HAL Id: hal-01259217
https://hal-univ-rennes1.archives-ouvertes.fr/hal-01259217
Submitted on 5 Sep 2016
HAL is a multi-disciplinary open access archive for the deposit and dissemination of sci-entific research documents, whether they are pub-lished or not. The documents may come from teaching and research institutions in France or abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est destinée au dépôt et à la diffusion de documents scientifiques de niveau recherche, publiés ou non, émanant des établissements d’enseignement et de recherche français ou étrangers, des laboratoires publics ou privés.
one-year nationwide program of the French Cooperative
Thoracic Intergroup (IFCT) for advanced non-small cell
lung cancer (NSCLC) patients.
Fabrice Barlesi, Julien Mazières, Jean-Philippe Merlio, Didier Debieuvre, Jean
Mosser, Hervé Léna, l’Houcine Ouafik, Benjamin Besse, Isabelle Rouquette,
Virginie Westeel, et al.
To cite this version:
Fabrice Barlesi, Julien Mazières, Jean-Philippe Merlio, Didier Debieuvre, Jean Mosser, et al.. Routine molecular profiling of cancer: results of a one-year nationwide program of the French Cooperative Thoracic Intergroup (IFCT) for advanced non-small cell lung cancer (NSCLC) patients.. Lancet, Elsevier, 2016, 287 (10026), pp.1415-1426. �10.1016/S0140-6736(16)00004-0�. �hal-01259217�
Title
Routine molecular profiling of cancer: results of a one-year nationwide program of the French Cooperative Thoracic Intergroup (IFCT) for advanced non-small cell lung cancer (NSCLC) patients.
Barlesi F,1 Mazieres J,2 Merlio JP,3 Debieuvre D,4 Mosser J,5 Lena H,6 Ouafik L,7 Besse B,8 Rouquette I,9 Westeel V,10 Escande F,11 Monnet I,12 Lemoine A,13 Veillon R,14 Blons H,15
Audigier-Valette C,16 Bringuier PP,17 Lamy R,18 Beau-Faller M,19 Pujol JL, 20 Sabourin JC,21 Penault-Llorca F,22 Denis MG,23 Lantuejoul S, 24 Morin F,25 Tran Q,25 Missy P,25 Langlais A,26 Milleron B, 27 Cadranel J,27 Soria JC,8 Zalcman G28; for the Biomarkers France contributors (listed at the end of the manuscript).
1- Aix Marseille University; Assistance Publique Hôpitaux de Marseille. Multidisciplinary Oncology & Therapeutic Innovations Dept.; Centre d’Investigation Clinique; Marseille, France.
2- Hôpital Larrey, Centre Hospitalier Universitaire, Université Paul Sabatier, Toulouse, France.
3- Centre Hospitalier Universitaire de Bordeaux, Pôle Biologie et Anatomie Pathologique, Pessac, France ; Université de Bordeaux, Histologie et Pathologie Moléculaires des Tumeurs, Bordeaux, France.
4- Hopital Emile Muller, Service de Pneumologie, Mulhouse, France.
5- Centre Hospitalier Universitaire de Rennes, Département de génomique et génétique moléculaire, plateforme INCA, Rennes, France.
6- Hopital Pontchaillou, Service de Pneumologie, Centre Hospitalier Universitaire, Rennes, France.
7- Aix Marseille University; Assistance Publique Hôpitaux de Marseille, Service de Transfert d’Oncologie Biologique, Marseille, France.
8- Gustave Roussy, Cancer Campus, Villejuif, France and University Paris-Sud, Châtenay-Malabry, France. 9- Institut Universitaire du Cancer de Toulouse, Oncopôle, Service d'Anatomie Pathologique, Toulouse,
France.
10- Université de Franche-Comté, EA3181, Centre Hospitalier Universitaire Jean Minjoz, Service de Pneumologie, Besançon, France.
11- Centre Hospitalier Universitaire de Lille, Département de biochimie et biologie moléculaire, Centre de Biologie Pathologie, Lille, France.
12- Centre Hospitalier Intercommunal de Créteil, Service de pneumologie et de pathologie professionnelle, Créteil, France.
13- Assistance Publique-Hôpitaux de Paris ; Groupe Hospitalier des Hôpitaux Universitaires Paris-Sud, Service d’oncogénétique – Oncomolpath; Université Paris 11; Villejuif, France.
14- Centre Hospitalier Universitaire de Bordeaux, Service des Maladies Respiratoires, Pessac, France.
15- UMR-S1147, INSERM; Université Paris Descartes ; Assistance Publique Hôpitaux de Paris Département de biologie, Hôpital Européen Georges Pompidou, Paris, France.
16- Centre Hospitalier Sainte Musse, Service de pneumologie, Toulon, France.
17- Hôpital Edouard Herriot, Service d’anatomie et de cytologie pathologique, Hospices Civils de Lyon, Université Claude Bernard Lyon 1, Lyon Cancer Research Center, UMR 1057 INSERM, Lyon, France. 18- Centre Hospitalier de Bretagne Sud, Service d’Oncologie Médicale, Lorient, France.
19- Centre Hospitalier Universitaire de Hautepierre, Laboratoire de Biochimie et de Biologie Moléculaire & Plate-forme de Génomique des Cancers, Strasbourg, France.
20- Centre Hospitalier Universitaire de Montpellier, Unité d'Oncologie Thoracique, Pôle Cœur Poumon, Hôpital Arnaud de Villeneuve, Montpellier, France.
21- Centre Hospitalier Universitaire de Rouen, Département d’anatomie et de cytologie pathologiques, Rouen, France.
22- Centre Jean Perrin, Département d’anatomie et de cytologie pathologique, Clermont-Ferrand, France. 23- Centre Hospitalier Universitaire de Nantes, Laboratoire de Biochimie, Nantes, France.
24- Centre Hospitalier Universitaire A Michallon, Département d'Anatomie et Cytologie Pathologiques DACP, Institut de Biologie et de Pathologie, Université Joseph Fourier- INSERM U 823, Institut Albert Bonniot, Grenoble, France.
25- Clinical Research Unit, French Cooperative Thoracic Intergroup (IFCT), Paris, France. 26- Biostatistics unit, French Cooperative Thoracic Intergroup (IFCT), Paris, France.
27- Assistance Publique Hôpitaux de Paris, Hôpital Tenon, Service de Pneumologie, Sorbonne Universités, UPMC Univ Paris 06, Paris, France.
28- Centre Hospitalier Universitaire de Caen, Service de Pneumologie et Oncologie Thoracique, Centre de Recherche Clinique, Université de Caen-Basse Normandie, Caen, France.
ABSTRACT
Background. The molecular profiling of advanced non-small cell lung cancer (NSCLC) patients for known oncogenic drivers is currently recommended during routine care. However, nationally, the feasibility and impact of this policy are unknown.
Methods. The characteristics, molecular profiles and clinical outcomes of advanced NSCLC patients, who were routinely screened for EGFR mutations and ALK rearrangements but also HER2, KRAS, BRAF, and PIK3CA mutations by certified regional genetics centers in France, were assessed consecutively during a one-year period.
Results. Overall, 18,679 molecular analyses of 17,664 NSCLC patients (median age, 64.5 years; male, 64.6%; smokers or former smokers, 81.2%; adenocarcinoma, 76%) were performed. The median interval between the initiation of analysis and the written result was 11 days (interquartile range: 7-16 days). A genetic alteration was found in 49.5% of the analyses: EGFR, HER2, KRAS, BRAF, or PIK3CA mutations or ALK rearrangement in 11.0, 0.8, 28.7, 1.9, 2.3 and 4.8% of the cases, respectively. The presence of a genetic alteration impacted the first-line treatment for 51.3% of the patients and was associated with a significant improvement in the overall response rate for first- (36.5 [95%CI: 34.7-38.2] vs. 32.6% [95%CI: 29.5-35.6], p=0.03) and second-line treatment (16.9 [95%CI: 15.0-18.8] vs. 9.3% [95%CI: 6.7-11.9], p<0.001), first-line progression-free survival (10.0 [95%CI: 9.2-10.7] vs. 7.1 [95%CI: 6.1-7.9] months, p<0.001) and overall survival (16.5 [95%CI: 15.0-18.3] vs. 11.8 months [95%CI: 10.1-13.5], p<0.001).
Conclusions. Routine nationwide molecular profiling of advanced NSCLC patients is feasible. The frequency of the genetic alterations, the acceptable turnaround times in obtaining the analysis results and the clinical advantage provided by the detection of a genetic alteration indicates that routine nationwide molecular profiling provides a clinical benefit.
RESEARCH IN CONTEXT
Evidence before this study.
Before undertaking this study, we did a systematic review of the scientific literature (English only) published up to December 2010 using PubMed and abstracts from ASCO and ESMO meetings (2007-2010) to identify studies assessing nationwide routine molecular profiling of advanced non small cell lung cancer patients for one or more genetic alterations known (or supposed) to be oncogenic drivers. Using the search terms “non small cell lung cancer”, “advanced” or “metastatic”, and “EGFR” or “ALK” or “BRAF” or “HER2” or “PIK3CA” or “KRAS” or “multiplex” or “sequencing” and “nationwide” or names of various countries around the world, we did not identify any published data.
Added value of this study.
This study demonstrates that a routine nationwide molecular profiling of advanced NSCLC patients is feasible within an acceptable turnaround time in obtaining the results. Even if looking at a limited number of genetic alterations (i.e. presently six genes), the frequency of these genetic alterations allows to potentially consider a targeted therapy (either commercially available for EGFR and ALK or within a clinical trial for the other alterations) for treating these patients. Finally, when a genetic alteration was detected, the outcome was a longer median OS indicating a possible prognostic advantage and/or a major change in the management paradigm for these advanced NSCLC patients.
Implications of all the available evidence.
In the meantime, the LCMC initiative (the largest multi-institutional study in the western world) suggested that molecular profiling helps to orient patients toward targeted therapies
and dedicated trials and individuals with drivers receiving a matched targeted agent lived longer. Our study extends LCMC results to a nation-wide approach and the current evidence indicates that routine nationwide molecular profiling provides a clinical benefit to advanced NSCLC patients.
INTRODUCTION
Lung cancer is among the most frequent types of cancer in Western countries and, with more than one million expected deaths per year, is the leading cause of cancer deaths.1 However, the molecular hallmarks of cancer are only currently understood.2 The treatment of lung cancer has entered a new era due to the discovery of epidermal growth factor receptor (EGFR)-activating mutations and anaplastic lymphoma kinase (ALK) gene rearrangements, which lead to changes in outcomes of many, but still a minority of, lung cancer patients.3,4
Moreover, lung cancer displays one of the highest rates of genetic alterations,5 some of which are actionable via the administration of drugs that have already been approved, are available off-label for other indications (dabrafenib or vemurafenib for BRAF,6 trastuzumab or afatinib for HER2 mutations,7 crizotinib for ROS1 rearrangements8) or are under investigation in clinical trials. Therefore, high expectations are placed on this personalized (also referred to as ‘stratified’ or ‘precision’) medicine.
In this context, many medical centers have been organized to provide lung cancer patients with routine assessments of EGFR mutations and ALK rearrangements. In some of these centers additional molecular alterations are tested, typically by research programs.9–12 The preliminary data obtained from these previous programs suggest that molecular profiling helps to orient patients toward targeted therapies and dedicated trials. However, the actual impact of broad molecular screening and subsequent personalized medicine has yet to be addressed in a prospective randomized trial.10 In addition, the characteristics and efficacy results reported by these programs are based on a limited series of selected patients. Therefore, there is a need for a wide overview in an unselected, ‘all-comer’ population to increase our understanding of the actual epidemiology of lung cancer biomarkers and of their potential impact on therapeutic strategies.
The French National Cancer Institute (i.e., INCa) funded a nationwide program for the systematic routine analysis of EGFR mutations and ALK rearrangements as well as of HER2, KRAS, BRAF and PIK3CACA mutations in advanced stage non-squamous non-small cell lung cancer (NSCLC) patients in 28 certified molecular genetics centers covering the entire French territory.13–15 Here, we report the results of the ‘Biomarkers France’ study, which assessed the characteristics, molecular profiles and clinical outcomes of 17,664 consecutive NSCLC patients who were screened during a one-year period by this program.
PATIENTS AND METHODS Patients
All consecutive NSCLC patients who were routinely screened for molecular alterations during a one-year period at one of the 28 certified molecular genetics centers were eligible for this study. The prescription of this routine molecular screening, mandatory for advanced non-squamous NSCLC, was solely the responsibility of the treating physician. Notably, national recommendations for screening for EGFR mutations (both activating and T790M), ALK rearrangements and four ‘emerging biomarkers’ (KRAS, BRAF, HER2 and PIK3CACA mutations) have been available since 2010.16 In addition, NSCLC patients exhibiting a less advanced stage or patients carrying other tumor types (i.e., mixed histology, especially, never-smoker patients, etc) could have been screened upon approval by their local multidisciplinary tumor board (MTB).
Ethics
This study was approved by a national ethics committee for observational studies (Comité d’Evaluation des Protocoles de Recherche Observationnelle, CEPRO) on 09/28/2011, by the French Advisory Committee on Information Processing in Material Research in the Field of Health (Comité Consultatif sur le Traitement de l'Information en Matière de Recherche dans le Domaine de la Santé, CCTIRS) on 09/22/2011 and by the National Commission of Informatics and Liberty (CNIL) on 12/18/2011, according to French laws, and was registered on the ClinicalTrials.gov website (NCT01700582).
Each clinician identified as the prescriber of a molecular analysis between April 2012 and April 2013 received written information describing the study protocol and the process for accessing the database as well as a confidential password to connect to the ‘Biomarkers France’ secured Web CRF.
All NSCLC patients included in this program received information from their institution or referring clinician, as recommended by competent authorities, that specified that, according to French laws, they were allowed to ask for complete access to/removal of their own collected data.
Study oversight
This study was sponsored by the French Cooperative Thoracic Intergroup (IFCT) and was funded by an unrestricted grant from INCa. The steering committee included representatives of the certified molecular genetics centers, INCa, and the IFCT. The feasibility and potential technical issues of this project were initially evaluated by analyzing patients screened at three certified molecular genetics centers over a 3-month period (November 2011 to January 2012); this analysis served as a test for the subsequent nationwide study. As no major difficulty was observed for these initial 346 patients, the steering committee decided to open the one-year national recruitment period in April 2012. The 28 molecular genetics centers had to send their results to the IFCT using a specific datasheet for each patient. Then, the data were recorded and monitored by the IFCT. The authors had full access to the de-identified data and analyses for the current report.
Molecular analyses
The molecular analyses of EGFR (NG_007726.3), HER2 (NG_007503.1), KRAS (NG_007524.1), BRAF (NG_007873.3), and PIK3CACA (NG_012113.2) mutations and of ALK (NG_009445.1) rearrangements were performed on a routine basis at 28 certified molecular genetics centers (supplementary Figure 1). The methodology for these analyses16 and the results of the prospective cross validation quality assessment studies were previously reported.17–19 Briefly, each molecular genetics center used either the Sanger sequencing method or a more sensitive validated allele-specific technique (generally to be confirmed by Sanger sequencing) to assess EGFR (exons 18-21)17-18, HER2 (exon 20), BRAF (exon 15),
KRAS (exon 2)17-19 and PIK3CACA (exons 9 and 20) mutations (supplementary Table 1). A certified break-apart fluorescence in situ hybridization assay was used to assess ALK rearrangements.20 In addition, each regional genetics center performed either concurrent analysis of all recommended molecular alterations in the six genes or a sequential approach in which the EGFR and ALK assessments were performed first, and then each of the other molecular alterations were assessed until a mutation was found.
Data collection
The results of the molecular assessments of EGFR, HER2, KRAS, BRAF, and PIK3CA mutations and ALK rearrangements as well as histological typing and the percentage of tumor cells by the referring pathologist and the turnaround time before obtaining the analysis results (from the date of tumor reception to the date of submission of the written molecular report to the clinician) were provided directly to the IFCT by the certified molecular genetics centers. The results regarding the median time until results were obtained were expressed as first and third quartiles (Q1 and Q3) to avoid excessive data dispersion. Simultaneously, the treating physician of each patient (n=3,831) was provided with secure access to her/his own patient’s data. Data on sex; ethnicity (Asian versus non-Asian); smoking history (never, former or current smoker); past familial medical history of cancer; ECOG PS (0-1 versus 2 or more); TNM stage, as defined by the seventh edition of the American Joint Committee on Cancer;21 pathological diagnosis, as defined by the 2011 International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society (IASLC/ATS/ERS) classification;22 and the modality of sample’s collection (bronchoscopy, thoracic transthoracic biopsy, thoracic surgery or other) were collected. The type of treatment (standard chemotherapy, type of chemotherapy, targeted therapy, or, if applicable, the clinical trial along with the type of treatment), the impact of the molecular results on the treatment decision, and outcomes (overall response rate, assessed – usually by RECIST - by treating
physician; ORR; first-line and second-line, when applicable, date(s) of progression and survival status) were collected and reported per investigator review. The patients were treated on a routine basis after a local MTB and in accordance with national and international guidelines.23–25 At the time the study was conducted, erlotinib and gefitinib were approved for the treatment of patients with EGFR mutations (including first-line) while crizotinib was available only for the second-line treatment of patients with ALK rearrangements. KRAS, BRAF, HER2 and PI3K mutations were targetable by drugs available through clinical trials. The connection to and completion of the database were voluntarily performed by the treating physicians.
Objectives
The primary objective of this study was to describe the frequency of the molecular alterations in six genes that were routinely screened via a nationwide approach in consecutive patients with NSCLC. The secondary objectives were to combine the clinical and biological databases, to document the turnaround time in obtaining the molecular results, to assess the ability of the treating physician to use these data to select an ad hoc therapy (on a standard basis or via inclusion in a clinical trial) and to measure the patients’ outcomes (progression-free survival, PFS; and overall survival, OS).
Statistics
Descriptive statistics, including median and range or quartiles for continuous variables or frequencies and percentages for categorical variables, were used. The median follow-up duration was defined as the time from the date of molecular analysis assessment to the closing date of the analysis. First-line PFS was defined as the time from the date of molecular analysis assessment to the date of the first progression or death due to any cause. Second-line PFS was defined as the time from second-line treatment initiation to the date of the second progression or death from any cause. The OS duration was defined as the date of the
molecular analysis assessment to the date of death or final follow-up. Survival curves were estimated globally and for groups of interest using the Kaplan-Meier method. We compared the groups of interest using the two-sided log-rank test. The characteristics (with or without mutation) of each biomarker of the patients were compared using the Chi-square test for qualitative variables or using the nonparametric test for quantitative variables. Univariate Cox models were applied to select the most promising prognostic variables (threshold p= 0.20). A multivariate Cox model was then applied to adjust for potential confounders (clinical or molecular characteristics associated with PFS or OS). Adjusted HRs with 95% CIs were calculated. All statistical tests were two-sided, and a P-value of less than 0.05 was considered statistically significant. All analyses were performed using SAS software, version 9.3 (SAS Institute).
Role of the funding source.
The INCa funded this study. One INCa representative participated in the steering committee of the study. The funding source has no role in study design, data collection, analysis, and interpretation, and preparation of this manuscript.
RESULTS
Timelines and flow chart
The study recruitment period was from April 2012 to April 2013, and the database was locked for the current analysis on July 23, 2014. Overall, 19,386 results of routine molecular analyses were recorded in the database. After review, 707 analyses (3.6%) were excluded, primarily because they were related to other types of solid tumors (Figure 1). Finally, 18,679 results, representing 17,664 NSCLC patients, were analyzed.
Patients and results of the molecular analyses
The primary characteristics of the 17,664 patients are summarized in Table 1. The number of samples analyzed per patient was typically one (n=16,696, 94.5%) but two or more samples were analyzed in 927 (5.2%) and 41 (0.2%) of the patients, respectively. The median interval between tissue specimen collection and the initiation of molecular analysis was 8 days (4-16 days), and the median interval from the initiation of molecular analysis to the final written report of the analysis (the results for EGFR mutation if the analyses were conducted sequentially) was 11 days (7-16 days).
The frequencies of EGFR, HER2, KRAS, BRAF, and PIK3CA mutations and of ALK rearrangements were 11.0, 0.8, 28.7, 1.9, 2.3 and 4.8%, respectively (Table 2 and supplementary Table 2). The frequencies of these molecular alterations in these six genes, overall and for three specific subgroups (i.e., adenocarcinoma, women, and never-smokers), are displayed in Figure 2. The screen failure rates varied from 1.4 to 4.3%. In 173 cases, two (n=170, 0.9%) or more (n=3, 0.01%) molecular alterations were identified in the same sample (supplementary Table 3).
Treatment
For 51.3% of the cases, the results of the routine molecular profiling were considered when making the decision for the first-line therapeutic strategy; however, in 22.6% of the cases, the
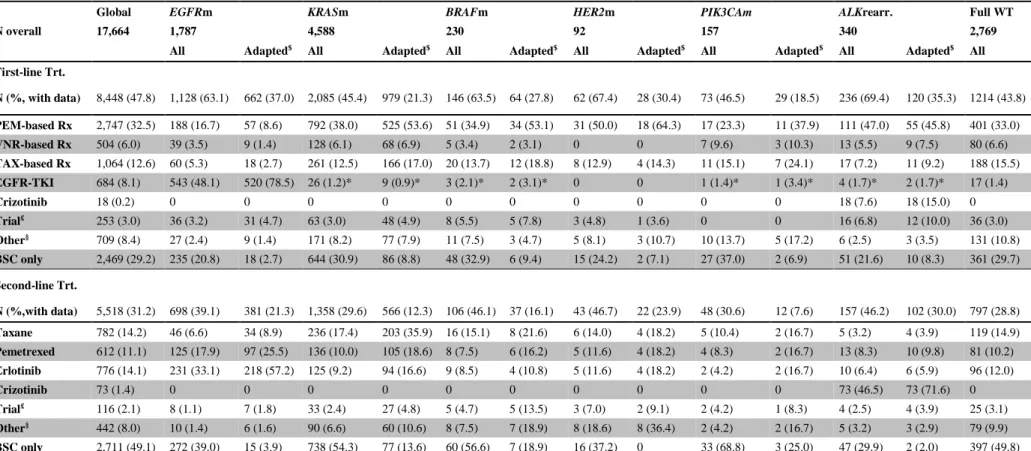
turnaround time in obtaining these results motivated the local MTB to decide the treatment strategy without knowing these results. The frequencies of genetic alterations in the patients who were managed without considering the results of the molecular analyses (data not shown) were comparable to that of the overall patient population, except for EGFR mutations (11.3 and 13.3%, respectively; p=0.003). The types of first- and second-line treatments are summarized in Table 3. As treatment-resistant EGFR T790M mutations were systematically found concomitantly with an activating EGFR mutation, the frequencies of these two mutations are jointly reported. Radiotherapy was performed to improve local thoracic control or as a palliative treatment in 10.1 and 25.7% of patients, respectively.
Clinical outcomes
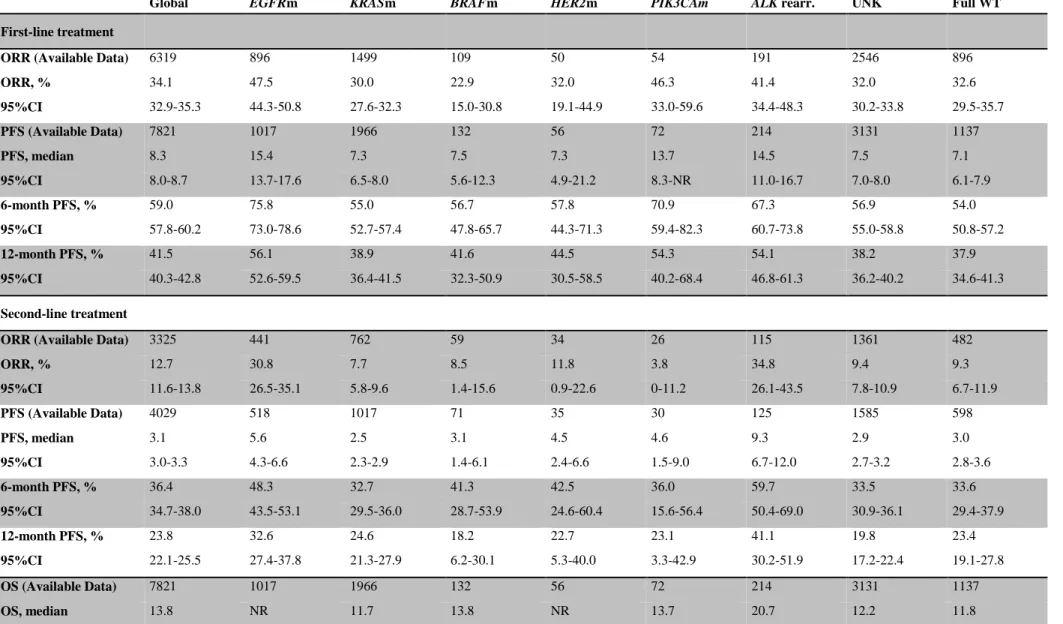
The median follow-up duration at the time of analysis was 24.9 months (95%CI: 24.8-25.0). The outcomes of the 17,664 patients for which data were available are summarized in Table 4 (and in supplementary Table 4 for advanced stage patients only). The presence of a genetic alteration was associated with a significantly higher ORR for the first-line treatments (36.5% [95%CI: 34.7-38.2] versus in the absence of a genetic alteration (32.6% [95%CI: 29.5-35.6]) (p=0.03) and for the second-line treatments (16.9% [95%CI: 15.0-18.8]) versus in the absence of a genetic alteration (9.3% [95%CI: 6.7-11.9]) (p<0.001). The presence of a genetic alteration was also associated with significantly longer first-line PFS and OS (Figure 3 and supplementary Figure 2). When excluding the patients carrying an EGFR mutation, the presence of a genetic alteration (versus absence) resulted in a non-significant difference in ORR in first-line (31.4% [95%CI: 29.4-33.5] versus 32.6% [95%CI: 29.5-35.7]; p=0.54) or second-line (11% [95%CI: 9.1-12.8] versus 9.3% [95%CI: 6.7-11.9]; p=0.34) and OS (13.3 months [95%CI: 12.1-14.3] versus 11.8 months [95%CI: 10.1-13.4]; p=0.37). Cox multivariate analysis confirmed that ALK rearrangements (HR=0.70 [95%CI: 0.5-0.9]),
EGFR (HR=0.53 [95%CI: 0.4-0.6]) and HER2 mutations (HR=0.60 [95%CI: 0.4-1.0]) had a favorable prognostic impact (supplementary Table 5).
DISCUSSION
One challenge of personalized medicine is the provision of all cancer patients with an assessment of molecular alterations that are related to the management of their disease. The results reported in this study illustrate the success of the nationwide, INCa-organized program in this setting. The molecular screening performed in the current program, which involved more than 20,000 advanced NSCLC patients per year, enabled the detection with an acceptable turnaround time of at least one potentially actionable molecular alteration in almost 50% of the analyses and impacted the treatment decisions for 51.3% of the included patients. When a genetic alteration was detected, the primary outcome was a 4.7 months longer median OS indicating a possible prognostic advantage and/or a major change in the management paradigm for these lung cancer patients.
The successful implementation of molecular profiling of lung cancer patients at a single institution or in a consortium of institutions has been reported previously.8-11 However, the number of examined patients was frequently small, as the largest number reported previously was 1007 patients. The LCMC initiative was the largest multi-institutional study in the western world. Our study follows LCMC but aims at broadening the number of centers able to provide molecular profiling with a clear ambition to be a nation-wide approach. Considering that 39,000 new lung cancer cases (any stage and histology) are reported each year in France,26 18,000 advanced non-squamous NSCLC patients represent the number of patients which is expected to be screened for EGFR mutations and ALK rearrangements according to the current guidelines.23-25 With 17,664 patients, the results reported here not only involved the largest sample but also were unlikely to have been influenced by the patients’ selection of a specific institution or participation in a given clinical trial or research program.27
The frequency of some molecular alterations might appear to be lower than previously reported (11% of EGFR mutations compared with 17% for LCMC in the US),9 but the
present results most likely more closely reflect the characteristics of a global population, particularly in Western countries. To the best of our knowledge, no other study at a nationwide level has been published to date. Therefore, these results provide solid evidence for clinical trials or routine programs of molecular screening of lung cancer patients, especially considering the HER2, BRAF and PIK3CA data because very rare genetic alterations (0.8, 1.9 and 2.3%, respectively) could still represent a large population given the high incidence of lung cancer worldwide.
This study attempted to collect data on a common cancer population from daily practice during a one-year period. Based on this objective, a relatively simple case report form was selected and more than 3,800 treating physicians were approached for the study. This design implied the acceptance of some degree of missing data, which was a drawback of this study. Another limitation of the study is related to the molecular alterations that were screened. Several potential actionable targets for lung cancer have been described in recent years, and some of these targets were not included in the program reported here. The molecular alterations screened in this program were selected in 2009 and this strategy was primarily a success, as the data for BRAF6 and HER27 targeting are now robust. On the other hand, other emerging biomarkers, such as KRAS mutations, remained uncertain.28 PIK3CA mutations are no longer routinely assessed, while ROS1 assessment is now part of the routine molecular testing at certified molecular genetics centers in France.29 More importantly, the results reported here do not suggest an improvement in the inclusion rate of clinical trials. As only 3% of the patients in the national database have been enrolled in clinical trials while being assessed for molecular alterations that were actionable only using experimental compounds in clinical development, this objective of this national program remains to be met.29
In conclusion, this national program broadly (and very exhaustively) screened lung cancer patients for genetic alterations in six genes, including four emerging genetic alterations, to identify actionable targets that improved the survival of approximately 50% of these patients, although at a non-negligible financial cost.16 Therefore, these results encourage all ongoing worldwide initiatives to provide cancer patients with access to personalized medicine and provide robust information to organize these initiatives.
The Biomarkers France contributors (listed here are the representatives of each regional molecular genetics center and/or the pathologist who collaborated in this project and the treating physicians who provided data for 40 patients or more) not included in the list of authors:
Al Freijat F (Centre Hospitalier de Belfort-Montbéliard, Service de Pneumologie, Belfort, France)
Baala L (UF Biologie Moléculaire, Structure de Génétique, CHRO & UMR7355, INEM CNRS et Université Orléans, France)
Beaudoux O (Laboratoire de Biologie Médicale, Institut Jean-Godinot, Reims, France)
Bigay-Game L (Hôpital Larrey, Centre Hospitalier Universitaire, Service de Pneumologie, Toulouse, France)
Bonnette P (Hôpital Foch, Service de Chirurgie Thoracique, Suresnes, France)
Boyer JC (Plateforme de génétique moléculaire des cancers, Laboratoire de Biochimie, CHU Nîmes, Nîmes, France)
Chinet T (Hôpital Ambroise Paré, Assistance Publique Hôpitaux de Paris, Service de Pneumologie, Boulogne-Billancourt, France)
Coudert B (Centre Georges François Leclerc, Service d’Oncologie Médicale, Dijon) Daniel C (Institut Curie, Service d’Oncologie Médicale, Paris, France)
De Fraipont F (Institut de Biologie et Pathologie, Centre Hospitalier Universitaire, Service Biochimie des Cancers et Biothérapies, La Tronche, France
Descourt R (Hôpital Morvan, Centre Hospitalier Régional Universitaire, Service d’Oncologie Médicale, Brest, France)
Dubrez J (Clinique Lafourcade, Service Chirurgie Thoracique et Cardiovasculaire, Bayonne, France)
Fabre E (Hôpital Européen Georges Pompidou, Assistance Publique Hôpitaux de Paris, Paris, France)
Filaire M (Centre Jean Perrin, Service de Chirurgie Thoracique, Clermont-Ferrand, France) Forest F (Centre Hospitalier Universitaire, Service Anatomie et Cytologie pathologiques, Saint Etienne, France)
Foucher P (Bocage Central, Centre Hospitalier Universitaire, Hôpital de Semaine Cardiologie Pneumologie, Dijon, France)
Gervais R (Centre Francois Baclesse, Service de pneumologie, Caen, France) Gossot D (Institut Montsouris, Département Chirurgie Thoracique, Paris, France)
Gury JP (Centre Hospitalier Intercommunal de la Haute-Saône, Service de Pneumologie, Vesoul, France)
Hofman P (Hôpital Pasteur, Centre Hospitalier Universitaire, Laboratoire de Pathologie Clinique et Expérimentale, Nice, France)
Huet I (Hôpital Larrey, Centre Hospitalier Universitaire, Service de Pneumologie, Toulouse, France)
Janicot H (Hôpital Gabriel Montpied, Centre Hospitalier Universitaire, Service de Pneumologie, Clermont-Ferrand, France)
Jougon J (Hôpital Haut-Lévêque, Centre Hospitalier Universitaire, Service de Chirurgie Thoracique, Pessac, France)
Karayan-Tapon L (Centre Hospitalier Universitaire, Service de Cancérologie, Poitiers, France)
Labrousse F (Hôpital Dupuytren, Centre Hospitalier Universitaire, Service d’Anatomie Pathologique, Limoges, France)
Lacave R (Hôpital Tenon, Assistance Publique Hôpitaux de Paris, Service d’Histologie, Embryologie, Cytogénétique biologie tumorale et génétique moléculaire, Paris, France)
Lacroix L (Institut Gustave Roussy, Département de Biologie et Pathologie Médicales, Villejuif, France)
Lafitte JJ (Hôpital Albert Calmette, Centre Hospitalier Régional Universitaire, Service d’Oncologie Thoracique et Pneumologie, Lille, France)
Le Chevalier T (Institut Gustave Roussy, Service de Médecine, Villejuif, Paris)
Le Gac G (Hôpital Morvan, Centre Hospitalier Universitaire, Laboratoire de Génétique Moléculaire et d’Histocompatibilité, Brest, France)
Lehmann-Che J (Hôpital Saint-Louis, Assistance Publique Hôpitaux de Paris, Molecular Oncology Unit and University Paris-Diderot, Paris)
Léotard B (Hôpital Brabois, Centre Hospitalier Universitaire, Service de Génétique, Vandoeuvre-Les-Nancy, France)
Leroy K (Hôpital Henri Mondor, Université Paris Est Créteil, Département de Pathologie, INSERM U955, équipe N°9, Créteil, France)
Marcq M (Les Oudairies, Centre Hospitalier Départemental, Roche-sur-Yon, France)
Martin L (Hôpital du Bocage, Centre Hospitalier Universitaire, Service de pathologie, Dijon, France)
Martin M (Centre Hospitalier d’Angoulême, Service d’Oncologie, Angoulême, France) Martinet Y (Hôpital Brabois, Centre Hospitalier Régional Universitaire, Vandoeuvre-lès-Nancy, France)
Merle P (Hôpital Gabriel Montpied, Centre Hospitalier Universitaire, Service de Pneumologie, Clermont-Ferrand, France)
Moreau L (Hôpital Pasteur, Hôpitaux Civils de Colmar, Service de Pneumologie, Colmar, France)
Morel A (Institut de Cancérologie de L’Ouest Site Paul Papin, Service d’Oncologie Biologie, Angers, France)
Moro-Sibilot D (Hôpital Michallon, Centre Hospitalier Universitaire, Service de Pneumologie, La Tronche, France)
Mougin C (Hôpital Jean Minjoz, Centre Hospitalier Régional Universitaire, Laboratoire de Biologie Cellulaire et Moléculaire, Besançon, France)
Nocent Ejanaini C (Centre Hospitalier Côte Basque, Service de Pneumologie, Bayonne, France)
Ouafi L (Institut Curie, Département de Biopathologie, Paris)
Pagès JC (Hôpital Trousseau, Centre Hospitalier Régional Universitaire, Service de Biochimie et Biologie Moléculaire, Tours, France)
Pedeutour F (Faculté de Médecine, Laboratoire de Génétique des Tumeurs Solides, Nice, France)
Poudenx M (Centre Antoine Lacassagne, Service de Pneumologie, Nice, France)
Pouessel D (Hôpital Saint Louis, Assistance Publique Hôpitaux de Paris, Service d’Oncologie Médicale, Paris, France)
Régnard JF (Hôpital Cochin, Assistance Publique Hôpitaux de Paris, Service de Chirurgie Thoracique, Paris, France)
Richard N (Hôpital de la Côte de Nacre, Centre Hospitalier Universitaire, Service de Génétique, Caen, France)
Rodier JM (Hôpital Bichat – Claude-Bernard, Assistance Publique Hôpitaux de Paris, Service de Cancérologie, Paris, France)
Solassol J (Hôpital Arnaud de Villeneuve, Centre Hospitalier Régional Universitaire, Université Montpellier 1, Laboratoire de Biopathologie, Montpellier, France)
Soubeyran I (Institut Bergonié, Service de Biopathologie, Bordeaux, France)
Souquet PJ (Centre hospitalier Lyon Sud, Hospices Civils de Lyon, Service de Pneumologie, Pierre-Bénite, France)
Thiberville L (Hôpital Charles Nicolle, Centre Hospitalier Universitaire, Service de Pneumologie, Rouen, France)
Thomas De Montpréville V (Centre Chirurgical Marie Lannelongue, Service d’Anatomie Pathologique, Le Plessis Robinson, France)
Acknowledgements:
Prof. D. Maraninchi (past President, Institut National du Cancer, Paris, France), Prof. A. Buzyn (President, Institut National du Cancer, Paris, France), Prof. F. Calvo (past Head of Research Department, Institut National du Cancer, Paris, France) Dr. F. Nowak (Institut National du Cancer, Paris, France) for their constant support;
A. Deroy (Intergroupe Francophone de Cancérologie Thoracique, Paris, France), S. Dos Santos (Intergroupe Francophone de Cancérologie Thoracique, Paris, France) for their participation to data collection, monitoring and computing.
Author contributions:
FB, JM, DD, FM, BM, QT, JC, JCS et GZ designed the study, all the authors participated in the data acquisition, FB, JM, FM, QT, PM, AL, JC, JCS, and GZ analyzed and interpret the data, FB, JM, FM, QT, PM, AL, JC, JCS, and GZ wrote the manuscript; the final version was approved by all the authors. No writing assistance other than copy editing was provided to the authors.
REFERENCES
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013;63:11–30.
2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74.
3. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39.
4. Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94.
5. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21.
6. Planchard D, Mazieres J, Riely GJ, et al. Interim results of phase II study BRF113928 of dabrafenib in braf V600E mutation–positive non-small cell lung cancer (NSCLC) patients. J Clin Oncol 2013;31(Suppl)8009.
7. Mazières J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol 2013;31:1997-2003. 8. Camidge DR, Ou SH, Shapiro G, et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). J Clin Oncol 2014;32 (Suppl):5s; [Abstr:8001)].
9. Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006.
10. Serizawa M, Koh Y, Kenmotsu H, et al. Assessment of mutational profile of Japanese lung adenocarcinoma patients by multitarget assays: a prospective, single-institute study. Cancer 2014;120:1471-81.
11. Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011;22:2616-24.
12. Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn 2011;13:74-84.
13. Andre F, Nowak F, Arnedos M, Lacroix L, Viens P, Calvo F. Biomarker discovery, development, and implementation in France: a report from the French National Cancer Institute and cooperative groups. Clin Cancer Res 2012;18:1555-60.
14. Nowak F, Soria JC, Calvo F. Tumour molecular profiling for deciding therapy-the French initiative. Nat Rev Clin Oncol 2012;9:479-86.
15. Nowak F, Calvo F, Soria JC. Europe does it better: molecular testing across a national health care system-the French example. AM SOC Clin Oncol Educ Book 2013:332-7.
16. Institut National du Cancer, ed. Synthèse de l’activité des plateformes hospitalières de génétique moléculaire des cancers en 2012, en vue d’optimiser leur évolution. Collection Bilans d’activité et d’évaluation, Boulogne-Billancourt, France: l’INCa, 2014.
17. Beau-Faller M, Degeorges A, Rolland E, et al. Cross-validation study for epidermal growth factor receptor and KRAS mutation detection in 74 blinded non-small cell lung carcinoma samples: a total of 5550 exons sequenced by 15 molecular French laboratories (evaluation of the EGFR mutation status for the administration of EGFR-TKIs in non-small cell lung carcinoma [ERMETIC] project--part 1). J Thorac Oncol 2011;6:1006-15.
18. Blons H, Rouleau E, Charrier N, et al.; MOKAECM Collaborative Group. Performance and cost efficiency of KRAS mutation testing for metastatic colorectal cancer in routine diagnosis: the MOKAECM study, a nationwide experience. PLOS ONE 2013;8:e68945.
19. Beau-Faller M, Blons H, Domerg C, et al. A multicenter blinded study evaluating EGFR and KRAS mutation testing methods in the clinical non-small cell lung cancer setting--IFCT/ERMETIC2 Project Part 1: Comparison of testing methods in 20 French molecular genetic National Cancer Institute platforms. J Mol Diagn 2014;16:45-55.
20. McLeer-Florin A, Moro-Sibilot D, Melis A, et al. Dual IHC and FISH testing for ALK gene rearrangement in lung adenocarcinomas in a routine practice: a French study. J Thorac Oncol 2012;7(2):348-54.
21. Goldstraw P, Crowley J, Chansky K, et al.. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol 2007;2:706-14.
22. Travis WD, Brambilla E, Riely GJ. New pathologic classification of lung cancer: relevance for clinical practice and clinical trials. J Clin Oncol 2013;31:992-1001.
23. Depierre A, Lagrange JL, Theobald S, et al. Summary report of the Standards, Options and Recommendations for the management of patients with non-small-cell lung carcinoma (2000). Br J Cancer 2003;89 (Suppl 1):S35-49.
24. Azzoli CG, Temin S, Aliff T, et al. 2011 Focused update of 2009 American Society of Clinical Oncology clinical practice guideline update on chemotherapy for stage iv non-small-cell lung cancer. J Clin Oncol 2011;29:3825-31.
25. Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25 (Suppl 3):iii27-39.
26. Institut National du Cancer, ed. Les cancers en France en 2013. Collection état des lieux et des connaissances. Boulogne-Billancourt, France: l’INCa, 2014.
27. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511(7511):543-50.
27. Martin P, Leighl NB, Tsao MS, Shepherd FA. KRAS mutations as prognostic and predictive markers in non-small cell lung cancer. J Thorac Oncol 2013;8:530-42.
28. Plateformes hospitalières de génétique moleculaire des cancers: faits marquants et synthèse d’activité 2013. Collection bilans d’activité et evaluation, ouvrage collectif édité par l’INCa, Boulogne-Billancourt, Novembre 2014
29. Barlesi F, Zalcman G, Mazieres J. How to promote and organize clinical research in lung cancer. In: Pass HI, Ball D, Scagliotti GV, eds. The IASLC Multidisciplinary Approach to Thoracic Oncology. Aurora, CO: International Association for the Study of Lung Cancer, 2014.
Table 1: Characteristics of the 17,664 patients whose results of routine molecular analyses were available in the database.
Data availability N %
N (%)
Age, years (median, range) 17,664 64.5 (18-98)
Sex 17,555 Male 11,346 64.6
Female 6,209 35.4
Ethnicity 7,350 Asian 96 1.3
Caucasian or other 7,254 98.7
Smoking history 8,619 Never 1,619 18.8
Former smoker 3,597 41.7
Current smoker 3403 39.5
ECOG PS 7,817 0/1 5,607 71.7
2 1,423 18.2
3-4 787 10.1
Previous cancer 7,848 Within family 961 12.2
TNM 2007 stage 8,637 I/II 1,392 16.1
III/IV/relapse 7,245 83.9
Histology 17,664 Adenocarcinoma 13,425 76.0
Large cell carcinoma 589 3.3
NOS or other histology 2,773 15.7
Sample collection 17,664 Bronchoscopy 5,038 28.5
CT-guided transthoracic biopsy 4,229 23.9
Surgery 4,712 26.7
Others/unknown 3,685 20.9
Samples analyzed per patient 17,664 1 16,696 94.5
>1 968 5.5
Turnaround time, days 18,679* From sample collection to initiation of analysis 8.0 8.0 (4.0-16.0) -
(median, Q1-Q3) From initiation of analysis to report of results 11.0 (7.0-16.0) -
Table 2: Results of the 18,679 molecular analyses stratified by clinical characteristics.
EGFR, N (%) KRAS, N (%) BRAF, N (%)
M R (T790M) WT UNK M WT UNK M WT UNK
N (%) 1,786 (9.5) 161 (0.9) 15,759 (84.4) 973 (5.2) 4,894 (26.2) 12,107 (64.8) 1,678 (9.0) 262 (1.4) 13,644 (73.0) 4,773 (25.6) Age (median) 68.4§ 65.7§ 64.5§ 65.8§ 63.3§ 65.4§ 66.6§ 65.9 64.7 65.7 Sex* Male 568 (31.8) § 49 (30.4) § 10,699 (67.9) § 631 (64.9) § 3,245 (66.3) § 7,698 (63.6) § 1,004 (59.8) § 160 (61.1) 8,881 (65.1) 2,906 (60.9) Female 1,208 (67.6) § 111 (68.9) § 4,963 (31.5) § 339 (34.8) § 1,621 (33.1) § 4,331 (35.8) § 669 (39.9) § 101 (38.5) 4,686 (34.3) 1,834 (38.4) Ethnicity Asian 49 (5.0) § 6 (7.1) § 46 (0.7) § 7 (1.6) § 15 (0.8) § 79 (1.5) § 14 (1.8) § 0 72 (1.2) 36 (1.9) Other 938 (95.0) § 79 (92.9) § 6,365 (99.3) § 421 (98.4) § 1,954 (99.2) § 5,101 (98.5) § 748 (98.2) § 150 (100) 5,800 (98.8) 1,853 (98.1)
Smoking history Never 683 (59.9) § 53 (57.0) § 939 (12.4) § 98 (20.4) § 138 (5.9) § 1,397 (23.0) § 238 (28.0) § 41 (25.0) 1,229 (18.0) 503 (22.0)
Former 316 (27.7) § 31 (33.3) § 3,305 (43.7) § 213 (44.3) § 1,104 (46.9) § 2,410 (39.7) § 351 (41.3) § 63 (38.4) 2,887 (42.3) 915 (40.1)
Current 142 (12.4) § 9 (9.7) § 3,312 (43.8) § 170 (35.3) § 1,113 (47.3) § 2,260 (37.3) § 260 (30.6) § 60 (36.6) 2,709 (39.7) 864 (37.9)
ECOG PS 0 or 1 784 (76.0) § 71 (78.0) § 4,916 (71.8) § 314 (70.4) § 1,526 (71.5) 4,028 (73.2) 531 (68.0) 109 (74.1) 4,538 (72.7) 1,438 (70.8)
>2 247 (24.0) § 20 (22.0) § 1,932 (28.2) § 132 (29.6) § 609 (28.5) 1,472 (26.8) 250 (32.0) 38 (25.9) 1,700 (27.3) 593 (29.2)
Previous cancer Family 148 (13.8) § 23 (24.7) § 834 (12.3) § 45 (9.8) § 267 (12.7) 703 (12.7) 80 (9.9) 19 (12.3) 775 (12.4) 256 (12.6)
Stage (TNM 2007) I/II 177 (15.4) 13 (13.4) 1,248 (16.5) 61 (12.5) 391 (16.7) 1,015 (16.7) 93 (10.9) 23 (13.9) 1,143 (16.7) 333 (14.6) III/IV/relapse 971 (84.6) 84 (86.6) 6,293 (83.5) 428 (87.5) 1,955 (83.3) 5,063 (83.3) 758 (89.1) 143 (86.1) 5,692 (83.3) 1,941 (85.4) Histology ADC 1,502 (84.1) § 145 (90.1) § 11,854 (75.2) § 742 (76.3) § 4,069 (83.1) § 8,845 (73.1) § 1,329 (79.2) § 228 (87.0) § 10,610 (77.8) § 3,405 (71.3) §
SCQ 23 (1.3) § 1 (0.6) § 838 (5.3) § 47 (4.8) § 47 (1.0) § 792 (6.5) § 70 (4.2) § 1 (0.4) § 708 (5.2) § 200 (4.2) §
LCC 25 (1.4) § 1 (0.6) § 557 (3.5) § 31 (3.2) § 131 (2.7) § 432 (3.6) § 51 (3.0) § 6 (2.3) § 471 (3.5) § 137 (2.9) §
NOS & Others 236 (13.2) § 14 (8.7) § 2,510 (15.9) § 153 (15.7) § 647 (13.2) § 2,038 (16.8) § 228 (13.6) § 27 (10.3) § 1,855 (13.6) § 1,031 (21.6) §
Turnaround time, days Coll./Lab. 7 (3-15) § 9 (3-16) § 8 (4-16) § 9 (5-27) § 8 (4-15) 8 (4-16) 10 (5-25) 9 (6-15) 8 (4-16) 7 (0-15)
M, mutated (i.e. activating mutation); R, resistant mutation; WT, wild type; UNK, unknown (not contributive or not done analysis); ECOG PS, ECOG performance status; ADC, adenocarcinoma; SCQ, squamous cell carcinoma; LCC, large cell carcinoma; NOS, not otherwise specified; Full WT, patients with an established molecular profile without an EGFR, KRAS, BRAF, HER2, or PIK3CA mutation or ALK rearrangement; *, in 109 cases, the sex was not specified in the report (0.6%); §, comparison between the population with the considered molecular alteration and the population with unknown or full WT is significantly different, p<0.05. See also supplementary Table 2 for percentage presented per row.
Table 2 (cont): Results of the 18,679 molecular analyses stratified by clinical characteristics.
HER2, N (%) PIK3CA, N (%) ALK, N (%) Full WT, N (%)
M WT UNK M WT UNK Rearranged WT UNK
N (%) 98 (0.5) 11,625 (62.3) 6,956 (37.2) 252 (1.4) 10,426 (55.8) 8,001 (42.8) 388 (2.1) 7,746 (41.5) 10,545 (56.4) 2,833 (15.2) Age (median) 66.2 64.7 65.3 67.9§ 64.6§ 65.3§ 61.2§ 65.0§ 65.1§ 64.8 Sex Male 40 (40.8) § 7,577 (65.2) § 4,330 (62.2) § 154 (61.1) 6,812 (65.3) 4,981 (62.3) 206 (53.1) § 5,016 (64.8) § 6,725 (63.8) § 2,033 (71.8) Female 58 (59.2) § 3,971 (34.2) § 2,592 (37.3) § 98 (38.9) 3,549 (34.0) 2,974 (37.2) 180 (46.4) § 2,675 (34.5) § 3,766 (35.7) § 775 (27.4) Ethnicity Asian 0 64 (1.3) 44 (1.6) 1 (1.0) 63 (1.3) 44 (1.4) 5 (2.1) 38 (1.1) 65 (1.5) 8 (0.7) Other 63 (100) 5,054 (98.7) 2,686 (98.4) 101 (99.0) 4,634 (98.7) 3,068 (98.6) 238 (97.9) 3,304 (98.9) 4,261 (98.5) 1,141 (99.3) Smoking history Never 42 (63.6) § 1,065 (17.6) § 666 (21.0) § 38 (30.9) § 987 (18.1) § 748 (20.3) § 116 (43.3) § 697 (18.6) § 960 (18.3) § 167 (13.1)
Former 16 (24.2) § 2,553 (42.3) § 1,296 (40.9) § 49 (39.8) § 2,292 (42.0) § 1,524 (41.3) § 92 (34.3) § 1,638 (43.6) § 2,135 (40.7) § 574 (45.1)
Current 8 (12.1) § 2,419 (40.1) § 1,206 (38.1) § 36 (29.3) § 2,178 (39.9) § 1,419 (38.4) § 60 (22.4) § 1,422 (37.8) § 2,151 (41.0) § 532 (41.8)
ECOG PS 0/1 51 (81.0) 3,921 (72.2) 2,113 (72.2) 80 (70.2) 3,610 (73.0) 2,395 (71.3) 205 (80.7) § 2,476 (71.2) § 3,404 (72.7) § 817 (69.2)
>2 12 (19.0) 1,506 (27.8) 813 (27.8) 34 (29.8) 1,334 (27.0) 963 (28.7) 49 (19.3) § 1,003 (28.8) § 1,279 (27.3) § 364 (30.8)
Previous cancer Family 8 (11.8) 731 (13.3) 311 (10.9) 14 (12.5) 633 (12.6) 403 (12.2) 28 (11.0) 457 (13.2) 565 (12.0) 156 (13.2) Stage I/II 5 (7.2) § 1,086 (18.0) § 408 (12.8) § 25 (19.8) 980 (17.9) 494 (13.4) 34 (12.5) 527 (14.0) 938 (17.9) 215 (16.8) III/IV/relapse 64 (92.8) § 4,931 (82.0) § 2,781 (87.2) § 101 (80.2) 4,480 (82.1) 3,195 (86.6) 238 (87.5) 3,228 (86.0) 4,310 (82.1) 1,061 (83.2) Histology ADC 90 (91.8) § 8,959 (77.1) § 5,194 (74.7) § 161 (63.9) § 8,079 (77.5) § 6,003 (75.0) § 331 (85.3) § 6,218 (80.3) § 7,694 (73.0) § 2,084 (73.6) SCQ 0§ 656 (5.6) § 253 (3.6) § 45 (17.9) § 577 (5.5) § 287 (3.6) § 4 (1.0) § 346 (4.5) § 559 (5.3) § 232 (8.2) LCC 0§ 436 (3.8) § 178 (2.6) § 8 (3.2) § 405 (3.9) § 201 (2.5) § 12 (3.1) § 262 (3.4) § 340 (3.2) § 140 (4.9) NOS or other 8 (8.2) § 1,574 (13.5) § 1,331 (19.1) § 38 (15.1) § 1,365 (13.1) § 1,510 (18.9) § 41 (10.6) § 920 (11.9) § 1,952 (18.5) § 377 (13.3) Turnaround time, days Coll. to Lab. 9 (5-23) 8 (5-16) 7 (1-15) 9 (5-17) 8 (5-16) 7 (1-16) 7 (3-13) § 7 (3-15) § 9 (5-17) § 7 (3-14)
Table 3: Patient treatment stratified by the line of therapy and by the results of routine molecular analyses.
Global EGFRm KRASm BRAFm HER2m PIK3CAm ALKrearr. Full WT
N overall 17,664 1,787 4,588 230 92 157 340 2,769
All Adapted$ All Adapted$ All Adapted$ All Adapted$ All Adapted$ All Adapted$ All
First-line Trt. N (%, with data) 8,448 (47.8) 1,128 (63.1) 662 (37.0) 2,085 (45.4) 979 (21.3) 146 (63.5) 64 (27.8) 62 (67.4) 28 (30.4) 73 (46.5) 29 (18.5) 236 (69.4) 120 (35.3) 1214 (43.8) PEM-based Rx 2,747 (32.5) 188 (16.7) 57 (8.6) 792 (38.0) 525 (53.6) 51 (34.9) 34 (53.1) 31 (50.0) 18 (64.3) 17 (23.3) 11 (37.9) 111 (47.0) 55 (45.8) 401 (33.0) VNR-based Rx 504 (6.0) 39 (3.5) 9 (1.4) 128 (6.1) 68 (6.9) 5 (3.4) 2 (3.1) 0 0 7 (9.6) 3 (10.3) 13 (5.5) 9 (7.5) 80 (6.6) TAX-based Rx 1,064 (12.6) 60 (5.3) 18 (2.7) 261 (12.5) 166 (17.0) 20 (13.7) 12 (18.8) 8 (12.9) 4 (14.3) 11 (15.1) 7 (24.1) 17 (7.2) 11 (9.2) 188 (15.5) EGFR-TKI 684 (8.1) 543 (48.1) 520 (78.5) 26 (1.2)* 9 (0.9)* 3 (2.1)* 2 (3.1)* 0 0 1 (1.4)* 1 (3.4)* 4 (1.7)* 2 (1.7)* 17 (1.4) Crizotinib 18 (0.2) 0 0 0 0 0 0 0 0 0 0 18 (7.6) 18 (15.0) 0 Trial£ 253 (3.0) 36 (3.2) 31 (4.7) 63 (3.0) 48 (4.9) 8 (5.5) 5 (7.8) 3 (4.8) 1 (3.6) 0 0 16 (6.8) 12 (10.0) 36 (3.0) Other§ 709 (8.4) 27 (2.4) 9 (1.4) 171 (8.2) 77 (7.9) 11 (7.5) 3 (4.7) 5 (8.1) 3 (10.7) 10 (13.7) 5 (17.2) 6 (2.5) 3 (3.5) 131 (10.8) BSC only 2,469 (29.2) 235 (20.8) 18 (2.7) 644 (30.9) 86 (8.8) 48 (32.9) 6 (9.4) 15 (24.2) 2 (7.1) 27 (37.0) 2 (6.9) 51 (21.6) 10 (8.3) 361 (29.7) Second-line Trt. N (%,with data) 5,518 (31.2) 698 (39.1) 381 (21.3) 1,358 (29.6) 566 (12.3) 106 (46.1) 37 (16.1) 43 (46.7) 22 (23.9) 48 (30.6) 12 (7.6) 157 (46.2) 102 (30.0) 797 (28.8) Taxane 782 (14.2) 46 (6.6) 34 (8.9) 236 (17.4) 203 (35.9) 16 (15.1) 8 (21.6) 6 (14.0) 4 (18.2) 5 (10.4) 2 (16.7) 5 (3.2) 4 (3.9) 119 (14.9) Pemetrexed 612 (11.1) 125 (17.9) 97 (25.5) 136 (10.0) 105 (18.6) 8 (7.5) 6 (16.2) 5 (11.6) 4 (18.2) 4 (8.3) 2 (16.7) 13 (8.3) 10 (9.8) 81 (10.2) Erlotinib 776 (14.1) 231 (33.1) 218 (57.2) 125 (9.2) 94 (16.6) 9 (8.5) 4 (10.8) 5 (11.6) 4 (18.2) 2 (4.2) 2 (16.7) 10 (6.4) 6 (5.9) 96 (12.0) Crizotinib 73 (1.4) 0 0 0 0 0 0 0 0 0 0 73 (46.5) 73 (71.6) 0 Trial£ 116 (2.1) 8 (1.1) 7 (1.8) 33 (2.4) 27 (4.8) 5 (4.7) 5 (13.5) 3 (7.0) 2 (9.1) 2 (4.2) 1 (8.3) 4 (2.5) 4 (3.9) 25 (3.1) Other§ 442 (8.0) 10 (1.4) 6 (1.6) 90 (6.6) 60 (10.6) 8 (7.5) 7 (18.9) 8 (18.6) 8 (36.4) 2 (4.2) 2 (16.7) 5 (3.2) 3 (2.9) 79 (9.9) BSC only 2,711 (49.1) 272 (39.0) 15 (3.9) 738 (54.3) 77 (13.6) 60 (56.6) 7 (18.9) 16 (37.2) 0 33 (68.8) 3 (25.0) 47 (29.9) 2 (2.0) 397 (49.8)
$, the treatment was selected considering the results of the molecular analyses (i.e., targeted therapy if an actionable alteration had been identified, chemotherapy for WT patients, etc); Trt, treatment; Rx, Regimen; PEM, pemetrexed; VNR, vinorelbine; TAX, taxanes; £, usually based on targeted agents; BSC, best supportive care; §, including, but not limited to, another type of chemotherapy, crizotinib via an expanded access program (ATU) before its registration, off-label targeted therapy, a non-registered combination of therapies, etc; *, patients with tumor displaying two molecular alterations including EGFR mutation.
Table 4: Global outcomes and outcomes stratified by line of therapy and by molecular alteration.
Global EGFRm KRASm BRAFm HER2m PIK3CAm ALK rearr. UNK Full WT
First-line treatment
ORR (Available Data) 6319 896 1499 109 50 54 191 2546 896
ORR, % 34.1 47.5 30.0 22.9 32.0 46.3 41.4 32.0 32.6 95%CI 32.9-35.3 44.3-50.8 27.6-32.3 15.0-30.8 19.1-44.9 33.0-59.6 34.4-48.3 30.2-33.8 29.5-35.7 PFS (Available Data) 7821 1017 1966 132 56 72 214 3131 1137 PFS, median 8.3 15.4 7.3 7.5 7.3 13.7 14.5 7.5 7.1 95%CI 8.0-8.7 13.7-17.6 6.5-8.0 5.6-12.3 4.9-21.2 8.3-NR 11.0-16.7 7.0-8.0 6.1-7.9 6-month PFS, % 59.0 75.8 55.0 56.7 57.8 70.9 67.3 56.9 54.0 95%CI 57.8-60.2 73.0-78.6 52.7-57.4 47.8-65.7 44.3-71.3 59.4-82.3 60.7-73.8 55.0-58.8 50.8-57.2 12-month PFS, % 41.5 56.1 38.9 41.6 44.5 54.3 54.1 38.2 37.9 95%CI 40.3-42.8 52.6-59.5 36.4-41.5 32.3-50.9 30.5-58.5 40.2-68.4 46.8-61.3 36.2-40.2 34.6-41.3 Second-line treatment
ORR (Available Data) 3325 441 762 59 34 26 115 1361 482
ORR, % 12.7 30.8 7.7 8.5 11.8 3.8 34.8 9.4 9.3 95%CI 11.6-13.8 26.5-35.1 5.8-9.6 1.4-15.6 0.9-22.6 0-11.2 26.1-43.5 7.8-10.9 6.7-11.9 PFS (Available Data) 4029 518 1017 71 35 30 125 1585 598 PFS, median 3.1 5.6 2.5 3.1 4.5 4.6 9.3 2.9 3.0 95%CI 3.0-3.3 4.3-6.6 2.3-2.9 1.4-6.1 2.4-6.6 1.5-9.0 6.7-12.0 2.7-3.2 2.8-3.6 6-month PFS, % 36.4 48.3 32.7 41.3 42.5 36.0 59.7 33.5 33.6 95%CI 34.7-38.0 43.5-53.1 29.5-36.0 28.7-53.9 24.6-60.4 15.6-56.4 50.4-69.0 30.9-36.1 29.4-37.9 12-month PFS, % 23.8 32.6 24.6 18.2 22.7 23.1 41.1 19.8 23.4 95%CI 22.1-25.5 27.4-37.8 21.3-27.9 6.2-30.1 5.3-40.0 3.3-42.9 30.2-51.9 17.2-22.4 19.1-27.8 OS (Available Data) 7821 1017 1966 132 56 72 214 3131 1137 OS, median 13.8 NR 11.7 13.8 NR 13.7 20.7 12.2 11.8
95%CI 13.3-14.4 10.6-13.1 8.5-21.9 8.7-NR 17.0-NR 11.5-13.0 10.1-13.5
6-month OS, % 70.0 84.2 64.5 67.8 80.6 74.0 80.2 68.4 67.6
95%CI 68.9-71.0 81.9-86.6 62.2-66.7 59.5-76.2 69.7-91.5 62.9-85.0 74.7-85.7 66.6-70.2 64.7-70.6
12-month OS, % 54.0 72.9 49.3 52.0 61.4 57.4 70.2 50.3 49.2
95%CI 52.7-55.3 69.8-75.9 46.6-51.9 42.4-61.6 47.0-75.7 43.4-71.4 63.6-76.8 48.2-52.4 45.7-52.7
ORR, overall response rate; PFS, progression free survival; OS, overall survival; UNK, Unknown representing the cases with at least one unknown result after the assessment of the six genes status, NR, not reached.
FIGURE LEGENDS
Figure 1: Flow-chart of the study (patients and molecular analyses performed).
Figure 2: Frequency of the genetic alterations in the six genes in the 18,679 analyzed samples (expressed as the percentage of positive samples for each molecular alteration relative to the number of available analyses, with UNK representing the cases with at least one unknown result after the assessment of the six genes status), global results (panel A), adenocarcinoma only (panel B), women only (panel C), and never-smokers only (panel D).
Figure 3: Outcomes of the 17,664 patients undergoing molecular analyses: first-line PFS for patients with and without genetic alteration (panel A); first-line PFS stratified by molecular profile (panel B); second-line PFS for patients with and without a genetic alteration (panel C); second-line PFS stratified by molecular profile (panel D); OS of patients with and without a genetic molecular alteration (panel E); and OS stratified by molecular profile (panel F). UNK in panel B, D and F represents the cases with at least one unknown result after the assessment of the six genes status.