Interaction between methionine synthase isoforms
and MMACHC: characterization in cblG-variant, cblG
and cblC inherited causes of megaloblastic anaemia
Ma’atem B. Fofou-Caillierez
1,{, Nadir T. Mrabet
1,{, Ce´line Che´ry
1, Natacha Dreumont
1,
Justine Flayac
1, Mihaela Pupavac
1,2, Justine Paoli
1, Jean-Marc Alberto
1, David Coelho
1,
Jean-Michel Camadro
4, Franc¸ois Feillet
1, David Watkins
2, Brian Fowler
3, David S. Rosenblatt
2and Jean-Louis Gue´ant
1,∗1
Inserm U954 Nutrition-Genetics-Environmental Risk Exposure and Reference Centre of Inborn Metabolism Diseases, Medical Faculty of Nancy University and University Hospital Centre, Nancy, France,2Department of Human Genetics, McGill University, Montreal, Quebec, Canada and3Metabolic Unit, University Children’s Hospital, Basel, Switzerland
4
Mitochondria, Metals and Oxidative Stress Group, Jacques Monod Institute, UMR7592 CNRS—Paris Diderot University, Paris, France
Received May 27, 2013; Revised May 27, 2013; Accepted June 25, 2013
The cblG and cblC disorders of cobalamin (Cbl) metabolism are two inherited causes of megaloblastic anaemia. In cblG, mutations in methionine synthase (MTR) decrease conversion of hydroxocobalamin (HOCbl) to methyl-cobalamin, while in cblC, mutations in MMACHC disrupt formation of cob(II)alamin (detected as HOCbl). Cases with undetectable methionine synthase (MS) activity are extremely rare and classified as ‘cblG-variant’. In four ‘cblG-variant’ cases, we observed a decreased conversion of cyanocobalamin to HOCbl that is also seen in cblC cases. To explore this observation, we studied the gene defects, splicing products and expression of MS, as well as MS/MMACHC protein interactions in cblG-variant, cblG, cblC and control fibroblasts. We observed a full-size MS encoded by MTR-001 and a 124 kDa truncated MS encoded by MTR-201 in cblG, cblC, control fibroblasts and HEK cells, but only the MTR-201 transcript and inactive truncated MS in cblG-variant cells. Co – immunoprecipi-tation and proximity ligation assay showed interaction between truncated MS and MMACHC in cblG-variant cells. This interaction decreased 2.2, 1.5 and 5.0-fold in the proximity ligation assay of cblC cells with p.R161Q and p.R206W mutations, and HEK cells with knock down expression of MS by siRNA, respectively, when com-pared with control cells. In 3D modelling and docking analysis, both truncated and full-size MS provide a loop anchored to MMACHC, which makes contacts with R-161 and R-206 residues. Our data suggest that the inter-action of MS with MMACHC may play a role in the regulation of the cellular processing of Cbls that is required for Cbl cofactor synthesis.
INTRODUCTION
Cobalamin (Cbl) exists in several forms in cells and body fluids, including hydroxocobalamin (HOCbl), methylcobalamin (MeCbl) and 5′-deoxyadenosylcobalamin (AdoCbl) (1). Me-thionine synthase (MS, EC 2.1.1.13) generates MeCbl during
the catalytic cycle of methionine synthesis and is deficient in the cblG defect (2). The MMACHC protein encoded by the 5-exon MMACHC gene mutated in the cblC defect (3) is a 31.7 kDa cytosolic Cbl trafficking chaperone that removes the upper axial ligand of exogenous Cbl (4,5). The generated cob(II)alamin in base-off conformation can be seen as HOCbl
†
The first two authors had equal contribution and should be regarded as joint First Authors.
∗To whom correspondence should be addressed at: INSERM U954, Nutrition-Genetics-Environmental Risk Exposure and Reference Centre of inborn
metabolism diseases, Faculte´ de Me´decine, 9, avenue de la Foreˆt de Haye, B.P. 184, 54505 Vandoeuvre-le`s-Nancy Cedex, France. Tel:+33 383683292; Fax:+33 383683279, Email: jean-louis.gueant@medecine.uhp-nancy.fr
#The Author 2013. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com
Human Molecular Genetics, 2013, Vol. 22, No. 22 4591–4601 doi:10.1093/hmg/ddt308
after extraction and HPLC analysis (5). MMACHC has a dual activity, catalysing either reductive decyanation when it encoun-ters cyanocobalamin (CNCbl) or dealkylation of alkylcobala-mins (e.g. MeCbl), which is reflected in its 3D structure (4). MTR has 33 exons and encodes the MS protein with a predicted molecular weight of 140.4 kDa (2,6,7).
Despite the prediction of transcripts in the www.ensembl.org database, experimental data on MTR splicing are lacking. Among the predicted transcripts, MTR-201 contains 31 exons and encodes a truncated protein with a molecular weight of 124.1 kDa and no activity. It differs from MTR-001 by lacking exons 16, 17 and 18 and by addition of a supplementary exon after exon 33 of MTR-001. The missing exons of MTR-201 belong to the postulated N5-methyltetrahydrofolate-(5-CH3-THF) and the Cbl-binding domains.
The cblG and cblC defects share a number of abnormalities in-cluding megaloblastic anaemia, neurological disturbances, hyper-homocysteinaemia and hypomethioninaemia (1,6–9). Cases of cblG with undetectable MS activity are very rare and are classified as ‘cblG-variant’ (10). We have observed in cultured fibroblasts from ‘cblG-variant’ cases a decreased conversion of CNCbl to HOCbl, similar to cells from cblC cases (11–13). To explain this paradoxical similarity, we studied the expression of MTR and MS, as well as the interaction of MS and MMACHC in cblG-variant, cblG and cblC fibroblast cell lines.
RESULTS MS activity
The fibroblast cell lines studied are four cblG-variant cases (in-cluding AE and three cases from Canada, MG#1, MG#2 and MG#3), one cblG case (KF) and two cblC cases (SA and WG4130). SA and WG4130 were children hospitalized in their first year of life, who presented with megaloblastic anaemia, dramatic increase of homocysteine, low methionine and serum vitamin B12levels within the reference range, as
pre-viously described (1,11,12). The serum methylmalonic acid level was within the reference range in cblG variants and cblG, but elevated in SA and WG4130 (11,12); cblG-variant fibro-blasts had no detectable functional MS activity (measured by the conversion of 5 – [14C]CH3– THF to [14C]-methionine). By
comparison, KF cells and control fibroblasts had functional MS activity of 0.17 and 1.87 nmol/h/mg protein, respectively. Cobalamin processing in skin fibroblasts from patients Labelled Cbl was incubated with cells for 92 h. The conversion of CNCbl to HOCbl was dramatically lower in fibroblasts from the 3 cblG-variant patients (MG#1, MG#2 and MG#3) than in those from 41 standard cblG cases evaluated in the Department of Human Genetics of McGill university, Canada (3.8 + 3.1 and 23.5 + 12.1%, respectively, P ¼ 0.008). In the cblG-variant fibroblasts of the fourth case, patient AE evaluated in Europe, only 14.3 + 3.6% of CNCbl was converted to HOCbl. This was intermediate between the 3.8 + 2.6 and 6.1 + 0.1% reported in cblC fibroblasts and the 21.6 + 8.0% reported in cblG fibroblasts, and much lower than the 66.7 + 13.0% observed in control fibroblasts (Fig.1A and B). Conversely, the conversion of CNCbl to AdoCbl was higher in cblG-variant
cells, compared with the other cells (Supplementary Material, Fig. S1 and S2). To investigate this unexpected cellular pheno-type of cblG variant, we performed an extensive molecular study of gene defects and gene expression, as well as of MS and MMACHC protein interactions in fibroblasts of AE (cblG variant), KF (cblG), SA and WG4130 (cblC).
Genomic DNA analysis
AE and KF cell lines had no mutations in the MMACHC gene. Conversely, SA and WG4130 were cblC cell lines with MMACHC mutations and no MTR mutations. A c.609+ 1088 G . A substitution in the MTR gene of AE was expected to create a new splice acceptor site (GTGG becoming GTAG) and the subsequent formation of two donor sites in intron 6 (using Alamutw software). This substitution was absent in the DNA of 70 controls, and was not reported in the 1000 Genome database. SA was a cblC case with compound heterozygosity for c.271dupA, p.R91 . GNsX14 and c.616C . T, p.R206W of MMACHC. WG4130 was a cblC case with compound hetero-zygosity for c.328delAACC, p.N110EfsX13 and 482G . A, p.R161Q of MMACHC.
Transcripts and protein analysis
RT – PCR analysis of amplicons of MTR-001 transcripts showed two additional larger bands in the amplicon that encompassed exons 6 and 7 in AE fibroblasts, but not in control, KF and SA cells (Fig.2A and B). Sequencing identified the two expected al-ternative splicing products. The largest product resulted from the insertion of a 129 bp sequence from intron 6 in the reading frame and the medium product from the insertion of a 78 bp sequence identical to the beginning of the 129 bp sequence. The 13th codon of both insertions was a stop codon (Fig.2A – C). KF was a cblG cell line with a MTR homozygous mutation c.3518C . T, p.P1173L. RT–PCR assays were performed to study the other splicing products of MTR in the five cell lines, according to the 2012 edition of www.ensembl.org database. The transcripts of MS predicted in this database are shown in Figure3A. All PCR amplicons were sequenced. MTR-201, MTR-003 and MTR-004 transcripts, but not MTR-202, were detected in all cell lines, whereas MTR-001 and MTR-002 were not detected in AE cells. The 78 bp sequence insertion of intron 6 was also detected in MTR-004, in AE fibroblasts (Fig.3B).
Western blot analysis of MS showed the expression of the 140 kDa full-size enzyme, translated by MTR-001 and a 124 kDa truncated protein, corresponding to the MTR-201 tran-script in control, cblG and cblC cell lines. In contrast, only the 124 kDa truncated isoform, corresponding to the MTR-201 tran-script was found in the AE cblG-variant fibroblasts (Fig.4A). The relative distribution of both MS isoforms varied as a func-tion of growth, in Caco2 TC7 cells. The relative intensity of the 124 kDa isoform increased, whereas the 140 kDa isoform decreased after cells reached confluence (Supplementary Mater-ial, Fig. S3). The 124 kDa band was shown not to be a proteolysis product since the addition of protease inhibitors to the protein extract of fibroblasts had no influence on the appearance of the 140 and 124 kDa isoforms, while addition of trypsin to the ex-traction buffer produced complete degradation of the isoforms (Supplementary Material, Fig. S4). We also confirmed that the
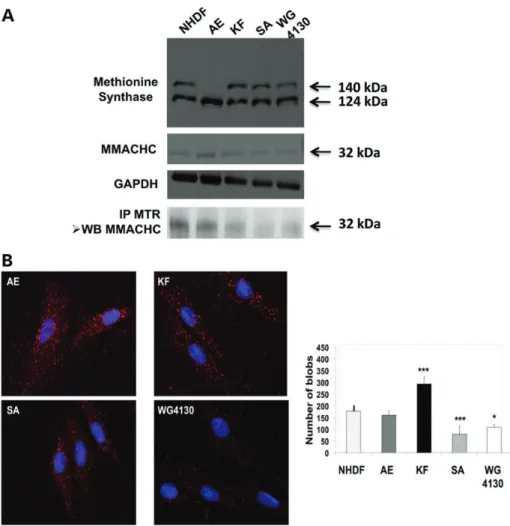
124 kDa protein band corresponded to MS by mass spectrometry analysis of cell lysate from NHDF control fibroblasts and fibro-blasts of AE, a cblG-variant case (Supplementary Material, Fig. 5S). We provide evidence that MS interacts with MMACHC in all cell lines using co-immunoprecipitation and quantification of the protein – protein interactions with the prox-imity ligation assay (PLA) (Duolinkw) (Fig. 4B). The co-immunoprecipitation of MMACHC by anti-MS antibody was much lower in the SA and WG4130 cblC cell lines, com-pared with cblG, cblG-variant and control fibroblasts (Fig.4A). In agreement with these findings, MMACHC – MS interaction evaluated by the PLA was 1.6-fold higher in the KF cell line and 1.5- and 2.2-fold reduced in the cblC cell lines expressing the p.R161Q and p.R206W mutations, respectively, compared with control fibroblasts (Fig.4B). We have investi-gated knock down of MS expression by siRNA transfection to obtain control data of the PLA, in human embryonic kidney (HEK) cells. The transfection of HEK cells with siRNA #24 (tar-geting the end of exon 24) increased the cell expression of the 124 kDa isoform and decreased the full-size MS, while transfec-tion with siRNA #16 (targeting exon 16) decreased both full-size MS and 124 kDa isoform (Fig.5A). Compared with the control condition, these transfection conditions decreased the inter-action of MS with MMACHC by 50%, in Duolink experiments (Fig. 5B). Transfection with siRNA #6 (targeting exon 6) decreased dramatically the expression of the full-size MS in western blot analysis and the interaction with MMACHC by 80%, in Duolink experiments (Fig.5B).
Modelling and docking analyses of interactions between MS and MMACHC
In 3D modelling of the MS – MMACHC interface, a loop present in both MS and MTR-201, is shown to be buried within a deep (over 15 A˚ ) invagination in MMACHC, thereby establishing
contacts with both Arg-161 and Arg-206. This loop corresponds to a fragment of 19 amino acids that extends from position 749 to position 767 and constitutes a significant part (883 A˚2; 66%) of the total contact surface area (Fig.6). We also analysed intermo-lecular hydrogen bonds between the docked partners. The results are illustrated using 2D-GraLab (14) in Figure7A and B. The interaction domain of MS with MMACHC essentially spans two linear sequence regions composed of residues 688 – 706 and 745 – 768. The persistent occurrence of these two fragments in the sequence of MTR-201 is in agreement with the interaction between MMACHC and truncated MS, which was shown by co-immunoprecipitation and PLA in cblG-variant AE fibro-blasts (Fig.4A and B). The interaction involved 43 residues from MMACHC and 38 from MS. The two p.R161Q and p.R206W mutations of MMACHC reported in SA and WG4130 respectively were considered in the interaction pre-dicted by the docking analysis (Fig. 7B). Hydrogen bonds of both residues are found to be equally stabilizing. In agreement with our experimental findings, it is clear, however, that the more conservative mutation R161Q is likely to be less deleteri-ous to the MMACHC – MS interaction, than the R206W muta-tion, in which the residue change is significantly different both in size and physico-chemical properties (Fig.7B).
DISCUSSION
In this study, we report a decreased conversion of CNCbl to HOCbl, in cblG-variant fibroblasts, similar to that observed in cblC fibroblasts (11–13). Conversely, the conversion of CNCbl to AdoCbl was higher in cblG-variant cells, compared with the other cells. To investigate the genetic and molecular mechanisms that may explain this unexpected finding, we per-formed a molecular genetic study of MTR and studied possible interactions of the MS and MMACHC proteins in fibroblasts of one cblG-variant case (AE), as compared with cblG, cblC Figure 1. Analyses of cyano-[57Co]Cbl conversion into Cbl isoforms in fibroblasts from AE cblG-variant, KF cblG, SA and WG4130 cblC cases and NHDF control cell line. The cells were incubated with labelled Cbl for 92 h in dark at 378C under 5% CO2atmosphere, as described in methods. (A) Identification and quantification of
[57Co]Cbl-labelled isoforms (OH-Cbl, CN-Cbl, Ado-Cbl, Me-Cbl) were performed by HPLC in fibroblast extracts and expressed as a percentage of intracellular cyano-[57Co]Cbl. (n ¼ 6, mean + standard error of the mean). The range of the relative distribution (%) of Cbl isoforms in control fibroblasts is 17 – 20, 17 – 30, 15 – 35 and 25 – 45%, for OH-Cbl, CNCbl, AdoCbl and Me-Cbl, respectively. (B) Ratios of intracellular percentage of OH-[57Co]Cbl versus intracellular percentage
of cyano-[57Co]Cbl.
and control fibroblast cell lines. The data show that alternative splicing in cells from the cblG variant produces an MTR-201 transcript that encodes a truncated MS, and a MTR-001 tran-script with two insertions containing stop codons. The transla-tion of truncated isoforms of MS through MTR alternative splicing has been also reported very recently in human brain tissues (15). The missing exons of MTR-201 belong to the 5-CH3-THF- and Cbl-binding domains, explaining the lack of
MS activity of the truncated 124 kDa protein (6,7). The decreased expression of MMACHC protein in the two cblC cases can be explained by the fact one of the two causal mutations results in a stop codon (11,12).
We show interaction between MMACHC and both full-size active MS and/or truncated MS by co-immunoprecipitation and PLA of fibroblasts. This was confirmed by the decreased ex-pression of either the full-size MS, the truncated MS or the two isoforms in HEK cells transfected with siRNA. The present data and the recent observation of protein – protein interaction
between MMACHC and MMADHC suggest that intracellular Cbl is processed through close protein – protein interactions, possibly to ensure substrate channelling and to allow only minimal exposure of Cbl to the environment (16,17). It has been shown that the interaction between MMADHC and MMACHC plays a role in the regulation of the balance between AdoCbl and MeCbl synthesis (16,18). In vitro, MMACHC has the ability to decyanate CNCbl to cob(II)alamin (13). The interaction between MMACHC and MS isoforms could therefore be part of the mechanisms regulating the MMACHC-dependent processing of Cbl. This could explain the decreased decyanation of Cbl observed in the cblG-variant fibroblasts. In normal conditions, the expression of both trun-cated and full-size enzyme results from an MTR splicing mech-anism that produces a regulatory mechmech-anism for directing the Cbl towards either MeCbl or AdoCbl pathways. In cblG-variant cells, the interaction of MMACHC with the truncated MS protein only may deregulate the cellular distribution of Cbl Figure 2. Molecular analyses of the cblG-variant (AE). (A) RT – PCR with primer pairs cMTRm_04 and cMTRm_05 (see Supplementary Material, Table SI) encom-passing exons 4 – 7 and 6 – 10, respectively. The upper and medium bands of AE mRNA contain the 129-bp and 78-bp inserts. (B) Alternative splicing products iden-tified by sequencing the amplicon that encompassed exons 6 and 7. (C) Sequencing of genomic DNA and splicing consequences of the G . A substitution in position c.609+ 1088. Inserts generated a TGA stop codon indicated by an arrow. Numbers 1, 2 and 3 in open squares indicate the position of the new splice acceptor site and the subsequent donor sites, respectively.
between MeCbl and AdoCbl. This mechanism displays similar-ity with the MMACHC – MMADHC interaction, which involves full size and truncated proteins (16,18).
In contrast to MMACHC, the N-terminal sequence of MMADHC contains a mitochondrial targeting sequence leading to both mitochondrial and cytoplasmic localization (16). Consistently, MMADHC has been found in both compart-ments in contrast to the cytosolic MMACHC (19). Therefore, these results and our data suggest that the interaction between MMACHC with MS occurs either at the same time or after the one between MMACHC and MMADHC.
MMADHC mutations affecting the translation of the C-terminal part of the protein lead to impairment of the remethy-lation pathway, with increased homocysteine, improved mito-chondrial targeting of MMADHC and subsequent increase of AdoCbl, with a concomitant decrease in MeCbl formation (16,18). This increased synthesis of AdoCbl is also observed in our cblG-variant patients. This suggests that the orientation of Cbl towards AdoCbl may also be influenced on the synthesis of MeCbl through MMACHC – MS. These recent data and our results therefore highlight the cross-point role of MMACHC for regulating the conversion of Cbl into either MeCbl or AdoCbl through interactions with the respective key proteins of these two pathways.
The 3D structure of human MMACHC (pdb ID ¼ 3SOM) has been resolved (20), but not that of human MS, except for a rather short C-terminal fragment. We, therefore, built a 3D-model of human MS, using the E. coli B12-dependent MS as a structural
template. The proposed MS – MMACHC interaction is based on an MS loop that anchors within a cleft made in MMACHC, in close vicinity to Arg-161 and Arg-206 residues. The model-ling analysis also agrees with previous data showing that MMACHC binds Cbl in the ‘base-off’ state required by MS (3–5,20). The C-terminal domain of MMACHC displays simi-larity to the C-terminal domain of TonB proteins, a protein involved in bacterial Cbl uptake, which directly interacts with proteins involved in Cbl import (3). The differences seen in the PLA between control, cblG-variant and cblC fibroblasts, re-spectively, suggest an influence of R161Q and R206W muta-tions on the MMACHC – MS interaction, which was consistent with the involvement of the R161 and R206 residues in the docking analysis. The cblC patient, SA, has a p.R206W change within the C-terminal domain of MMACHC, very close to the loop of five-residue 198 – 203 that deviates from the protein backbone of the superimposed TonB structure (3). It was predicted to be deleterious for MMACHC – MS inter-action in the docking analysis and was also in the cell line showing the least interaction between MS and MMACHC in Figure 3. MTR transcription in control and cblG-variant, cblG and cblC fibroblasts. (A) Prediction of MTR splicing, according to the www.ensembl.org/ database of EMBL-EBI and Sanger Centre (see Supplementary Material, Tables SII and SIII). Transcripts encoding functional proteins are shown in yellow, transcripts encoding proteins of unknown function in red, transcripts not encoding a protein in blue. The position of missing codons in the other transcripts is indicated by open ellipses, using the MTR-001 transcript as a reference. Horizontal bars depict amplicons produced in RT – PCR, for the spicing analysis. (B) RT – PCR of transcripts MTR-201, MTR-202, MTR-002 and MTR-003. MTR-203 was not detected. GAPDH was used as an internal standard.
the PLA. The model for the interaction of the two proteins iden-tifies contact sequences in MS that are present in both full-length and truncated protein. Hence, the absence of a part of the Cbl-binding domain may result in a truncated MS that is unable to bind Cbl donated by MMACHC despite a ‘productive’ interaction.
In conclusion, we found an interaction between MS and MMACHC, which suggests a regulatory role of MS in the intra-cellular metabolism of Cbl, through MTR splicing of two tran-scripts that encode the full-size enzyme and a non-functional truncated protein, respectively. This regulatory interaction was highlighted in cblG-variant fibroblasts with decreased HOCbl and increased AdoCbl, in which MTR encoded only the non-functional MS isoform.
MATERIALS AND METHODS Cell culture
The fibroblast cell lines were from the University Hospitals of Basel, Switzerland (AE, cblG-variant), Nancy University, France (KF, cblG and SA, cblC), McGill University, Canada
(MG#1, MG#2 and MG#3, 3 cblG-variant cases and WG4130, cblC) and ATCC (Manassas, Virginia, USA) (NHDF control fibroblasts). Fibroblasts were grown in Dulbecco’s modified Eagle medium supplemented with 10% heat-inactivated foetal calf serum (PAA), as described (20,21). HEK-293 and TC7 cells were maintained in Dulbecco’s modified Eagle medium with 10% foetal calf serum supplemented with antibiotics (21,22).
MS activity
MS activity was determined as described (21). The assay mixture contained, in a final volume of 100 ml; 100 mMpotassium
phos-phate buffer (pH 7.2) (Sigma-Aldrich), 25 mMof 5 – [14C] CH3–
THF (GE Healthcare), 25 mM dithiothreitol (Sigma-Aldrich),
25 mM ascorbate (Sigma-Aldrich), 0.2 mM SAM
(Sigma-Aldrich), 50 mM methylcobalamin (Sigma-Aldrich), 5 mM
homocysteine and cell extract containing 100 – 400 mg of protein. For blanks, the cell extract was replaced with 100 mg of bovine serum albumin. Samples were incubated from 20 min to 1 h at 378C. The reaction mixture was heated at 958C for 15 min, and cooled on ice for 10 min. The reaction Figure 4. Protein expression of MS and MMACHC in control and cblG-variant, cblG and cblC fibroblasts. (A) Western blot of methionine synthase (MS) and MMACHC proteins, and western blot of MMACHC immunoprecipitated with antibody against MS protein. GAPDH was used as an internal control of protein ex-pression. (B) Proximity ligation assays (Duolinkw) of interaction between MS and MMACHC. The quantification of linkage was performed by dot counting (blobs) using Matlab, as described previously (22,23). The intensity of interactions is shown in the histogram (∗P , 0.05;∗∗P , 0.01;∗∗∗P , 0,001).
mixture was centrifuged for 10 min at 12 000g at 48C and the pellet eliminated. The radioactivity was quantified by scintilla-tion counting in a Packard Tri Carb Model C1900 scintillascintilla-tion counter. MS activity was expressed as nanomoles of [14C] Me-thionine formed per milligram protein per hour.
Cobalamins assay in patient fibroblasts
For studies of Cbl uptake, fibroblasts were incubated with 31.25 pM [57Co]CNCbl (MP Biomedicals, Inc., Orangeburg,
NY, USA) for 92 h in dark at 378C under 5% CO2atmosphere.
The following steps were performed on ice under dim red light in order to prevent photolysis of Cbl cofactors (23). Cells were harvested by trypsinization and centrifuged to collect whole pellets. These pellets were resuspended in a presence of a solu-tion containing 10 mM HOCbl, CNCbl, AdoCbl and MeCbl
and 50 ml of Streptomyces griseus protease (Sigma, 80 U/ml, dissolved in TBS-calcium buffer) to digest the proteins. To achieve the digestion, the resuspended pellets were incubated 3 h at 378C. After the incubation, two volumes of acetone (2208C) were added to the digested cells and incubated over-night at 2208C. This step was followed by centrifugation at 48C to pellet the digested proteins and the supernatant was retained. The pellets were washed once again with 500 ml cold acetone, centrifuged and the collected supernatant was added to the previous collected supernatant. The supernatant was
evaporated in SpeedVac 2 h by the room temperature. The remaining aqueous samples were analysed by HPLC using a Merck column Resolve C-18 (E. Merck, Interchim, Montluc¸on) in a gradient of acetonitrile buffered with 85 mMof
triethylam-monium phosphate at pH 3. Genomic DNA analysis
DNA was extracted from cultured fibroblasts using an QIAmp DNA micro extraction kit (Qiagen, Courtaboeuf, France). PCR amplification of all exons of MTR and MMACHC was performed using 33 and 5 primers pairs, respectively (Supplementary Ma-terial, Table SI). PCR reaction was performed using OptimaseTM Polymerase (Transgenomic, Lanorville, France); products were purified using an QIAquick PCR purification kit (Qiagen). Direct sequencing was performed using the BigDye terminator v1.1 Cycle Sequencing Kit (Applied Biosystems, Villebon sur Yvette, France). The sequences were compared with the MTR and MMACHC reference sequences.
Transcript analysis
Total RNA was isolated from cultured fibroblast cells using the RNeasy Plus Mini extraction kit (Qiagen, Courtaboeuf, France). First strand cDNA was synthesized from 2 mg of total RNA using oligo(dT)12 – 18 primers with SuperScriptw II reverse Figure 5. Transfection of HEK cells with siRNA targeting junction of exons 12 – 13, end of exon 24, exon 16, exon 6 and exon 13 of MTR, respectively (see Supple-mentary Material, Table SIV). (A) Western blots of cells transfected at Day 3. The proteins were extracted after 48 h, as described in methods. GAPDH was used as an internal control of protein expression. (B) Proximity ligation assays (Duolinkw) of interaction between MS and MMACHC, after transfection with siRNA targeting junction of exons 12 – 13, end of exon 24, exon 16 and exon 6, respectively (∗P , 0.05;∗∗∗P , 0.001).
transcriptase (Invitrogen, Mantes la Jolie, France) according to the manufacturer’s protocol. PCR reactions were performed using PCR buffer (1× as final concentration), MgCl2(1.5 mM
as final concentration), dNTP (0.2 mMas final concentration),
23 pairs of specific primers (0.5 pmol/ml as final concentration), cDNA (2 ml) and Taq DNA Polymerase (1 U as final concentra-tion) in an i-cycler thermal cycler (BioRad). All reagents were from Invitrogen. Twenty-three primers were used for the amplification of the whole open-reading frame of gene MTR. Their sequences are given in Supplementary Material, Table SI. For the study of MTR splicing, PCR was performed using four primers pairs (MTR201-, MTR202-, MTR003- and MTR004-primers), according to the splicing prediction of www.ensem bl.org/ database of EMBL-EBI (2012 edition) and Sanger Centre (primer sequences are given in Supplementary Material, Table SII and the transcript nomenclature in Supplementary Material, Table SIII), the same reagents, final concentrations and equipment as mentioned above.
Inhibition of MS expression by siRNA transfection in HEK-293 cells
For siRNA experiments, HEK-293 cells were transfected at 40% confluency with Lipofectamine RNAiMax (Life Technologies) according to the manufacturer’s recommendations using 50 nM
of each siRNA. The design of siRNA to target different exons of MTR isoforms is given in Supplementary Material, Table SIV. The negative control siRNA was purchased from Life Technologies and the sequences of the other siRNAs are as follows: siMTR#1: 5′-AUAGUUUCCUGCCAUGAUG-3′, siMTR#2: 5′-GGAGGAGCAACCACUUCAA-3′, siMTR#3:
5′-GGACUGGAAUGGAGGAACA-3′, siMT810: 5′- GGGUC GACAUCUUACUCAU-3′ and siMTR811: 5′- GGAUUGG ACCAUACACCAA-3′; siRNA were designed to target junction of exons 12 – 13, end of exon 24, exons 16 – 18, exon 6 and exon 13 of MTR isoforms, as described in Supplementary Material, Table SIII. After 48 h, proteins were extracted in a buffer con-taining 50 mMTris – HCl pH 6.8, 20 mMEDTA and 5% SDS,
sonicated briefly and analysed by SDS – PAGE and western blotting.
Protein analysis
Whole cell lysates (50 mg) from cultured fibroblasts were pre-pared in the presence of 5% (v/v) of protease inhibitor cocktail (PIC, Sigma, P8340), separated and transferred in nitrocellulose membranes (Millipore) as described (21). The membranes were incubated overnight at 48C with primary antibodies diluted in TBS buffer containing 5% non-fat dried milk, as follows: anti-MS (goat polyclonal, Abcam), anti-MMACHC (rabbit poly-clonal, Abcam) and anti-GAPDH antibody (mouse monopoly-clonal, Santa Cruz Biotechnology) at a dilution of 1 : 1000. Appropriate secondary antibodies conjugated to horse radish peroxidase were used for detection with an ECL Detection kit (Amersham). Co-immunoprecipitation experiments were performed by means of a Piercew Co-Immunoprecipitation commercial kit (Thermo Scientific Pierce). This optimized method enables iso-lation of native protein complexes from a lysate by directly im-mobilizing purified antibodies onto an agarose support, as described (24).
We performed the PLA (Duolink in situ PLA reagents; Olink Bioscience; Eurogentec, Angers, France) to visualize and quantify interactions of MMACHC and MS in situ, in fibroblasts and HEK cells. Since the procedure is based on a stoichiometric reaction, each dot corresponds to a close interaction between the two pro-teins. The experimental conditions for the use of primary antibodies for this technique were identical to those used for the other immu-nohistochemistry experiments. A pair of oligonucleotide-labelled secondary antibodies (PLA probes) was used according to the man-ufacturer’s instructions to bind to the primary antibodies (25,26). This pair of secondary antibodies generates a signal only when the two probes are in close proximity (,4 nm). After amplifica-tion, the signal from each detected pair is visualized as an individual fluorescent dot. The PLA signals were counted and assigned to a specific subcellular location based on microscopy images (BX51WI microscope, Olympus, Tokyo, Japan, with Blob-Finder/MatLab freeware from the Centre for Image Analysis, Uppsala University, Uppsala, Sweden).
Modelling and docking analyses of the interactions between MS and MMACHC
Although there is an existing 3D structure model for human MMACHC (pdb ID ¼ 3SOM), none is yet available for human MS, except for a rather short C-terminal fragment (2O2 K; resi-dues 926 – 1265). Docking prediction required therefore model-ling a plausible 3D structure for human MS. This endeavour made use of MolIDE (27). Accordingly, homology modelling was performed using, as a structural template, the E. coli Cbl-dependent MS (3IVA; residues 663 – 1264; 52% identity, 67% positive, 4% gap, alignment length: 604, hit length: 579), Figure 6. Modelling analysis of the interface between MS and MMACHC.
Hom-ology modelling of human MS was performed using, as a structural template, the E. coli B12-dependent methionine synthase, as described in methods section. The final model obtained is named MTR_3_3IVAA_0.pdb and has eight insertions and one deletion. This and an existing 3D structure model for human MMACHC (pdb ID ¼ 3SOM) were further optimised with the BRUGEL package for modelling the interface between MS and MMACHC. The model illu-strated, using Rasmol (detailed information is provided in methods) corresponds to the most stable interactions involving the MMACHC pair, Arg-161 and Arg-206. MS is shown as an orange ribbon. MMACHC is displayed as a model and coloured according to secondary structures with a-helices in magenta, b-sheets in yellow, turns in pale blue and random in white; Arg-161 and Arg-206 are shown in CPK dots and labelled.
which, along with the C-terminal activation domain, also con-tains the Cbl-binding domain. Full hydrogen atoms structures of homology-modelled human MS and MMACHC were docked using the ClusPro 2.0 server (http://cluspro.bu.edu/ login.php) (28). The model illustrated, using Rasmol (Windows Version 2.7.5) (http://www.rasmol.org) (29,30) corresponds to the most stable interactions involving the MMACHC pair, Arg-161 and Arg-206. Sequence alignment was adjusted so as to exclude the occurrence of indels within
secondary structure elements. Loops, which then included even-tual indels, were modelled with LOOPY (31). The final model obtained was named MTR_3_3IVAA_0.pdb and has eight inser-tions and one deletion. This and 3SOM structures were further optimized with the BRUGEL molecular modelling package (32) following two successive rounds each consisting of hydro-gen atoms construction/optimization with heavy atoms con-strained and 2000 steps of unrecon-strained steepest-descent energy minimization, followed by two rounds of unrestrained Figure 7. Docking analyses of the bonds between MS and MMACHC. (A) MMACHC – MS total interface analysis. The figure was generated using 2D-GraLab (14) and BRUGEL (26) softwares. The distances between bonding partners are in Angstroms and measured between H atoms and the corresponding hydrogen-bond acceptor atom. Arg-161 and Arg-206 are emphasized with a red star. (B) MMACHC Arg-161 and Arg-206 hydrogen hydrogen-bonds in the complex with MS. The figure was generated with 2D-GraLab (14), but all geometric data are those of BRUGEL. All distances are in Angstroms. Dual distance values separated by a slash (/) between hydrogen-bond partners correspond to the presence of two hydrogen bonds.
conjugated-gradient minimization (500 steps, each). The result-ing model structure was then analysed in BRUGEL for the contact surface area in the complex (1297 A˚2, a value at the lower edge of measured interface areas, consistent with the re-versibility of the complex). Contact data were generated with 2D-GraLab for total interface analysis (14).
Statistical analysis
Results were expressed as means + standard error of the mean. Comparison between cell phenotypes was performed by one-way ANOVA.
SUPPLEMENTARY MATERIAL
Supplementary material is available at HMG online. AUTHOR CONTRIBUTIONS
J.-L.G. designed and coordinated research, analysed data and wrote the paper, M.B.F.-C. and N.T.M. performed research (N.T.M. designed all molecular modelling research), analysed data and wrote part of the paper, C.C., M.P., J.P., J.-M.A. per-formed research and analysed data, D.C., F.F., D.W., B.F. and D.S.R. analysed data and revised the paper.
ACKNOWLEDGEMENTS
We thank B. Woldseth and T. Rootwelt from the Department of Medical Biochemistry, Rikshospitalet, Oslo, Norway, who sent us fibroblasts from one case.
Conflict of Interest statement. The authors have no conflict of interest to declare.
FUNDING
This work was supported by grants from the French minister of Health (for the Reference Centre of inborn metabolism diseases) and by Inserm and the Region Lorraine (for Inserm U954). B.F. received a grant from the Swiss National Foundation.
REFERENCES
1. Watkins, D. and Rosenblatt, D.S. (2011) Inherited disorders of folate and cobalamin transport and metabolism. In Valle, D., Beaudet, A.L., Vogelstein, B., Kinzler, K.W., Antonarakis, S.E. and Ballabio, A. (eds), Scriver ’s Online Metabolic and Molecular Bases of Inherited Disease. 2. Chen, Z., Chakraborty, S. and Banerjee, R. (1995) Demonstration that
mammalian methionine synthases are predominantly cobalamin-loaded. J. Biol Chem., 270, 19246– 19249.
3. Lerner-Ellis, J.P., Tirone, J.C., Pawelek, P.D., Dore´, C., Atkinson, J.L., Watkins, D., Morel, C.F., Fujiwara, T.M., Moras, E., Hosack, A.R. et al. (2006) Identification of the gene responsible for methylmalonic aciduria and homocystinuria, cblC type. Nat. Genet., 38, 93 – 100.
4. Koutmos, M., Gherasim, C., Smith, J.L. and Banerjee, R. (2011) Structural basis of multifunctionality in a vitamin B12-processing enzyme. J. Biol. Chem., 286, 29780– 29787.
5. Hannibal, L., Axhemi, A., Glushchenko, A.V., Moreira, E.S., Brasch, N.E. and Jacobsen, D.W. (2008) Accurate assessment and identification of
naturally occurring cellular cobalamins. Clin. Chem. Lab. Med., 46, 1739– 1746.
6. Leclerc, D., Campeau, E., Goyette, P., Adjalla, C.E., Christensen, B., Ross, M., Eydoux, P., Rosenblatt, D.S., Rozen, R. and Gravel, R.A. (1996) Human methionine synthase, cDNA cloning and identification of mutations in patients of the cblG complementation group of folate/cobalamin disorders. Hum. Mol. Genet., 5, 1867 – 1874.
7. Li, Y.N., Gulati, S., Baker, P.J., Brody, L.C., Banerjee, R. and Kruger, W.D. (1996) Cloning, mapping and RNA analysis of the human methionine synthase gene. Hum. Mol. Genet., 5, 1851 – 1858.
8. Wilson, A., Leclerc, D., Saberi, F., Campeau, E., Hwang, H.Y., Shane, B., Phillips, J.A. III, Rosenblatt, D.S. and Gravel, R.A. (1998) Functionally null mutations in patients with the cblG-variant form of methionine synthase deficiency. Am. J. Hum. Genet., 63, 409 – 414.
9. Watkins, D., Ru, M., Hwang, H.Y., Kim, C.D., Murray, A., Philip, N.S., Kim, W., Legakis, H., Wai, T., Hilton, J.F. et al. (2002) Hyperhomocysteinemia due to methionine synthase deficiency, cblG, structure of the MTR gene, genotype diversity, and recognition of a common mutation, P1173L. Am. J. Hum. Genet., 71, 143 – 153.
10. Sillaots, S.L., Hal, l.C.A., Hurteloup, V. and Rosenblatt, D.S. (1992) Heterogeneity in cblG, differential retention of cobalamin on methionine synthase. Biochem. Med. Metab. Biol., 47, 242 – 249.
11. Morel, C.F., Lerner-Ellis, J.P. and Rosenblatt, D.S. (2006) Combined methylmalonic aciduria and homocystinuria (cblC), phenotype-genotype correlations and ethnic-specific observations. Mol. Genet. Metab., 88, 315 – 321.
12. Lerner-Ellis, J.P., Anastasio, N., Liu, J., Coelho, D., Suormala, T., Stucki, M., Loewy, A.D., Gurd, S., Grundberg, E., Morel, C.F. et al. (2009) Spectrum of mutations in MMACHC, allelic expression, and evidence for genotype-phenotype correlations. Hum. Mutat., 30, 1072 – 1081.
13. Kim, J., Gherasim, C. and Banerjee, R. (2008) Decyanation of vitamin B12by
a trafficking chaperone. Proc. Natl. Acad. Sci. USA., 105, 14551 – 14554. 14. Zhou, P., Tian, F. and Shang, Z. (2008) 2D Depiction of nonbonding
interactions for protein complexes. J. Comput. Chem., 30, 940 – 951. 15. Muratore, C.R., Hodgson, N.W., Trivedi, M.S., Abdolmaleky, H.M.,
Persico, A.M., Lintas, C., De la Monte, S. and Deth, R.C. (2013) Age-dependent decrease and alternative splicing of methionine synthase mRNA in human cerebral cortex and an accelerated decrease in autism. PLoS One, 8, e56927.
16. Stucki, M., Coelho, D., Suormala, T., Burda, P., Fowler, B. and
Baumgartner, M.R. (2012) Molecular mechanisms leading to three different phenotypes in the cblD defect of intracellular cobalamin metabolism. Hum. Mol. Genet., 21, 1410 – 1418.
17. Plesa, M., Kim, J., Paquette, S.G., Gagnon, H., Ng-Thow-Hing, C., Gibbs, B.F., Hancock, M.A., Rosenblatt, D.S. and Coulton, J.W. (2011) Interaction between MMACHC and MMADHC, two human proteins participating in intracellular vitamin B metabolism. Mol. Genet. Metab., 102, 139 – 148. 18. Coelho, D., Suormala, T., Stucki, M., Lerner-Ellis, J.P., Rosenblatt, D.S.,
Newbold, R.F., Baumgartner, M.R. and Fowler, B. (2008) Gene identification for the cblD defect of vitamin B12metabolism.
N. Engl. J. Med., 358, 1454– 1464.
19. Mah, W., Deme, J.C., Watkins, D., Fung, S., Janer, A., Shoubridge, E.A., Rosenblatt, D.S. and Coulton, J.W. (2012) Subcellular location of MMACHC and MMADHC, two human proteins central to intracellular vitamin B12 metabolism. Mol Genet Metab., 108, 112 – 118.
20. Froese, D.S., Krojer, T., Wu, X., Shrestha, R., Kiyani, W., von Delft, F., Gravel, R.A., Oppermann, U. and Yue, W.W. (2012) Structure of MMACHC reveals an arginine-rich pocket and a domain-swapped dimer for its B12 processing function. Biochemistry, 51, 5083– 5090.
21. Forges, T., Chery, C., Audonnet, S., Feillet, F. and Gueant, J.L. (2010) Life-threatening methylenetetrahydrofolate reductase (MTHFR) deficiency with extremely early onset, characterization of two novel mutations in compound heterozygous patients. Mol. Genet. Metab., 100, 143 – 148. 22. Pons, L., Battaglia-Hsu, S., Orozco-Barrios, C., Ortiou, S., Chery, C., Alberto, J.M., Arango-Rodriguez, M.L., Dumas, D., Martinez-Fong, D., Freund, J.N. et al. (2009) Anchoring secreted proteins in endoplasmic reticulum by plant oleosin, the example of vitamin B12 cellular sequestration by transcobalamin. Plos One, 4, e6325.
23. Pons, L., Guy, M., Lambert, D., Hatier, R. and Gueant, J.L. (2000) Transcytosis and coenzymatic conversion of [(57)Co]cobalamin bound to either endogenous transcobalamin II or exogenous intrinsic factor in caco-2 cells. Cell Physiol. Biochem., 10, 135 – 148.
24. Pooya, S., Blaise, S., Moreno Garcia, M., Giudicelli, J., Alberto, J.M., Gue´ant-Rodriguez, R.M., Jeannesson, E., Gueguen, N., Bressenot, A., Nicolas, B. et al. (2012) Methyl donor deficiency impairs fatty acid oxidation through PGC-1a hypomethylation and decreased ER-a, ERR-a, and HNF-4a in the rat liver. J. Hepatol., 57, 344 – 351.
25. So¨derberg, O., Gullberg, M., Jarvius, M., Ridderstra˚le, K., Leuchowius, K.J., Jarvius, J., Wester, K., Hydbring, P., Bahram, F., Larsson, LG. et al. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods, 3, 995 – 1000.
26. Akchiche, N., Bossenmeyer-Pourie´, C., Kerek, R., Martin, N., Pourie´, G., Koziel, V., Helle, D., Alberto, J.M., Ortiou, S., Camadro,, J.M. et al. (2012) Homocysteinylation of neuronal proteins contributes to folate
deficiency-associated alterations of differentiation, vesicular transport, and plasticity in hippocampal neuronal cells. FASEB J., 26, 3980 – 3992. 27. Canutescu, A.A. and Dunbrack, R.L. Jr. (2005) MolIDE (molecular
integrated development environment): a homology modeling framework you can click with. Bioinformatics, 21, 2914 – 2916.
28. Kozakov, D., Hall, D.R., Beglov, D., Brenke, R., Comeau, S.R., Shen, Y., Li, K., Zheng, J., Vakili, P., Paschalidis, ICh. et al. (2010) Achieving reliability and high accuracy in automated protein docking: cluspro, PIPER, SDU, and stability analysis in CAPRI rounds 13 – 19. Proteins, 78, 3124 – 3130.
29. Sayle, R. and Milner-White, E.J. (1995) Rasmol: biomolecular graphics for all. Tr. Biochem. Sci., 20, 374.
30. Bernstein, H.J. (2000) Recent changes to RasMol, recombining the variants. Tr. Biochem. Sci., 25, 453 – 455.
31. Xiang, Z., Soto, C.S. and Honig, B. (2002) Evaluating conformational free energies: The colony energy and its application to the problem of protein loop prediction. Proc. Natl Acad. Sci. USA, 99,
7432 – 7437.
32. Delhaise, P., Bardiaux, M., De Maeyer, M., Prevost, M., Van B Custem, D., Alard, P. and Wodak, S.J. (1988) The BRUGEL package - toward computer-aided-design of macromolecules. J. Mol. Graph., 6, 219. Human Molecular Genetics, 2013, Vol. 22, No. 22 4601
![Figure 1. Analyses of cyano-[ 57 Co]Cbl conversion into Cbl isoforms in fibroblasts from AE cblG-variant, KF cblG, SA and WG4130 cblC cases and NHDF control cell line](https://thumb-eu.123doks.com/thumbv2/123doknet/14886141.646917/3.918.241.705.112.404/figure-analyses-cyano-conversion-isoforms-fibroblasts-variant-control.webp)