(con) lacZ2 fis::Cm, pelD::uidA Km This work
A1828 lmrT(con) lacZ2 pelE::uidA Km Hugouvieux-Cotte-Pattat et al. (1992) A4548 lmrT(con) lacZ2 fis::Cm, pelE::uidA Km This work
A4610 lmrT(con) lacZ2 kdgR::Sm, pelB::uidA Km This work A4611 lmrT(con) lacZ2 fis::Cm, kdgR::Sm, pelB::uidA Km This work A4612 lmrT(con) lacZ2 kdgR::Sm, pelD::uidA Km This work A4613 lmrT(con) lacZ2 fis::Cm, kdgR::Sm, pelD::uidA Km This work A4614 lmrT(con) lacZ2 kdgR::Sm, pelE::uidA Km This work A4615 lmrT(con) lacZ2 fis::Cm, kdgR::Sm, pelE::uidA Km This work Plasmids
pGEM-T Cloning vector, AprlacZ′ Promega
pBluescript Cloning vector, AprlacZ′ Stratagene
pNB4 Cloning vector, ApruidA Bardonnet and Blanco (1992)
pSR1321 pBluescript with the 357 bp fragment containing the pelA regulatory region Nasser et al. (1994) pN906 pBluescript with the 470 bp fragment containing the pelB regulatory region Nasser et al. (1994) pWN2481 pBluescript with the 318 bp fragment containing the pelD regulatory region Rouanet et al. (1999) pSR1175 pBluescript with the 470 bp fragment containing the pelE regulatory region Nasser et al. (1994) pTL4 pNB4 with the EcoRI-HindIII fragment containing the pelD regulatory region from
the pWN2481
This work pTL5 pNB4 with EcoRI-HindIII fragment containing the pelE regulatory region from the
pSR1175
This work Phages
PhiEC2 General transducing phage of Erwinia chrysanthemi Résibois et al. (1984) Oligonucleotides
AW158 5′-TGACCACCCAGCCATCCTTC-3′ Applied Biosystems bla3B4 5′-CAGGAAGGCAAAATGCCGC-3′ Castang et al. (2006) DM152 5′-CATGTCAAATTTCACTGCTTCATCC-3′ Applied Biosystems
pelD qPCR f 5′-GACAGAAGCAGCGTCAACTG-3′ This work
pelD qPCR r 5′-TCTGATCGTCAAAGCTGGTG-3′ This work
pelE qPCR f 5′-AGCGAATTCAAAGCAGCACT-3′ This work
pelE qPCR r 5′-GGCGTTTCGATGTACAGGTT-3′ This work
rsmA qPCR f 5′-GAGTTGGCGAAACCCTCAT-3′ This work
rsmA qPCR r 5′-GCTGAGACTTCTCTGCCTGAA-3′ This work
uidAdeb 5′-CTGGTCAACCTTTAATCTG-3′ Castang et al. (2006)
a. Genotype symbols are according to Berlyn (1998). lmrT(con) indicates that the transport system encoded by the gene lmrT, which mediates
the entry of lactose, melibiose and raffinose into the cells, is constitutively expressed. lacZ′ indicates that the 3′ end of this gene is truncated. Km, kanamycin; Cm, chloramphenicol; Ap, ampicillin; Sm, streptomycin.
the Qiagen Rneasy Mini kit procedure (Qiagen). RNA was quantified using the Nanodrop spectrophotometer and then
checked on 1%-agarose gel containing 0.5mg ml-1ethidium
bromide (BrET). cDNAs were synthesized with 0.5–2mg of
RNA by using the SuperScriptTMfirst-strand synthesis system
for RT-PCR (Invitrogen) in the presence of 50 ng random
hexamersmg-1
of RNA. The reaction was incubated at 25°C
(10 min), 42°C (50 min) and 70°C (15 min). 0.1–1ml of the
reaction mixture obtained was used for qPCR reactions in
10ml using the LightCyclerR faststart DNA masterplusSYBR
Green I kit from Roche (Roche Applied Science). The real-time PCR reaction was performed in a Roche LightCycler 480. Reactions were performed at 95°C for 10 min and 35 cycles of 95°C for 15 s, 55°C for 15 s and 72°C for 20 s. Target gene expression is defined by the method described by Pfaffl (2001). The rsmA gene, for which similar expression was observed in the parental strain and in its fis derivative
throughout growth, was used as a reference for
normalization. 2.5¥ 105 copies of GeneAmplimer pAW 109
RNA (Applied Biosystems) were added to the reverse transcription reaction and used as a control for the retro-transcription efficiency (Wisniewski and Rogowsky, 2004).
In vitro DNA/protein interaction
Band-shift assay and DNase I footprinting were performed as previously described (Nasser et al., 1997). The regulatory region of the pelA, pelB, pelD, pelE DNA fragments was recovered from plasmids pSR1321, pN906, pWN2481, pSR1175 respectively, by a EcoRI-HindIII digestion for pelA, pelD and pelE and NsiI-HindIII for pelB. The DNA fragments obtained were further end-labelled by filling up the HindIII
extremities in the presence of (a-32P)dCTP (3000 Ci mmol-1,
GE HealthCare) and the Klenow fragment of DNA
polymerase. The labelled DNA fragments were purified after electrophoresis on agarose gel using the Qiagen gel extrac-tion kit. The signals obtained were detected by autoradiogra-phy on Amersham MP film.
Potassium permanganate reactivity assay
The reactions for the potassium permanganate (KMnO4) reactivity assay were performed with supercoiled templates. Five hundred nanograms of plasmids pTL4 (containing the pelD regulatory region) and pTL5 (containing the regulatory region of pelE) and the proteins, as indicated, was incubated
in 50ml of a buffer containing 10 mM Tris-HCl pH 8, 10 mM
MgCl2, 150 mM KCl, 0.2 mM dithiothreitol and 0.1% (v/v) Nonidet P-40 (Roche). After incubation at 30°C for 15 min,
0.1 vol. of 100 mM KMnO4solution was added for 15 s to the
reaction mixtures containing DNA and proteins. The reactions
were stopped by the addition of 0.1 vol. of 14 M
b-mercaptoethanol, 40 mg of glycogen (Roche, Mannheim, Germany) and sodium acetate to 0.3 M, precipitated with 3 vols of ice-cold ethanol, and washed twice with 70% ethanol. The reaction products were solubilized in water and were divided into equal parts. Both parts were used as a template for five cycles of amplification by Taq polymerase
with the 5′ radio-labelled primer uidAdeb for the detection of
modified bases at the pelD and pelE promoters and bla3B4
(Table 1) for the detection of modified bases at the bla promoter. The amplification products were analysed on a 6% sequencing gel. The signals obtained were detected using
Cyclone PhosphoImager (Packard). E. coli s70RNA
poly-merase was from Amersham Biosciences, the protein molar-ity was determined based on the concentration of the batches (mg ml-1
).
In vitro transcription
Supercoiled plasmids pTL4 and pTL5, containing the pelD and pelE regulatory regions, were used for in vitro transcrip-tion and primer extension reactranscrip-tions according to Lazarus and Travers (1993). The mRNA obtained after in vitro transcrip-tion was divided into equal parts and used for primer exten-sion by reverse transcriptase (Moloney murine leukaemia virus reverse transcriptase, RNase H minus, Invitrogen) with radioactively end-labelled primers uidAdeb for pelD and pelE mRNAs and bla3B4 for the bla transcript (Table 1). The extension with primers uidAdeb and bla3B4 yields 156 bp for pelD, 96 bp for pelE and 100 bp for bla.
Acknowledgements
We are grateful to Valerie James for the English corrections, to A. Buchet for critical reading of the manuscript and to our colleagues G. Condemine, S. Reverchon, V. Shevchik and N. Hugouvieux-Cotte-Pattat for their support and advice. We thank C. Burau and G. Effantin for technical assistance. This work was supported by grants from the Centre National de la Recherche Scientifique (CNRS), and from the Programme de Microbiologie 2003 (ACIM-2-17).
References
Ball, C.A., Osuna, R., Ferguson, K.C., and Johnson, R.C. (1992) Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol 174: 8043–8056.
Bardonnet, N., and Blanco, C. (1992) ‘uidA-antibiotic-resistance cassettes for insertion mutagenesis, gene fusions and genetic constructions. FEMS Microbiol Lett
72: 243–247.
Berlyn, M.K. (1998) Linkage map of Escherichia coli K-12, edition 10, the traditional map. Microbiol Mol Biol Rev 62: 814–984.
Bourson, C., Favey, S., Reverchon, S., and Robert-Baudouy, J. (1993) Regulation of the expression of a pelA::uidA fusion in Erwinia chrysanthemi and demonstration of the synergistic action of plant extract with polygalacturonate on pectate lyase synthesis. J Gen Microbiol 139: 1–9. Browning, D.F., and Busby, S.J. (2004) The regulation of
bacterial transcription initiation. Nat Rev Microbiol 2: 57–65.
Burr, T., Barnard, A.M., Corbett, M.J., Pemberton, C.L., Simpson, N.J., and Salmond, G.P. (2006) Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol Microbiol 59: 113–125.
Castang, S., Reverchon, S., Gouet, P., and Nasser, W. (2006) Direct evidence for the modulation of the activity of
the Erwinia chrysanthemi quorum-sensing regulator ExpR by acylhomoserine lactone pheromone. J Biol Chem
281: 29972–29987.
Castillo, A., and Reverchon, S. (1997) Characterization of the pecT control region from Erwinia chrysanthemi 3937. J Bacteriol 179: 4909–4918.
Condemine, G., Dorel, C., Hugouvieux-Cotte-Pattat, N., and Robert-Baudouy, J. (1992) Some of the out genes involved in the secretion of pectate lyases in Erwinia chrysanthemi are regulated by KdgR. Mol Microbiol 6: 3199–3211. Dorman, C.J., and Deighan, P. (2003) Regulation of gene
expression by histone-like proteins in bacteria. Curr Opin Genet Dev 13: 179–184.
Expert, D. (1999) Withholding and exchanging iron: interac-tions between Erwinia spp. and their plant hosts. Annu Rev Phytopathol 37: 307–334.
Falconi, M., Prosseda, G., Giangrossi, M., Beghetto, E., and Colonna, B. (2001) Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroin-vasive Escherichia coli. Mol Microbiol 42: 439–452. Franza, T., Sauvage, C., and Expert, D. (1999) Iron
regula-tion and pathogenicity in Erwinia chrysanthemi 3937: role of the Fur repressor protein. Mol Plant Microbe Interact
12: 119–128.
Franza, T., Michaud-Soret, I., Piquerel, P., and Expert, D. (2002) Coupling of iron assimilation and pectinolysis in Erwinia chrysanthemi 3937. Mol Plant Microbe Interact
15: 1181–1191.
He, S.Y., Schoedel, C., Chatterjee, A.K., and Collmer, A. (1991) Extracellular secretion of pectate lyase by the Erwinia chrysanthemi out pathway is dependent upon Sec-mediated export across the inner membrane. J Bacteriol
173: 4310–4317.
Hugouvieux-Cotte-Pattat, N., and Robert-Baudouy, J. (1985) Lactose metabolism in Erwinia chrysanthemi. J Bacteriol
162: 248–255.
Hugouvieux-Cotte-Pattat, N., Dominguez, H., and Robert-Baudouy, J. (1992) Environmental conditions affect tran-scription of the pectinase genes of Erwinia chrysanthemi 3937. J Bacteriol 174: 7807–7818.
Hugouvieux-Cotte-Pattat, N., Condemine, G., Nasser, W., and Reverchon, S. (1996) Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol 50: 213–257. Jones, S., Yu, B., Bainton, N.J., Birdsall, M., Bycroft, B.W.,
Chhabra, S.R., et al. (1993) The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J 12: 2477–2482.
Kelly, A., Goldberg, M.D., Carroll, R.K., Danino, V., Hinton, J.C., and Dorman, C.J. (2004) A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology
150: 2037–2053.
Lazarus, L.R., and Travers, A.A. (1993) The Escherichia coli FIS protein is not required for the activation of tyrT transcription on entry into exponential growth. EMBO J
12: 2483–2494.
Lautier, T., and Nasser, W. (2007) The DNA nucleoid-associated protein Fis co-ordinates the expression of the main virulence genes in the phytopathogenic bacterium Erwinia chrysanthemi. Mol Microbiol, 66, in press.
Lenz, D.H., and Bassler, B.L. (2007) The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol Microbiol 63: 859–871.
Login, F.H., and Shevchik, V.E. (2006) The single transmem-brane segment drives self-assembly of OutC and the for-mation of a functional type II secretion system in Erwinia chrysanthemi. J Biol Chem 281: 33152–33162.
Maes, M., and Messens, E. (1992) Phenol as grinding mate-rial in RNA preparations. Nucleic Acids Res 20: 4374. Miller, J.H. (1972) Experiment in Molecular Genetics. New
York: Cold Spring Harbor Laboratory Press.
Moran, F., Nasuno, S., and Starr, M.P. (1968)
Oligogalactu-ronide trans-eliminase of Erwinia carotovora. Arch
Biochem Biophys 125: 734–741.
Nasser, W., and Reverchon, S. (2002) H-NS-dependent acti-vation of pectate lyases synthesis in the phytopathogenic bacterium Erwinia chrysanthemi is mediated by the PecT repressor. Mol Microbiol 43: 733–748.
Nasser, W., Reverchon, S., and Robert-Baudouy, J. (1992) Purification and functional characterization of the KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi. Mol Microbiol 6: 257–265.
Nasser, W., Reverchon, S., Condemine, G., and Robert-Baudouy, J. (1994) Specific interactions of Erwinia chry-santhemi KdgR repressor with different operators of genes involved in pectinolysis. J Mol Biol 236: 427–440. Nasser, W., Robert-Baudouy, J., and Reverchon, S. (1997)
Antagonistic effect of CRP and KdgR in the transcription control of the Erwinia chrysanthemi pectinolysis genes. Mol Microbiol 26: 1071–1082.
Nasser, W., Bouillant, M.L., Salmond, G., and Reverchon, S. (1998) Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol Microbiol 29: 1391–1405.
Nasser, W., Faelen, M., Hugouvieux-Cotte-Pattat, N., and Reverchon, S. (2001a) Role of the nucleoid-associated protein H-NS in the synthesis of virulence factors in the phytopathogenic bacterium Erwinia chrysanthemi. Mol Plant Microbe Interact 14: 10–20.
Nasser, W., Schneider, R., Travers, A., and Muskhelishvili, G. (2001b) CRP modulates fis transcription by alternate formation of activating and repressing nucleoprotein complexes. J Biol Chem 276: 17878–17886.
Nasser, W., Rochman, M., and Muskhelishvili, G. (2002) Transcriptional regulation of fis operon involves a module of multiple coupled promoters. EMBO J 21: 715–724. Nasser, W., Reverchon, S., Vedel, R., and Boccara, M.
(2005) PecS and PecT coregulate the synthesis of HrpN and pectate lyases, two virulence determinants in Erwinia chrysanthemi 3937. Mol Plant Microbe Interact 18: 1205– 1214.
Nomura, K., Nasser, W., Kawagishi, H., and Tsuyumu, S. (1998) The pir gene of Erwinia chrysanthemi EC16 regu-lates hyperinduction of pectate lyase virulence genes in response to plant signals. Proc Natl Acad Sci USA 95: 14034–14039.
Nomura, K., Nasser, W., and Tsuyumu, S. (1999) Self-regulation of Pir, a regulatory protein responsible for hyper-induction of pectate lyase in Erwinia chrysanthemi EC16. Mol Plant Microbe Interact 12: 385–390.
Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res
29: e45.
Pirhonen, M., Flego, D., Heikinheimo, R., and Palva, E.T. (1993) A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J 12: 2467–2476.
Praillet, T., Nasser, W., Robert-Baudouy, J., and Reverchon, S. (1996) Purification and functional characterization of PecS, a regulator of virulence-factor synthesis in Erwinia chrysanthemi. Mol Microbiol 20: 391–402.
Praillet, T., Reverchon, S., and Nasser, W. (1997) Mutual control of the PecS/PecM couple, two proteins regulating virulence-factor synthesis in Erwinia chrysanthemi. Mol Microbiol 24: 803–814.
Résibois, A., Pato, M., Higgins, P., and Toussaint, A. (1984)
Replication of bacteriophage mu and its mini-mu
derivatives. Adv Exp Med Biol 179: 69–76.
Reverchon, S., Nasser, W., and Robert-Baudouy, J. (1991) Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol Microbiol 5: 2203– 2216.
Reverchon, S., Nasser, W., and Robert-Baudouy, J. (1994) pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol Microbiol
11: 1127–1139.
Reverchon, S., Expert, D., Robert-Baudouy, J., and Nasser, W. (1997) The cyclic AMP receptor protein is the main activator of pectinolysis genes in Erwinia chrysanthemi. J Bacteriol 179: 3500–3508.
Reverchon, S., Bouillant, M.L., Salmond, G., and Nasser, W. (1998) Integration of the quorum-sensing system in the regulatory networks controlling virulence factor synthesis in Erwinia chrysanthemi. Mol Microbiol 29: 1407–1418. Robert-Baudouy, J., Nasser, W., Condemine, G., Reverchon,
S., Shevchik, V.E., and Hugouvieux-Cotte-Pattat, N. (2000) Pectic enzymes of Erwinia chrysanthemi, regulation and role in pathogenesis. In Plant-Microbe Interactions. Stacey, G., and Keen, N.T. (eds). St. Paul Minnesota: The American Phytopathological Society, vol. 5, pp. 221–268. Rodionov, D.A., Gelfand, M.S., and Hugouvieux-Cotte-Pattat, N. (2004) Comparative genomics of the KdgR regulon in Erwinia chrysanthemi 3937 and other gamma-proteobacteria. Microbiology 150: 3571–3590.
Rouanet, C., Nomura, K., Tsuyumu, S., and Nasser, W. (1999) Regulation of pelD and pelE, encoding major
alka-line pectate lyases in Erwinia chrysanthemi: involvement of the main transcriptional factors. J Bacteriol 181: 5948– 5957.
Rouanet, C., Reverchon, S., Rodionov, D.A., and Nasser, W. (2004) Definition of a consensus DNA-binding site for PecS, a global regulator of virulence gene expression in Erwinia chrysanthemi and identification of new members of the PecS regulon. J Biol Chem 279: 30158–30167. Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989)
Molecu-lar cloning: A Laboratory Manual. New York, NY, USA: Cold Spring Harbor Laboratory Press.
Schneider, R., Travers, A., Kutateladze, T., and Muskhelish-vili, G. (1999) A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli. Mol Microbiol 34: 953–964.
Schneider, R., Lurz, R., Luder, G., Tolksdorf, C., Travers, A., and Muskhelishvili, G. (2001) An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res 29: 5107–5114.
Sepulchre, J.A., Reverchon, S., and Nasser, W. (2007) Mod-eling the onset of virulence in a pectinolytic bacterium. J Theor Biol 244: 239–257.
Slany, R.K., and Kersten, H. (1992) The promoter of the tgt/sec operon in Escherichia coli is preceded by an upstream activation sequence that contains a high affinity FIS binding site. Nucleic Acids Res 20: 4193–4198. Surgey, N., Robert-Baudouy, J., and Condemine, G. (1996)
The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J Bacteriol 178: 1593–1599.
Wisniewski, J.P., and Rogowsky, P.M. (2004) VacuolarH+-translocating inorganic pyrophosphatase (Vpp1) marks partial aleurone cell fate in cereal endosperm develop-ment. Plant Mol Biol 56: 325–337.
Supplementary material
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/ j.1365-2958.2007.06010.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials sup-plied by the authors. Any queries (other than missing mate-rial) should be directed to the corresponding author for the article.