HAL Id: hal-03094535
https://hal.sorbonne-universite.fr/hal-03094535
Submitted on 4 Jan 2021
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
site-dependent phosphorylation in TrkA initiated
MAPK/ERK signaling
Shu Zhao, Jia Shi, Guohua Yu, Dali Li, Meng Wang, Chonggang Yuan,
Huihui Zhou, Amirabbas Parizadeh, Zhenlin Li, Min-Xin Guan, et al.
To cite this version:
Shu Zhao, Jia Shi, Guohua Yu, Dali Li, Meng Wang, et al.. Photosensitive tyrosine analogues unravel
site-dependent phosphorylation in TrkA initiated MAPK/ERK signaling. Communications Biology,
Nature Publishing Group, 2020, 3 (1), pp.706. �10.1038/s42003-020-01396-0�. �hal-03094535�
Tyrosine kinase A (TrkA) is a membrane receptor which, upon ligand binding, activates several pathways including MAPK/ERK signaling, implicated in a spectrum of human pathologies; thus, TrkA is an emerging therapeutic target in treatment of neuronal diseases and cancer. However, mechanistic insights into TrKA signaling are lacking due to lack of site-dependent phosphorylation control. Here we engineer two light-sensitive tyrosine analogues, namelyp-azido-L-phenylalanine (AzF) and the caged-tyrosine (ONB), through amber codon suppression to optically manipulate the phosphorylation state of individual intracellular tyr-osines in TrkA. We identify TrkA-AzF and ONB mutants, which can activate the ERK pathway in the absence of NGF ligand binding through light control. Our results not only reveal how TrkA site-dependent phosphorylation controls the defined signaling process, but also extend the genetic code expansion technology to enable regulation of receptor-type kinase activation by optical control at the precision of a single phosphorylation site. It paves the way for comprehensive analysis of kinase-associated pathways as well as screening of compounds intervening in a site-directed phosphorylation pathway for targeted therapy.
1Shanghai Key Laboratory of Brain Functional Genomics, Ministry of Education, East China Normal University (ECNU), Shanghai 200062, China.2Shanghai
Key Laboratory of Regulatory Biology, Institute of Biomedical Sciences and School of Life Sciences, East China Normal University (ECNU), Shanghai 200062, China.3Department of Anesthesiology, State Key Laboratory of Cardiovascular Disease, Fuwai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100037, China.4Division of Medical Genetics and Genomics, the Children’s Hospital, Institute of Genetics, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China.5Brain Cognition and Brain Disease Institute (BCBDI), Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions, Guangdong Provincial Key Laboratory of Brain Connectome and Behavior, CAS Key Laboratory of Brain Connectome and Manipulation, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China.6UMR 7238 (LCQB unit), Centre National de la Recherche Scientifique (CNRS), Institut National de la Sante et de la Recherche Medicale (INSERM), Institute of Biology, Paris-Seine (IBPS), Sorbonne University, 75005 Paris, France.7UMR 8256 (B2A unit), Centre National de la
Recherche Scientifique (CNRS), Institut National de la Sante et de la Recherche Medicale (INSERM), Institute of Biology, Paris-Seine (IBPS), Sorbonne University, 75005 Paris, France.8Present address: INSERM U1195 unit, University of Paris-Saclay, 94276 Le Kremlin Bicêtre, France.9These authors
I
ntroducing light-sensitive moieties into proteins provides a powerful approach to investigate molecular mechanisms, as well as biological functions with high-temporal and spatial resolutions1,2. An attractive strategy to engineer lightrespon-siveness relies on photoreactive unnatural amino acids (Uaas), allowing site-specific incorporation in a protein target. Uaas with unique light-sensitivity have been successfully incorporated into membrane receptors, GPCRs and ion channels, contributing to our understanding of receptor function3,4. However, this
approach has not yet been applied to the kinase receptors. Here, we present a light-sensitive tyrosine kinase A (TrkA) design through the genetic incorporation of photoreactive Uaas.
Kinase phosphorylation regulates diverse biological processes in eukaryotic systems5,6and has implications in various diseases, including neuronal pathologies and cancer7. Over 500 protein kinases have been identified in humans, producing a plethora of phosphorylation sites on tyrosine, threonine, and serine residues. Each kinase family has a distinct mechanism for the activation and propagation of signals6. Up to now, detailed maps and atlases
documenting thousands of phosphorylation sites. However, the complete description of modification at each phosphorylation site under any condition has certainly not been elucidated. One of the reasons that protein phosphorylation has been studied so exten-sively is the fact that Trk receptor family (including TrkA, TrkB, and TrkC) plays a central role in a very broad range of cellular responses and is implicated in a spectrum of human pathologies, including Alzheimer’s, Parkinson’s diseases, pain, and notably cancer8–13. Here, we have utilized the TrkA of neuronal growth
factor (NGF) as a model membrane kinase to investigate the site-dependent signaling transduction process. The Trk kinase family is expressed in a host of cells throughout the mammalian body, including neurons8,9,13,14. TrkA in particular was first to be
identified as a specific receptor for NGF and is activated upon NGF binding15,16. Activation of TrkA upon NGF binding at the cell surface leads to phosphorylation of tyrosine residues in the cytoplasmic domain, which in turn recruits signaling molecules and activates multiple signaling pathways involving AKT, MAPK/ ERK, and PI3K13. Single-molecule tracking shows that NGF
binding induces TrkA dimerization or oligomerization17 to
facilitate the recruitment of signaling transduction.
The cytoplasmic region of monomeric TrkA consists of five conserved tyrosine residues Y670, Y674 and Y675 within the tyrosine kinase domain, and Y490 and Y785 outside the kinase domain. Phosphorylation of Y490 requires the recruitment of adaptors Shc or Frs2 for activation of the AKT and MAPK/ERK pathways13,18while phosphorylated Y785 recruits PLC-γ113,18,19.
The MAPK/ERK pathways activated by TrkA include ERK1/ ERK2, and ERK5, which phosphorylate and activate downstream transcription factors to regulate target gene expression con-tributing to neuronal differentiation20. Phosphorylation on Y670, Y674, and Y675 are less well understood. Based on the conven-tional mutagenesis, it has been shown that phosphorylation in the activation loop of the kinase domain on Y670, Y674, and Y675 enhances the kinase activity21. However, a specific pathway after
phosphorylation for each site remains unaddressed.
To understand the pathway, it requires a strategy to control the phosphorylation state of the individual tyrosine with high-spatial and temporal resolution. We have focused on the applications of genetic code expansion technique to reprogram proteins in response to light22,23. A significant advance has been made through the development of genetic code expansion involving the readthrough of an amber (UAG) codon in an mRNA by a sup-pressor tRNA aminoacylated with the unnatural amino acid (Uaa)24,25. Numerous Uaas with chemical, physical, and
biolo-gical properties not present in the natural amino acids have been incorporated into proteins in eukaryotic systems, particularly the
mammalian systems by the introduction of engineered aminoacyl-tRNA synthetase (aaRS)/suppressor tRNA pairs that aminoacylate the suppressor tRNA in vivo with the desired Uaa. The light-sensitive Uaas have been demonstrated to switch pro-tein function after light stimulation23,26. Light is a unique signal input with three major advantages for studying biological func-tions: (1) it has high-temporal and spatial resolution to control a biological reaction; (2) reaction kinetics are faster and more specific than small molecule-based controls that rely on molecule diffusion; (3) cooperative mechanisms can be elucidated by combining controls by light and other molecules including ligand27.
Our approach takes advantage of the recent development of the genetic code expansion in mammalian cells. We were mainly focused on the introduction of a light-switch to regulate the phosphorylation state of the tyrosine side chain in TrkA. Two types of ultraviolet (UV) light-sensitive tyrosine analogs have been exploited: (1) caged-tyrosine (ONB), which releases a pho-tolabile moiety from the side chain upon light activation28,29; (2)
photo-cross-linking p-azido-L-phenylalanine (AzF), which crosslinks to a nearby molecule within the distance of 3–4 Å upon light activation30. The purposes of the present study were
several-fold. The first was to devise a suitable light-controlled metho-dology for the concept that the phosphorylation state of tyrosine can be specifically turned “on” to identify the downstream sig-naling process involved with the ligand binding in a kinase receptor. The second was, in the case of the TrkA receptor, to determine how three tyrosine residues in the tyrosine kinase domain initiate ERK signaling. We also sought to exploit our biological screens of light-induced ERK function to identify TrkA receptor that can be switched on without the ligand NGF. Finally, we have applied our approach to neuronal SH-SY5Y cell lines, providing evidence for the transferability of light-sensitive TrkA to control neurite growth. By embedding the methodology into the fast-growing field of molecular optogenetics, we highlight both the potential and limitations of this technique, and provide future perspectives for its use in neurophysiology.
Results
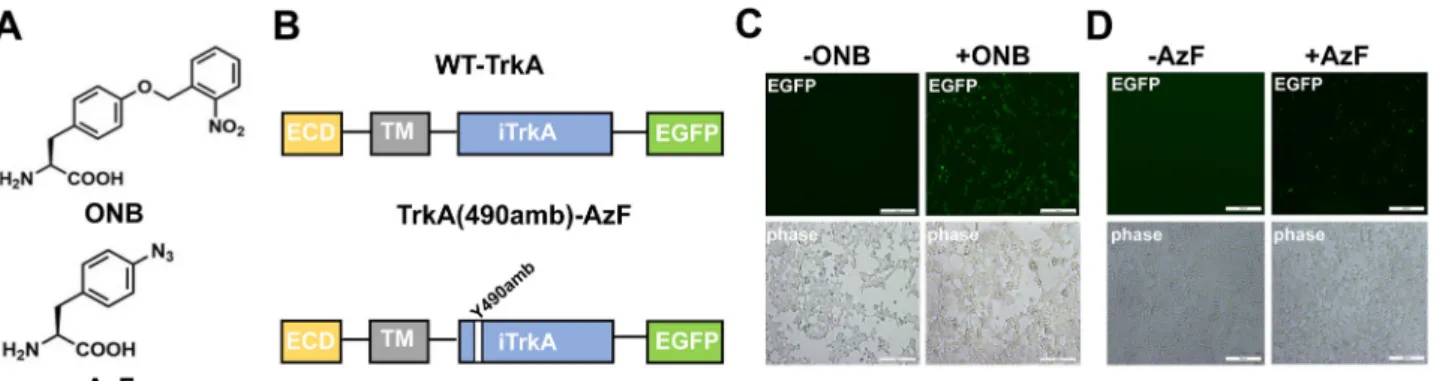
ONB and AzF site-specific incorporation into TrkA-Y490. To establish the site-specific incorporation of ONB and AzF (Fig.1a) into TrkA, wefirst constructed EGFP tagged TrkA receptor (wt) and the amber codon mutated TrkA receptor (Y490amb) (Fig.1b), to detect their expression in mammalian cells using the fluorescent signal. Based on this wt version, the amber mutation (UAG) was introduced into the Y490 position to generate the TrkA(490amb). When co-expressed the TrkA(490amb) with the plasmid encoding pONBYRS/U6-PyltRNA29orthogonal to ONB
(Fig. 1c) or the plasmid encoding AzF-RS/Bst-tRNACUA31
orthogonal to AzF (Fig.1d) in the presence of 1 mM Uaa in the cell media, fluorescent signals can be observed, suggesting suc-cessful incorporation of ONB or AzF in each condition. This AzF-RS/Bst-tRNACUA construct was compared with plasmids
separately encoding EcoliAzF-RS and Bst-tRNACUATyr, showing
similar expression levels quantified by the dual EGFP-mCherry reporter31(Supplementary Fig. 1). In the absence of AzF, there is
a minimal level offluorescent signal, suggesting the EcoliAzF-RS and Bst-tRNACUATyris orthogonal both in HEK293T and PC12
cells with a minimal level of misincorporation in the stop codon. To quantify the expression level of TrkA-EGFP, we first established the quantification procedure using a regular soluble EGFPamb reporter construct31, which only gives full-length expression protein when the amber mutation at position Y39 is suppressed (Supplementary Fig. 2a), with 44% of cells co-transfected with EGFPamb and AzF-RS/Bst-tRNACUA showing
fluorescent when adding AzF, compared to 100% of cells transfected with wt EGFP. In TrkA(490amb)-EGFP, there was 52.6% of cells showing fluorescent when adding AzF, compared to 100% of cells transfected with TrkA-EGFP (Fig. 1d and Supplementary Table 1), indicating that a remarkable yield of the membrane receptor TrkA-EGFP, which is comparable to the water-soluble EGFP.
Use HEK293T cells for TrkA site-dependent ERK activation. NGF binding to TrkA induces AKT or MAPK/ERK signaling cascades, and the Y490 site has been associated with these two pathways inferred by the mass-spectrometry analysis18(Fig.2a).
To determine the TrkA dependent pathway activations, we pro-bed the endogenous TrkA activation of MAPK/ERK in HEK293T and PC12 cell line. HEK293 cells are popular cell hosts for gene expression and analysis of the expressed protein due to its transferability by the various techniques. In comparison, the PC12 cells are useful as a model system for NGF neuronal dif-ferentiation and neurosecretion. We determined MAPK/ERK activation by the immunoblot analysis of phosphorylated ERK (p-ERK1/2, Thr202 and Tyr204), phosphorylated Akt and phos-phorylated creb (p-creb, Ser133) (Fig. 2b). In both cases, HEK293T cells and PC12 cells were treated with NGF for 10 minutes. Following ligand application, cells were immediately lysed and probed for TrkA, p-erk, p-akt, p-creb and actin as a loading control. In HEK293 cells where little endogenous TrkA was detected (with a faint band observed), there is no detectable
p-erk. In contrast, PC12 cells showed a high level of TrkA expression, it showed strong p-erk. For both cells, similar levels of p-creb and p-Akt were observed. HEK293T cells transfected with TrkA demonstrated p-erk signal, demonstrating a clear contrast from the non-transfected cells (Fig.2c).
To directly compare the expression of Uaa incorporated TrkA in HEK293T or PC12 cells, we co-transfected the low-differentiating PC12 cells using the same protocol as in HEK293T cells keeping the TrkA-Y490amb and AzF-RS/Bst-tRNACUAat same gene dosages. We observed that the yield in
PC12 cells is significantly lower (3% of the WT-TrkA) (Supplementary Figs. 3 and 4) than in HEK293 cells (52.6% of the WT-TrkA) presumably due to the challenge in lipofectamine mediated transient transfection in the neuronal cell line. We further characterized TrkA-Y490AzF induced ERK activation in HEK293 cells (Fig. 2). The expression of TrkA-Y490AzF in HEK293T cells was analyzed byfluorescent imaging and western blotting using wild-type TrkA as a control (Supplementary Fig. 5), which showed general agreement. TrkA-Y490AzF was synthe-sized only in those cells co-expressing EcoliAzF-RS and Bst-tRNACUATyr in the presence of exogenously added AzF
(Supplementary Fig. 5a–c, lane 4). In the absence of AzF, we observed no expression of TrkA by western blotting analysis (Supplementary Fig. 5a–c, lane 3) and only negligible EGFP signal (Supplementary Fig. 5a, b, lanes 3), confirming the stop of translation. The measurement of EGFP signals allowed us to quantitate the level of TrkA-Y490AzF expression, which was consistently found to be 52.6% of wild-type TrkA expression
Fig. 2 ERK activation by TrkA. a Schematic diagram of TrkA signaling pathway. b Western blot analysis of PC12 and HEK293T cells due to endogenous TrkA in the presence of ligand NGF for p-erk (phosphorylated ERK at Thr202 and Tyr204), phosphorylated akt, phosphorylated CREB.β-actin was used as loading control.c Western blot analysis of HEK293T cells in non-transfected (Ø) and transfected with wt-TrkA (wt) probing for endogenous TrkA, p-erk and total ERK.β-actin was used as loading control. d Western blot analysis of HEK293T cells expressing wt-TrkA, without (−) or with (+) NGF, and without (−) or with (+) UV light, for pERK. β-actin was used as loading control.
Fig. 1 The ONB and AzF incorporation in TrkA. a Structure of ONB and AzF. b Schematic diagram of the constructed TrkA receptor tagged with EGFP at the C-term. The full-length wt-TrkA receptor consists of an extracellular domain (ECD), a transmembrane domain (TM), an intracellular domain (iTrkA) and EGFP. TrkA(Y490amb) has an amber mutation (UAG) introduced into the Y490 position in the iTrkA domain.c, d Fluorescent images of live cells co-transfected with RS/tRNA4xat one site out of tyrosine kinase domain Y490, in the presence (+) or absence (−) of 1 mM AzF (c) and 1 mM ONB (d).
levels. Thus, we made the use of EGFP to visualize the expression of TrkA. We then studied the p-erk activation of TrkA-Y490AzF. Interestingly, immunoblot analysis of p-ERK showed that TrkA-Y490AzF mutant could not initiate p-erk (Supplementary Fig. 5c), revealing the replacement of azido group at Y490 site inhibited the autophosphorylation. In summary, these results showed that HEK293T is a convenient cellular model compared to PC12 cells to express TrkA site-specifically incorporated with a UAA and study the MAPK/ERK signaling. Unlike PC12 with a high level of endogenous TrkA expression, HEK293T has a low level of endogenous TrkA, it exhibits clear-cut p-erk activation only when exogenous TrkA was expressed.
Incorporation of AzF and ONB in tyrosine kinase domain sites. Using the Y490 site as a reference, we next studied the incorporation of AzF and ONB into TrkA at three different sites in the tyrosine kinase domain, Y670, Y674, and Y675, respectively (Fig. 3a). Fluorescent imaging analysis showed that TrkA-AzF mutants were produced to ~50% (ranged from 33.4 to 52.6%) of the WT-TrkA (Fig.3b, Supplementary Fig. 6a, and Supplemen-tary Table 1). In comparison, TrkA-ONB mutants all had a slightly lower level of expression than those containing AzF, ranged from 29.1 to 35.8% of the WT-TrkA (Fig. 3c, Supple-mentary Fig. 6b, and SuppleSupple-mentary Table 1). The average yields of TrkA mutants were calculated (Supplementary Table 1). The differences in the yields measured byfluorescent imaging overall suggest that we observe efficient amber suppression at various target sites and consequently efficient synthesis of receptor pro-teins containing AzF and ONB. In addition, WT-TrkA and its mutant variants localized primarily on the plasma membrane (Supplementary Fig. 7a, b).
UV initiated site-dependent ERK signaling. Wefirst tested the UV-stimulated activation of p-erk with the mutant TrkA-Y490ONB. Under UV light stimulation, ONB releases the caged group that restores the tyrosine, and the hydroxyl-group on tyrosine undergo autophosphorylation (Supplementary Fig. 8a and Supplementary Fig. 11a, b). To evaluate whether ONB photo-activation at Y490 site produces functional changes in ERK sig-naling, we applied UV light on live-cells expressing the mutant and by immunoblot analysis of p-erk post-UV treatment. HEK cells were transfected with the TrkA-Y490ONB and subjected to continuous UV light stimulation (40w) for 5 min in the presence
of ligand NGF. Following light illumination, cells were immedi-ately lysed and probed for p-erk, total erk, and actin as a loading control. In the absence of UV light, no p-erk was detected in TrkA-Y490ONB due to the presence of the caged group in ONB. UV illumination induced the clear appearance of p-erk compared to the control TrkA-Y490ONB (Fig.4a). In summary, our results successfully demonstrated the feasibility of p-erk activation through the light control at a single phosphorylation site in TrkA, in which the Uaa ONB serves as a robust light-switch to control the phosphorylation state.
We then studied whether incorporation of ONB into any of other three tyrosines could also initiate p-erk signaling in TrkA-670, 674, and 675 (Fig. 4a and Supplementary Fig. 11). In the absence of UV treatment, no p-erk was detected for any of the mutants. After UV treatment, two mutants (TrkA-Y674ONB and TrkA-Y675ONB) showed robust p-erk activation similar to what has been observed in the TrkA-Y490ONB mutant. Interestingly, the TrkA-Y670ONB mutant showed no detectable p-erk with or without UV treatment. To quantify the extent of p-erk, the intensity of p-erk signal was measured and normalized to the total ERK signal among all the mutants either in dark or under UV illumination (Supplementary Fig. 9a and Table1). After UV light stimulation, Y490ONB initiated 36.2% of p-erk, TrkA-Y674ONB initiated 44.6% and TrkA-Y675ONB initiated 32.0%, significantly more than their absence of UV controls (16.3%, 7.3%, and 4.4%, respectively) and the light-insensitive mutant TrkA-Y670ONB (absence of UV: 3.1%, presence of UV: 3.6%). As an additional control, we have made conventional mutants namely: TrkA-Y490F, Y670F, Y674F, Y675F, Y490D, Y670D, Y674D, and Y675D. The phenylalanine (F) is considered to abolish the phosphorylation, whereas the aspartic acid (D) may mimic the phosphorylated state. For all the F mutants, we have observed a significant reduction of the p-erk in compared to the wild-type receptors (Supplementary Fig. 10). These results demonstrated that ONB as a light-switch is sufficient for the gain of function to induce a downstream ERK signaling provided with the incorporation at a proper phosphorylation regulatory site (Fig.4b). Among the three tyrosines (Y670, Y674, and Y675) considered being within the activation loop of the TrkA kinase domain, Y674 and Y675 (but not Y670) may share the same mechanisms as the Y490 to initiate pERK signaling by autophosphorylation.
We next examined among all four sites the light-induced effect of AzF, which undergoes different photochemistry than ONB.
Fig. 3 Expression of TrkA mutants containing AzF or ONB at positions 490, 670, 674, and 675, respectively. a Schematic diagram of the full-length TrkA with ECD, TM and intracellular TrkA domains. Ligand NGF binds at the ECD domain. Four phosphorylated tyrosine sites subjected to mutation are highlighted.b The incorporation efficiency of TrkA AzF mutants in the absence (gray) or presence (dark) of AzF. c The incorporation efficiency expression of TrkA ONB mutants in the absence (gray) or presence (dark) of ONB. The incorporation efficiency was described as the percentage ratio of average fluorescence intensity in cells expressing Uaa-incorporating receptors divided by the average fluorescence intensity in cells expressing the WT receptors. n = 6 independent experiments. Error bars show s.d.
Upon UV excitation, AzF usually generates a nitrene radical, which may reduce to amine or forms a covalent linkage with the nearby atom of a protein at a distance of 3–4 Å30,32
(Supple-mentary Fig. 8b). Strikingly, except the TrkA-490AzF mutant showing no detectable p-erk after UV light stimulation (absence of UV: 11.4%, presence of UV: 7.8%), all three mutants
TrkA-Y670AzF, TrkA-Y674AzF, and TrkA-Y675AzF demonstrated UV-controlled p-erk activation (34.1%, 29.7%, and 37.4%, respectively) (Fig. 4c, Supplementary Fig. 9a, and Table 1), significantly more than their controls treated without UV (12.4%, 12.5%, and 10.1%, respectively). Interestingly, the Y674D and Y675D mutants also demonstrated different levels of reduction of Table 1 All sites were detected under the same conditions as wt, and were repeated more than three times.
Relative level of p-erk (%)(+NGF) ONB AzF
−UV +UV −UV +UV
(a) wt 65.1 ± 0.2 56.8 ± 0.1 TrkA-Y490 16.3 ± 0.1 36.2 ± 0.2 11.4 ± 0.2 7.8 ± 0.09 TrkA-Y670 3.1 ± 0.004 3.6 ± 0.009 12.4 ± 0.07 34.1 ± 0.09 TrkA-Y674 7.3 ± 0.002 44.6 ± 0.2 12.5 ± 0.03 29.7 ± 0.02 TrkA-Y675 4.4 ± 0.005 32.0 ± 0.1 10.1 ± 0.05 37.4 ± 0.2 (b) wt 16.4 ± 0.009 10.4 ± 0.01 TrkA-Y490 18.5 ± 0.2 16.9 ± 0.2 12.5 ± 0.04 8.6 ± 0.008 TrkA-Y670 9.7 ± 0.07 16.7 ± 0.2 8.4 ± 0.08 17.6 ± 0.2 TrkA-Y674 10.6 ± 0.08 34.5 ± 0.1 26.6 ± 0.1 80.4 ± 0.3 TrkA-Y675 3.7 ± 0.01 37.1 ± 0.2 7.9 ± 0.05 39.0 ± 0.2
Table 1(a) list all conditions treated with NGF (+NGF) and 1(b) without UV treatment (−NGF). For each measurement, the intensity of phosphorylated ERK to total ERK was measured, and the ratio of p-ERK/total ERK was calculated and presented as the relative level of p-erk (%). Error show s.d.
Fig. 4 UV light activates MAPK/ERK signaling pathway by site-specifically incorporated AzF and ONB in TrkA. a Western blot analysis of HEK293T cells expressing ONB mutants for phosphorylated ERK (Thr202 and Tyr204) in the presence of ligand NGF without (−) or with (+) UV light. b Binding of the NGF drives TrkA dimerization, autophosphorylation and initiates intracellular MAPK/ERK. In the presence of ligand NGF, ONB incorporated at the Y490, Y674, and Y675 (solid circles) sites in TrkA, respectively, after UV treatment initiate intracellular MAPK/ERK signaling (arrows). ONB incorporated in Y670 (empty circle) site does not initiate intracellular MAPK/ERK signaling (inhibited arrow)c Western blot analysis of HEK293T cells expressing AzF mutants for phosphorylated ERK (Thr202 and Tyr204) without (−) or with (+) UV light. UV photolysis was done with 365 nm UV cross-linker CL-1000L (Analytik Jena) at 40w for 5 min at 4 °C. TrkA andβ-actin were used as loading control. d Binding of the NGF drives TrkA dimerization, autophosphorylation and initiates intracellular MAPK/ERK. In the presence of ligand NGF, AzF incorporated at the Y670, Y674, and Y675 (solid circles) sites in TrkA, respectively, after UV treatment initiate intracellular MAPK/ERK signaling (arrows) presumably due to the crosslinking with a putative interactive protein. AzF incorporated in Y490 (empty circle) site does not initiate intracellular MAPK/ERK signaling (inhibited arrow).
p-erk. The Y490D and Y670D mutants maintained the same level of pERK activation compared to the WT. Taken together, these results indicate that AzF can serve as a light-switch to activate downstream ERK signaling in TrkA receptors, presumably through the AzF mediated photo-cross-linking mechanism (Fig. 4d) that is different from the photochemistry, which generated phenylamino side chain (Supplementary Fig. 8a, b). NGF independent light-controlled ERK signaling. Next, we tested whether UV light stimulation in TrkA could directly activate ERK without NGF (Fig. 5a and Supplementary Fig. 11j–m). All mutants were expressed and assayed in the absence and presence of NGF, and wt-TrkA serves as a control with no detectable p-erk neither without nor with UV treatment, confirming that the UV treatment per se does not cause sig-nificant functional changes. Interestingly, among four ONB mutants, in the absence of NGF, both Y674ONB and TrkA-Y675ONB showed UV-induced p-erk activation compared to the control without UV treatment (80.4% and 39.0%, respectively) (Fig.5b, Supplementary Fig. 9b, and Table1). The UV dependent activation was similar to the controls treated in the presence of NGF. In contrast, the TrkA-Y490ONB only demonstrated UV light-induced p-erk activation in the presence of NGF, whereas the TrkA-Y670ONB showed no detectable p-erk activation under all conditions (Fig.5b, c). We repeated the same experiment with AzF mutants, similar to the Y674ONB and TrkA-Y675ONB, TrkA-Y674AzF and TrkA-Y675AzF had NGF inde-pendent UV-induced p-ERK activation (34.5% and 37.1%, respectively) (Fig. 5d, Supplementary Fig. 9b, and Table 1). Whereas the TrkA-Y490AzF showed no detectable p-erk activa-tion under all condiactiva-tions, and the TrkA-Y670AzF only demon-strated UV light-induced p-ERK activation in the presence of NGF. All these results validated that light-induced activation through AzF or ONB incorporation of p-erk is highly effective in mimicking NGF function at Y674 and Y675 sites (Fig.5d, e).
To confirm if TrkA AzF mutant crosslinks to some proteins under UV treatment, we implement the photo-cross-linking experiment. We tested WT-TrkA, Y670AzF, TrkA-Y674AzF and TrkA-Y675AzF in the presence of NGF and absence of NGF, without and with UV (Supplementary Fig. 12). For all conditions, TrkA monomer could be detected as a characteristic 180 kDa band. Upon UV exposure, we observed higher molecular bands for TrkA-Y674AzF and TrkA-Y675AzF mutants (~300 kDa) in both +NGF and –NGF conditions, but not in WT or TrkA-Y670AzF.
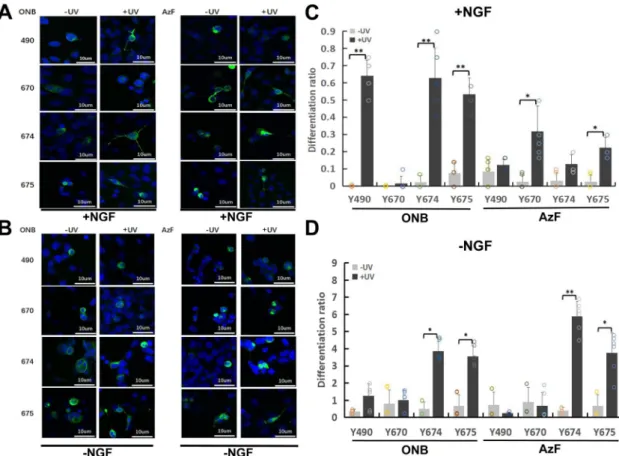
Expressing light-sensitive TrkA in neuronal SH-SY5Y cell line and controlling neuronal differentiation. Finally, we sought to test light-sensitive TrkA mutants in neurons. We decided tofirst concentrate on the establishment of the neurite growth control by light in SH-SY5Y cells, which do not have the endogenous TrkA that would obscure the functional effect from the mutant TrkA, as well as from the light activation. For that, we have tested whether the TrkA-ONB or AzF mutants could be successfully expressed. Following transient transfections of the constructs and supple-mentation of the culture media with Uaa (1 mM), treated exactly the same as for HEK293 cells and PC12 cells, we observed a large number of bright greenflorescent cells (Supplementary Fig. 7b), which were absent in controls where Uaa was not added. This demonstrated proper genetic encoding of the Uaa through the rescue of the amber stop codon by the orthogonal tRNA/aaRS pair. Following this successful implementation of light-sensitive TrkA Uaa mutants in SH-SY5Y cells, we then aimed at assaying the neurite growth upon activation of each tyrosine with UV light and compared their differentiation in the presence and absence of NGF (Fig. 6). SH-SY5Y cells do not project neurites with NGF stimulation. However, when they are expressed with TrkA and treated with NGF, cells project neuritis (Supplementary Fig. 7c), which is a process referred to as differentiation. We proceeded to
Fig. 5 UV light activates MAPK/ERK signaling pathway independent of ligand NGF. Western blot analysis of HEK293T cells expressing wt-TrkA (a), ONB mutants (b) including Y490ONB, Y670ONB, Y674ONB, Y675ONB, and AzF mutants (d) including Y490AzF, TrkA-Y670AzF, TrkA-Y674AzF, TrkA-Y675AzF; without (−) or with (+) NGF, and without (−) or with (+) UV light. TrkA and β-actin were used as loading control. In the absence of ligand NGF, ONB (c) or AzF (e) incorporated at the Y674 and Y675 (solid circles) sites in TrkA, respectively, after UV treatment initiate intracellular MAPK/ERK signaling (arrows). In the case of AzF, the signaling activation is presumably due to photo-cross-linking of an interactive protein in the MAPK/ERK signaling complex. ONB (c) or AzF (e) incorporated in Y490 and Y670 (empty circle) site does not initiate intracellular MAPK/ ERK signaling (inhibited arrow).
determine whether TrkA-Uaa mutants could achieve a similar cellular outcome with UV light stimulation.
SH-SY5Y cells expressing wt-TrkA showed robust differentia-tion when treated with 100 ng/mL NGF, indicating that exogenous TrkA retained ligand sensitivity and signaling activity (Supplementary Fig. 7c). UV light did not significantly alter the differentiation. No differentiation was observed for non-transfected cells. To determine the functional role of Y490, Y670, Y674 and Y675 in differentiation, we transfected SH-SY5Y cells with mutant constructs and illuminated them for 30 min as previously described for western blotting experiments (Fig.6) and compared their neurite growth to the WT-TrkA. In the presence of NGF, cells expressing Y490ONB, Y674ONB, Y675ONB, Y670AzF and Y675AzF showed significant UV-induced differ-entiation compared with UV untreated condition (Differdiffer-entiation ratios: 64.2, 62.8, 53.3, 31.9, and 22.2, respectively) (Fig.6a, c and Supplementary Table 2). In the absence of NGF, Y674ONB, Y675ONB, Y674AzF and Y675AzF showed UV-induced differ-entiation compared with UV untreated condition (Differdiffer-entiation ratios: 3.9, 3.6, 5.9, and 3.8, respectively) (Fig. 6b, d and Supplementary Table 2). Overall, there is a general agreement between ERK activity and cell differentiation.
Discussion
The common concept of TrkA signaling activation is that the binding of ligand NGF from the extracellular domain promotes
homo-oligomerization of TrkA on the cell membrane, which subsequently activates downstream signaling pathways. However, because of its intricate downstream pathway associated with multiple phosphorylation sites within the intracellular segment, the TrkA signaling is not yet fully elucidated. Particularly, the three sites Y670, Y674, and Y675 located in the active region of tyrosine kinase, are closely related to enzyme activation. Previous findings indicated that mutation of any of these three activation loop tyrosines leads to diminished TRK function21,33, however,
specific signaling process related to MAPK/ERK could not be elucidated due to the lack of site-controllable phosphorylation.
We were particularly interested in developing a framework for light-controlled protein function through site-specific control, which requires the availability of homogeneously expressed pro-teins with a light-switch installed at a specific site within the protein. In the two past decades, optogenetics based on natural light-sensitive proteins has evolved as a powerful strategy to study cellular functions with the precision of protein-specificity34.
Despite the power of manipulation of biological functions with cell-type and protein specificity, the regulation of a protein function with site-specificity by light remains a major challenge in the optogenetic approach. Although site-specific cysteine/mal-eimide labeling has been applied to render protein light-sensitive by attaching photo-switching ligands, the method can not be generalized to control functions associated with post-translation modifications such as phosphorylation. Here our approach by exploiting the genetic code expansion technology23–25, in
Fig. 6 TrkA-ONB and AzF mutants promote SH-SY5Y cell differentiation in response to UV Stimulation with and without NGF. a Representative fluorescence images depicting SH-SY5Y cells expressing TrkA-ONB (two columns on the left) or AzF (two columns on the right) mutants incorporated at indicated sites before (-UV) and after (+UV) treatment, with medium supplied with 100 ng/mL NGF for 30 min and kept in the dark for 24 h prior to imaging.b Representativefluorescence images depicting SH-SY5Y cells expressing TrkA-ONB (two columns on the left) or AzF (two columns on the right) mutants incorporated at indicated sites before (-UV) and after (+UV) treatment with medium supplied without NGF. c Differentiation ratios were calculated for various mutants expressed in SH-SY5Y cells in the presence of NGF with or without UV light.d Differentiation ratios calculated for SH-SY5Y cells in the absence of NGF expressing wt-TrkA (WT) or various mutants with or without UV light. Differentiation ratios were calculated as described in the method. Values represent the mean ± SD of six biological replicates (n = 6) with >200 cells counted per replicate. Scale bars, 10 µm.
combination with the light-sensitive Uaas and site-directed mutagenesis to optically activate membrane kinase TrkA with the site-precision provides a novel concept and tool-kit to investigate the NGF/TrkA signal some with clear-cut readout.
Our technical advantages over classical phosphomimetic mutations to aspartic acid and glutamic acid are that both ONB and AzF are chemically different from tyrosine so to block phosphorylation, and each undergoes different photo-chemistries. After UV light treatment, ONB releases the caging group to recapitulate the molecular and phenotypic consequences of Tyr. Light-dependent quantitative analysis for “gain-of-function” phenotypes lends strong support to the advantage of light-control of signaling process via Uaa to analyze each tyrosine’s contribu-tion to the specific pathway activacontribu-tion, which convencontribu-tional mutagenesis cannot afford. The two different Uaas provide unique photochemical mechanisms to control TrkA function by light, an advantage that the light-sensitive protein-based opto-genetic approach cannot offer. The remarkable tolerance of all tested sites for ONB and AzF incorporation can be attributed to the orthogonality of aaRS/suppressor tRNA pairs extensively optimized in mammalian systems22,29,31,35,36. The extent of incorporation of AzF was found to vary from ~52.6 to 33.4%, compared to ONB varying from 29.1 to 35.8%. We found that the mammalian HEK293T and SH-SY5Y cells are better cellular models than the neuronal PC12 cell line in studying TrkA sig-naling due to two main reasons: (1) minimal interference from the endogenous TrkA, (2) higher yield in amber suppression by the orthogonal aaRS/suppressor tRNA pairs. Between the two types of tyrosine analogs AzF and ONB introduced into four phosphorylation sites, we identified three categories of mutants: (1) UV-insensitive mutants (Y670ONB and TrkA-Y490AzF), (2) UV-sensitive mutants in the presence of ligand NGF (TrkA-Y490ONB and TrkA-Y670AzF), (3) UV-sensitive mutants independent of ligand binding (Y674ONB, TrkA-Y675ONB, TrkA-Y674AzF and TrkA-Y675AzF).
Their responses of ERK activation indicate two points of interest: (1) differences of ONB and AzF in Y490 and Y670 sites, and (2) ligand-independency observed for Y674 and Y675 sites. The interpretation of differential light activation between ONB and AzF in site Y490 is the most straightforward. The activation of the ERK pathway by irradiation cannot occur unless the caged compound ONB on Y490 has been released. The difference between Y490ONB being ligand-dependent for light-controlled p-erk activation may reflect the ligand-induced state-dependent activation process. When mutating Y490 to AzF, before light treatment, it is similar to conventional site-directed mutagenesis such as phenylalanine, which leads to the inactivation of MAPK/ ERK signaling. The lack of ERK activation after UV treatment confirms that the AzF used different photochemistry to ONB and the photo-product of AzF was unable to be phosphorylated. However, for the site of Y670, surprisingly we have observed that the introduction of AzF resulted in a light-sensitive ERK activa-tion but not in ONB mutaactiva-tion. This represents the first obser-vation that a photo-product of AzF leads to the actiobser-vation of a signaling pathway. Our interpretation is the following: unlike Y490, the Y670 locates in the activation loop that has been pro-posed to undergo significant conformational changes upon ligand binding21. Although we observed Y670 as a site in which ONB
and AzF showed clear differences in light-sensitivity, more direct structural data is required to infer information about the func-tional difference. It is probable that the nitrobenzyl-caging group in the ONB obstructs the loop conformation and induces an unfavorable conformation for kinase activity both before and after light stimulation. When AzF was introduced, due to the smaller size of the side chain more closely mimicking the tyrosine,
such obstruction did not occur. After light stimulation, the acti-vation of ERK signaling suggests that AzF may undergo cross-linking to an interactive counterpart that triggers the ERK signaling cascade (Fig. 5c, e).
We provide here a step in the development of light-controlled kinase receptors in a ligand-independent manner. Our results in Y674 and Y675 sites incorporated with ONB or AzF all demon-strate that the ERK signaling is independent of ligand binding. Previously, it has been shown that adaptor proteins Grb2, SH2B, and rAPS can bind to the phospho-tyrosines at Y670, Y674, and Y67537,38. Therefore, our results may suggest all three Y670, Y674, and Y675 sites are using protein–protein interactions to activate MAPK/ERK pathway, presumably through one or more of those adaptor proteins. AzF as a photocrosslinking Uaa has been previously applied in probing protein-ligand30,31,39,40 and protein–protein3,41,42 interactions. Our crosslinking results on
the TrkA-AzF mutants demonstrate the potential of the approach to investigate the light-dependent signaling mechanisms43,44, and opens the door for us to investigate the putative binding partners of the TrkA receptors. By coupling with mass-spectrometry, we foresee identifying the docking partners of TrkA docking at specific sites. We also note that AzF is a versatile Uaa with multiple applications, including serving as an infrared probe22or
chemical handle for bioorthogonal conjugations45. This is a unique advantage of the AzF incorporation that other methods cannot afford. Taken together other works on photocaged tyr-osines and lysines29,46,47in light-controlling intracellular kinase activity, the availability of multiple Uaas with different chemical properties open new avenues for using genetically encoded Uaas to signaling transduction, an approach likely to extend to other membrane kinase receptors such as epidermal growth factor receptor (EGFR), p75NTR (low-affinity NGF binding receptor), insulin-like growth factor receptor (IGFR), which are all struc-turally and functionally related to TrkA13,21. The genetic encoding technique is also well-suited for biological applications as it allows for both receptor-subtype selectivity and cell-type specificity.
In conclusion, our results demonstrate that Uaas can be effi-ciently introduced into membrane kinases to render them light-sensitive. The broad applicability of light-sensitive Uaas in molecular engineering and cellular physiology emphasizes its significance in the fields of optogenetic pharmacology and optical monitoring of protein conformational changes. A wealth of structural and functional information can be gained using this technology in vitro on recombinant proteins, while, in vivo, it has the potential to address fundamental questions regarding native receptor biology. Given its broad utility and functionally rich implementation, Uaa insertion is poised to become increasingly popular for investigating protein structure, function and dynamics. By scanning through different phosphorylation sites, we managed to identify light-sensitive TrkA independent of ligand NGF. Compared to the recent demonstration of ligand-independent sensitive TrkA based on the fusion of light-sensitive protein Arabidopsis cryptochrome 248–50, our TrkA
relies on the light activation of a specific phosphorylation site activating a specific signaling pathway using the two kinds of orthogonal tyrosyl-tRNA and pyrrolysyl- tRNA synthetase/ tRNACUA pairs to incorporate light-sensitive Uaas. The
intro-duction of Uaas to TrkA creates a much smaller change (10-100 Dalton) to the receptor than the light-sensitive proteins (~100 K Dalton); therefore, it is likely to maintain the function of the receptors, including ligand binding, protein–protein interactions and oligomerization. Since both pairs can be used to site-specifically incorporate Uaas in the multicellular model organ-isms, including zebrafish and mice we27 and others
Materials. p-azido-L-phenylalanine (AzF) was purchased from Chem-Impex International (Wood Dale, IL) and ONB was synthesized29. Standard E. coli genetic
techniques were performed53. E. coli strains XL-10 gold strain (TetrΔ (mcr A)183 Δ
(mcr CB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F’ proAB lacIqZΔM15 Tn10 (Tetr) Amy Camr] from ZEYE and TOP10 [F- mcrAΔ
(mrrr-hsdRMS-mcrBC)φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG] from Invitrogen were used for plasmid propagation and isolation. Oligonucleotides were obtained from Eurofins and Gene Synthesis Technologies. Plasmid DNA was purified using standard Maxi Prep Kits from Generay.
Plasmids for expression of EGFP, EGFP-mCherry, and TrkA-EGFP in mam-malian cells. The plasmids encoding wild-type EGFP (EGFP.wt), a variant thereof carrying a Y39am mutation (EGFP.Y39am) and EGFP-mCherry carrying an amber mutation were described earlier31. Plasmid PLNCX carrying the gene for wild-type
rat TrkA has been described54. An EGFP insert was prepared by PCR reaction and
subcloned into PLNCX-TrkA between the ClaI and StuI sites. This ligation created an inframe fusion between the TrkA carboxyl terminus truncating the last 4 amino acids and the EGFP amino terminus. Amber mutations were introduced into TrkA (TrkA.amb) at positions Y490, Y670, Y674, and Y675 using Quikchange muta-genesis (Stratagene). All plasmid constructs were confirmed by automated DNA sequencing.
Plasmids for expression of suppressor tRNA and mutant aaRS genes in mammalian cells. Plasmid AzF-RS(tRNA)4Xcarrying the gene encoding the
mutant E. coli Tyr tRNA-synthetase for AzF and the amber suppressor tRNA derived from B. stearothermophilus Tyr-tRNACUAhas been described
previously31,36. The plasmid carrying the gene encoding the mutant
Methano-sarcina barkeri pyrrolysine tRNA synthetase for ONB and the amber suppressor tRNA PylRS/tRNACUApair constructed as previously described29.
Cell culture, transfection, and incorporation of AzF and ONB. HEK293T cells (ATCC) were maintained in medium containing Dulbecco’s modified Eagle med-ium (DMEM) (4500 mg/L of glucose; Gibco, Cellgro) supplemented with 10% fetal bovine serum (Biogen). PC12 neuronal cells (ATCC) were maintained in medium containing 1640 (Thermo Fisher), 10% fetal bovine serum (Biogen). SH-SY5Y cells (ATCC) were maintained in medium containing DMEM/F12 (4500 mg/L of glu-cose; Gibco, Cellgro) supplemented with 10% fetal bovine serum (Biogen) and 1x Penicillin–Streptomycin solution). All cells were cultured at 37 °C in a 5% CO2
atmosphere. Cells were washed with phosphate-buffered saline, detached using trypsin/EDTA-solution, re-suspended in fresh growth medium and seeded into cell culture dishes. The cells were routinely tested for mycoplasma contamination. Cells were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. For transfection in a single well of a 6-well plate, 2μg of AzF-RS(tRNA)4X, 2μg of PylRS/tRNACUA, 2μg of TrkA.amb (or
EGFPamb, or EGFP-mCherry) were mixed with Lipofectamine 2000 and added to cells at 70–80% confluence according to the manufacturer’s protocol. Four hours after transfection, cells were fed with an equal volume of DMEM supplemented with 10% fetal bovine serum. The unnatural amino acid AzF was supplied at afinal concentration of 1 mM four hours post-transfection while o-nitrobenzyl (ONB) was supplied at afinal concentration of 50 μM 4 h post-transfection. Cells were har-vested 24–48 h post-transfection and fluorescence imaged or analyzed by western blotting. Suppression of the stop codon in EGFP, EGFP-mCherry, or TrkA results in full-length EGFP, whosefluorescence signal reports the incorporation of AzF or ONB at the TAG stop codon. In the presence of 1 mM AzF in the growth media, EGFPfluorescence intensity of cells, as measured by fluorescence microscopy, increased in comparison to the absence of AzF, indicating the incorporation of AzF or ONB into EGFP, EGFP-mCherry or TrkA.
Microscopy. Forty-eight hours post-transfection, medium was replaced with fresh DMEM and cells analyzed with a Laser-scanning confocal microscope using Bio-Rad Bio-Radiance 2100 system mounted on a Nikon Eclipse TE300 inverted micro-scope equipped with a Plan Fluor EL WD 20x/0.45 objective. Leica DMI 4000B
geQuant. Equal amounts of total protein were analyzed per lane.
NGF and UV treatment. Before NGF stimulation and photolysis experiments, medium was replaced with fresh Dulbecco’s phosphate-buffered saline without AzF or ONB, and cells incubated for 2 h at 37 °C in 5% CO2atmosphere. Stimulation was
achieved by adding human NGF (Millipore) to afinal concentration of 50 ng/ml. As control, an equal amount of NGF-free medium was used. Cells incubated for 15 min, and then they were either washed with ice-cold PBS directly for directly western analysis or for UV treatment. UV photolysis was done with 365 nm UV cross-linker CL-1000L (Analytik Jena) at 40w for 5 min. Afterwards, cells were washed with ice-cold PBS, lysed with RIPA buffer (Sigma) supplemented with phosphatase inhibitors (Pierce) and subjected to western analysis.
SH-SY5Y cell differentiation assay. Cells were harvested 48 h post-transfection and treated without or with NGF, and without or with UV as mentioned above. Differentiation ratios for each mutant at various conditions were calculated at various conditions: Differentiation ratios= (Nm/Nmt)/(NWT/NWTt).
Nm= Number of green fluorescing cells expressing a specific mutant with
neurite length longer than the cell body diameter
Nmt= Number of total green fluorescing cells expressing a specific mutant
NWT= Number of green fluorescing cells expressing WT-TrkA with neurite
length longer than the cell body diameter
NWTt= Number of total green fluorescing cells expressing WT-TrkA
Quantification and statistical analysis. Quantification of western blots was carried out using ImageJ. Details of all statistical datasets are included in the correspondingfigure legends.
List of primers. TrkA-Y490ambF: 5′-GGGAGCAAACAGGATTAGATACCCT-3′ TrkA-Y490ambR: 5′-GAGGTTAACAGAGTGACAGATGGTGCA-3′ TrkA-Y670ambF: 5′-GCATGAGCAGGGACATC-3′ TrkA-Y670ambR: 5′-TAGAGCACAGACTACTACCG-3′ TrkA-Y674ambF: 5′-GACTACAGCACAGAC-3′ TrkA-Y674ambR: 5′-TAGTACCGTGTGGGAGGTC-3′ TrkA-Y675ambF: 5′-CTACAGCACAGACTAC-3′ TrkA-Y675ambR: 5′-TAGCGTGTGGGAGGTCGG-3'
Statistical and reproducibility. All statistical analyses were performed using Microsoft Office Excel. Data are expressed as means ± S.E. Means of two groups were compared using Student′s t-test (unpaired, two-tailed), with P < 0.05 was considered to be statistically significant. All the experiments were performed at least three times with similar results.
Data availability
All data are available in the main and supplementaryfiles or available from authors upon reasonable request. Source Data underlyingfigures and tables are available as Supplementary Data 1. Repeats for gel experiments are shown in Supplementary Information.
Received: 10 May 2019; Accepted: 14 October 2020;
References
1. Fehrentz, T., Schönberger, M. & Trauner, D. Optochemical genetics. Angew. Chem. Int. Ed. Engl. 51, 12156–12182 (2011).
2. Szobota, S. & Isacoff, E. Y. Optical control of neuronal activity. Annu. Rev. Biophys. 39, 329–348 (2010).
3. Pless, S. A. & Ahern, C. A. Unnatural amino acids as probes of ligand-receptor interactions and their conformational consequences. Annu. Rev. Pharm. Toxicol. 53, 211–229 (2013).
4. Huber, T. & Sakmar, T. P. Chemical biology methods for investigating G protein-coupled receptor signaling. Chem. Biol. 9, 1224–1237 (2014). 5. Manning, G., Plowman, G. D., Hunter, T. & Sudarsanam, S. Evolution of
protein kinase signaling from yeast to man. Trends Biochem. Sci. 10, 514–520 (2002).
6. Kholodenko, B. N., Hancock, J. F. & Kolch, W. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 3, 165–176 (2006).
7. Kolch, W., Halasz, M., Granovskaya, M. & Kholodenko, B. N. The dynamic control of signal transduction networks in cancer cells. Nat. Rev. Cancer 9, 515–527 (2015).
8. Huang, E. J. & Reichardt, L. F. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 72, 609–642 (2003).
9. Marlin, M. C. & Li, G. Biogenesis and function of the NGF/TrkA signaling endosome. Int. Rev. Cell Mol. Biol. 314, 239–257 (2015).
10. Hirose, M., Kuroda, Y. & Murata, E. NGF/TrkA signaling as a therapeutic target for pain. Pain. Pract. 2, 175–182 (2016).
11. Sofroniew, M. V., Howe, C. L. & Mobley, W. C. Nerve growth factor signaling, neuroprotection, and neural repair. Annu. Rev. Neurosci. 24, 1217–1281 (2001).
12. Adriaenssens, E. et al. Nerve growth factor is a potential therapeutic target in breast cancer. Cancer Res. 68, 346–351 (2008).
13. Demir, I. E., Tieftrunk, E., Schorn, S., Friess, H. & Ceyhan, G. O. Nerve growth factor & TrkA as novel therapeutic targets in cancer. Biochim. Biophys. Acta 1866, 37–50 (2016).
14. Kaplan, D. R. & Stephens, R. M. Neurotrophin signal transduction by the Trk receptor. J. Neurobiol. 25, 1404–1417 (1994).
15. Kaplan, D. R., Martin-Zanca, D. & Parada, L. F. Tyrosine phosphorylation and tyrosine kinase activity of the trk proto-oncogene product induced by NGF. Nature 350, 158–160 (1991).
16. Klein, R., Jing, S. Q., Nanduri, V., O’Rourke, E. & Barbacid, M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell 65, 189–197 (1991). 17. Marchetti, L. et al. Ligand signature in the membrane dynamics of single TrkA
receptor molecules. J. Cell Sci. 126, 4445–4456 (2013).
18. Biarc, J., Chalkley, R. J., Burlingame, A. L. & Bradshaw, R. A. Dissecting the roles of tyrosines 490 and 785 of TrkA protein in the induction of downstream protein phosphorylation using chimeric receptors. J. Biol. Chem. 288, 16606–16618 (2013).
19. Obermeier, A. 1 Tyrosine 785 is a major determinant of Trk-substrate interaction. EMBO J. 12, 933–941 (1993).
20. Riccio, A., Ahn, S., Davenport, C. M., Blendy, J. A. & Ginty, D. D. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science 286, 2358–2361 (1999).
21. Cunningham, M. E. & Greene, L. A. A function-structure model for NGF-activated TRK. EMBO J. 17, 7282–7293 (1998).
22. Ye, S. et al. Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature 464, 1386–1389 (2010).
23. Chen, Y., Lu, L. & Ye, S. Genetic code expansion and optoproteomics. Yale J. Biol. Med. 90, 599–610 (2017).
24. Liu, C. C. & Schultz, P. G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 (2010).
25. Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
26. Klippenstein, V., Mony, L. & Paoletti, P. Probing ion channel structure and function using light-sensitive amino acids. Trends Biochem. Sci. 43, 436–451 (2018).
27. Gautier, A. et al. How to control proteins with light in living systems. Nat. Chem. Biol. 10, 533–541 (2014).
28. Miller, J. C., Silverman, S. K., England, P. M., Dougherty, D. A. & Lester, H. A. Flash decaging of tyrosine sidechains in an ion channel. Neuron 20, 619–624 (1998).
29. Arbely, E., Torres-Kolbus, J., Deiters, A. & Chin, J. W. Photocontrol of tyrosine phosphorylation in mammalian cells via genetic encoding of photocaged tyrosine. J. Am. Chem. Soc. 134, 11912–11915 (2012). 30. Grunbeck, A. & Sakmar, T. P. Probing G protein-coupled receptor-ligand
interactions with targeted photoactivatable cross-linkers. Biochemistry 52, 8625–8632 (2013).
31. Chen, Y. et al. Heritable expansion of the genetic code in mouse and zebrafish. Cell Res. 27, 294–297 (2017).
32. Reddington, S., Watson, P., Rizkallah, P., Tippmann, E. & Jones, D. D. Genetically encoding phenyl azide chemistry: new uses and ideas for classical biochemistry. Biochem. Soc. Trans. 41, 1177–1182 (2013).
33. Cunningham, M. E., Stephens, R. M., Kaplan, D. R. & Greene, L. A. Autophosphorylation of activation loop tyrosines regulates signaling by the TRK nerve growth factor receptor. J. Biol. Chem. 272, 10957–10967 (1997).
34. Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 9, 1213–1225 (2015).
35. Zhu, S. et al. Genetically encoding a light switch in an ionotropic glutamate receptor reveals subunit-specific interfaces. Proc. Natl Acad. Sci. USA 111, 6081–6086 (2014).
36. Ye, S. et al. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J. Biol. Chem. 283, 1525–1533 (2008).
37. Meakin, S. O., MacDonald, J. I., Gryz, E. A., Kubu, C. J. & Verdi, J. M. The signaling adapter FRS-2 competes with Shc for binding to the nerve growth factor receptor TrkA. A model for discriminating proliferation and differentiation. J. Biol. Chem. 274, 9861–9870 (1999).
38. MacDonald, J. I., Gryz, E. A., Kubu, C. J., Verdi, J. M. & Meakin, S. O. Direct binding of the signaling adaptor protein Grb2 to the activation loop tyrosines on the nerve growth factor receptor tyrosine kinase, TrkA. J. Biol. Chem. 275, 18225–18233 (2000).
39. Tian, M. & Ye, S. Allosteric regulation in NMDA receptors revealed by the genetically encoded photo-cross-linkers. Sci. Rep. 6, 34751 (2016). 40. Rannversson, H. et al. Genetically encoded photo-crosslinkers locate the
high-affinity binding site of anti-depressant drugs in the human serotonin transporter. Nat. Commun. 7, 11261 (2016).
41. Ray-Saha, S., Huber, T. & Sakmar, T. P. Antibody epitopes on G protein-coupled receptors mapped with genetically encoded photoactivatable cross-linkers. Biochemistry 53, 1302–1310 (2014).
42. Coin, I. et al. Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR. Cell 155, 1258–1269 (2013).
43. Poulsen, M. H., Poshtibana, A., Klippensteina, V., Ghisia, V. & Plesteda, A. J. R. Gating modules of the AMPA receptor pore domain revealed by unnatural amino acid mutagenesis. Proc. Natl Acad. Sci. 116, 13358–13367 (2019). 44. Klippenstein, V., Ghisi, V., Wietstruk, M. & Plested, A. J. R. Photoinactivation
of glutamate receptors by genetically encoded unnatural amino acids. J. Neurosci. 34, 980–991 (2014).
45. Huber, T. & Sakmar T. P. Chemical biological methods for investigating G protein-coupled receptor signaling. Chem. Biol. 21, 1224–1237 (2014). 46. Gautier, A., Deiters, A. & Chin, J. W. Light-activated kinases enable temporal
dissection of signaling networks in living cells. J. Am. Chem. Soc. 133, 2124–2127 (2011).
47. Luo, J. et al. Genetically encoded optical activation of DNA recombination in human cells. Chem. Commun. (Camb.). 52, 8529–8532 (2016).
48. Duan, L. et al. Optical activation of TrkA signaling. ACS Synth. Biol. 7, 1685–1693 (2018).
49. Brown, W., Liu, J. & Deiters, A. Genetic code expansion in animals. ACS Chem. Biol. 13, 2375–2386 (2018).
50. Han, S. et al. Expanding the genetic code of Mus musculus. Nat. Commun. 8, 14568 (2017).
51. Khamo, J. S., Krishnamurthy, V. V., Chen, Q., Diao, J. & Zhang, K. Optogenetic delineation of receptor tyrosine kinase subcircuits in PC12 cell differentiation. Cell Chem. Biol. 26, 400–410.e3 (2019).
52. Chang, K. Y. et al. Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat. Commun. 5, 4057 (2014).
53. Maniatis, T., Sambrook, J., & Fritsch, E. F. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor: Cold Spring Harbor Laboratory, 1989). 54. Jullien, J. et al. Trafficking of TrkA-green fluorescent protein chimerae during
nerve growth factor-induced differentiation. J. Biol. Chem. 278, 8706–8716 (2002).
Acknowledgements
We thank Dr. Tian He, Dr. Michel Labouesse, Prof. Xiahang Yang, Dr. Andrew Plested, and Dr. Iva Lucic for suggestions. We also thank Prof. Peng R. Chen at the Peking University for the generous gift of the PylRS/tRNACUAconstruct and ONB. We are
grateful to the Proteomic Resource Center at the Sorbonne University for providing technical resources. We acknowledge Shanghai Ruihui Co. for improving clarity of the writing. Financial support was provided by the Agence Nationale de la Recherche of France (ANR-JCJC grant to S.Y.), French CNRS, National Natural Science Foundation of China (31528007 to S.Y. and 31671108 to H.Z.) and the Scientific Instrument Innovation Team of Chinese Academy of Sciences (GJJSTD20180002 to H. Z.).
Author contributions
S.Z., J.S., G.Y., M.W., and S.Y. performed experiments. H.Z., D.L., C.Y., J.S., Z.L., M-X.G., and S.Y. designed experiments. S.Z. and H.Z., A.P., S.Y. performed data analyses of fluorescent imaging data. S.Z. performed data analyses of western blotting data. S.Z., G.Y., M.-X.G., and S.Y. wrote the manuscript. A.P., J.S., and H.Z. edited the manuscript.