Publisher’s version / Version de l'éditeur:
Plant Signaling & Behavior, 8, 11, pp. e26115:1-e26115:4, 2013-08-29
READ THESE TERMS AND CONDITIONS CAREFULLY BEFORE USING THIS WEBSITE. https://nrc-publications.canada.ca/eng/copyright
Vous avez des questions? Nous pouvons vous aider. Pour communiquer directement avec un auteur, consultez la première page de la revue dans laquelle son article a été publié afin de trouver ses coordonnées. Si vous n’arrivez pas à les repérer, communiquez avec nous à PublicationsArchive-ArchivesPublications@nrc-cnrc.gc.ca.
Questions? Contact the NRC Publications Archive team at
PublicationsArchive-ArchivesPublications@nrc-cnrc.gc.ca. If you wish to email the authors directly, please see the first page of the publication for their contact information.
NRC Publications Archive
Archives des publications du CNRC
This publication could be one of several versions: author’s original, accepted manuscript or the publisher’s version. / La version de cette publication peut être l’une des suivantes : la version prépublication de l’auteur, la version acceptée du manuscrit ou la version de l’éditeur.
For the publisher’s version, please access the DOI link below./ Pour consulter la version de l’éditeur, utilisez le lien DOI ci-dessous.
https://doi.org/10.4161/psb.26115
Access and use of this website and the material on it are subject to the Terms and Conditions set forth at
ScFKBP12 bridges rapamycin and AtTOR in Arabidopsis
Zhang, Rui; Meng, Zhigang; Zhou, Tao; Deng, Yong; Feng, Li; Wang, Yuan;
Sun, Guoqing; Guo, Sandui; Ren, Maozhi
https://publications-cnrc.canada.ca/fra/droits
L’accès à ce site Web et l’utilisation de son contenu sont assujettis aux conditions présentées dans le site LISEZ CES CONDITIONS ATTENTIVEMENT AVANT D’UTILISER CE SITE WEB.
NRC Publications Record / Notice d'Archives des publications de CNRC:
https://nrc-publications.canada.ca/eng/view/object/?id=e5e0add2-81ce-4ab2-b292-daec9df6f17d https://publications-cnrc.canada.ca/fra/voir/objet/?id=e5e0add2-81ce-4ab2-b292-daec9df6f17dShort CommuniCation
Plant Signaling & Behavior 8:11, e26115; november 2013; © 2013 Landes Bioscience
Short CommuniCation
From the earliest times, people have been looking for the med-icine to achieve immortality. Both eastern and western alchemists were obsessed with creating an elixir of life. Unfortunately almost all attempts have been miserable failures. 2009, rapamycin as the first drug to significantly extend lifespan in mammals is scientifi-cally proven,1 which is selected by Science as one of the Top 10
Scientific Achievements. This is of vital importance for the well-being of human, especially for the Gereatrics and Health Care because no drug agents is able to extend lifespan untill rapamycin was found so far.2 Rapamycin and its targets, therefore, have been
attracted more and more extensive attentions and becoming the focus of study in current Gereatrics and Preventive Medicine.2
Rapamycin is originally found for a macrolide antifungal drug in the soil Streptomyces hygroscopicus from Easter Island in 1970s.3 Shortly after this discovery, its potent
immunosup-pressive and antiproliferative properties were detected in 1970s.4,5
Thereafter it has been widely used as an immunosuppressant drug to prevent rejection in organ transplantation, especially in kidney transplants.6-8 After the 1980s, many studies showed that
rapamycin has the anti-cancer activities.9-11 Its deritivatives,
tem-sirolimus (CCI-779), everolimus (RAD001), and ridaforolimus (AP-23573) have been approved in clinical trails for several can-cers by FDA (Food and Drug Adiministration).12 Additionally,
rapamycin is a very useful agent for studying signal transduction in eukaryotic systems and able to block many signal transduc-tion pathways from yeast to mammalians.13 It was found that
rapamycin is able to inhibit cellular proliferation and cell cycle
progression mimicking the nutrient or energy starvation from yeast to mammals.14
The evolutionary distance between yeast and mammalian is more than 1 billion years, but their lifespan can be effectively extended by feeding rapamycin.15 This suggests that there are
evo-lutionarily conserved proteins targeted by rapamycin, existing in all eukaryotic species from yeast to mammalian. The targets of rapamycin didn’t change even going through 1 billion years evolu-tion and were identified as TOR1 (Target Of Rapamycin1) and TOR2 (Target Of Rapamycin1) in yeast in 1990s.16 The TOR
proteins are the phosphatidylinositol protein kinase and are highly conserved from yeast to mammalian.15 Most detected eukaryotic
species contain a single TOR while yeast has TOR1 and TOR2 and three TOR genes were recently found in Leishmania major.16-18
TOR is critical for organisms growth and development. Loss func-tion of TOR leads to lethal in all examined organisms from yeast to mammals.14 The protein kinase activity of TOR displayed by
TORC1 (TOR complex1) and TORC2 (TOR complex2). TORC1 and TORC2 are distinguished by RAPTOR and RICTOR, respectively, but LST8 is the constitutive member in both com-plexes.14-16 TOR complexes are the hub of various signaling
cas-cades through mediating upsteam signals including nutrition, energy, growth factor and stress signals.14,15 TOR phosphorylate
a series of downstream substrate proteins including S6K, 4E-BP, AKT to spatiotemporally regulate cell growth and division.14,15
FKBP12 (FK506 binding protein 12) bridged the interaction between rapamycin and TOR complexes.19 FKBP12 encodes a
*Correspondence to: Maozhi Ren; Email: renmaozhi@cqu.edu.cn Submitted: 08/06/13; Accepted: 08/12/13
Citation: Zhang R, Meng Z, Zhou T, Deng Y, Feng L, Wang Y, Sun G, Guo S, Ren M. ScFKBP12 bridges rapamycin and AtTOR in Arabidopsis. Plant Signaling & Behavior 2013; 8:e26115; PMID: 23989449; http://dx.doi.org/10.4161/psb.26115
ScFKBP12 bridges rapamycin and AtTOR
in Arabidopsis
rui Zhang1, Zhigang meng1, tao Zhou1, Yong Deng1, Li Feng2, Yuan Wang1, Guoqing Sun1, Sundui Guo1, and maozhi ren1,2,*
1Biotechnology research institute; Chinese academy of agricultural Sciences; Beijing, Pr China; 2School of Life Sciences; Chongqing university; Chongqing, Pr China
Keywords: yeast FKBP12, rapamycin, FK506, Arabidopsis growth
Abbreviations: ScFKBP12, Saccharomyces cerevisiae FK506 binding protein 12Kd; BP12, P35S:ScFKBP12 trans-genic Arabidopsis plants; TOR, target of rapamycin; TORC1, TOR complex1; TORC2: TOR complex2
FKBP12 encodes a prolyl isomerase and highly conserved in eukaryotic species. in yeasts and animals, FKBP12 can interact with rapamycin and FK506 to form rapamycin-FKBP12 and FK506-FKBP12 complex, respectively. in higher plants, FKBP12 protein lost its function to bind rapamycin and FK506. Early studies showed that yeast and human FKBP12 pro-tein can restore the rapamycin sensitivity in Arabidopsis, but the used concentration is 100–1000 folds higher than that in yeast and animals. high concentration of drugs would increase the cost and cause the potential secondary effects on plant growth and development. here we further discovered that BP12 plants generated in our previous study are hyper-sensitive to rapamycin at the concentration as low as that is effective in yeast and animals. it is surprising to observe that Wt and BP12 plants are not sensitive to FK506 in normal growth condition. these findings advance the current under-standing of rapamycin-tor signaling in plants.
peptidyl-prolyl cis-trans isomerase and highly conserved from yeast to human. In the presence of rapamycin, FKBP12 is able to bind FRB domain of TOR protein to form ternary complex.19,20
Rapamycin-FKBP12 disrupts the association between RAPTOR and TOR and therefore inhibited TOR protein kinase activi-ties and block TOR signaling cascades, leading to G1 cell cycle arrest.21 Human FKBP12 enables to complement yeast fkbp12
mutant strain, suggesting that FKBP12 is a highly conserved pro-tein in eukaryotes.20
In contrast to yeasts and mammals, studies with land plants consistently showed that plants are insensitive to rapamycin since plants FKBP12 proteins loss the function to bind rapamycin and failed to inhibit land plants growth.22,23 Several independent
studies showed that plant FKBP12 cann’t interact with rapamy-cin and TOR in vivo or in vitro by using either the yeast-two-hybrid method or direct binding assay.22-26 The protein structure
analysis showed that only plant FKBP12s has the unique struc-tural features with a disulfide bond.22 This unique structural
feature disrupted FKBP12 protein to bind rapamycin in higher plants. In the presence of yeast or human FKBP12, rapamycin is able to block TOR signaling in plant cells, but the used con-centration is 100–1000 folds (10μM) higher than that in yeast and animals.22-27 High concentration of drugs would increase the
cost and cause the potential secondary effects on plant growth and development. It should be important that plant TOR kinase activity can be effectively regulated by rapamycin at a concentra-tion as low as that in yeast and animals.
In our early study, we generated the Arabidopsis BP12 plants containing yeast FKBP12 and found that BP12 plants treated with rapamycin significantly reduces TOR kinase activity, slow down the growth rate, extend the lifespan and failed to response the nutrition and light signals through using genetics, transcrip-tomic, and metabolomic approaches.27 We observed that the
rapamycin still effectively inhibited the growth of BP12 plants at 500 nM concentration in our early study.27 In this study, we
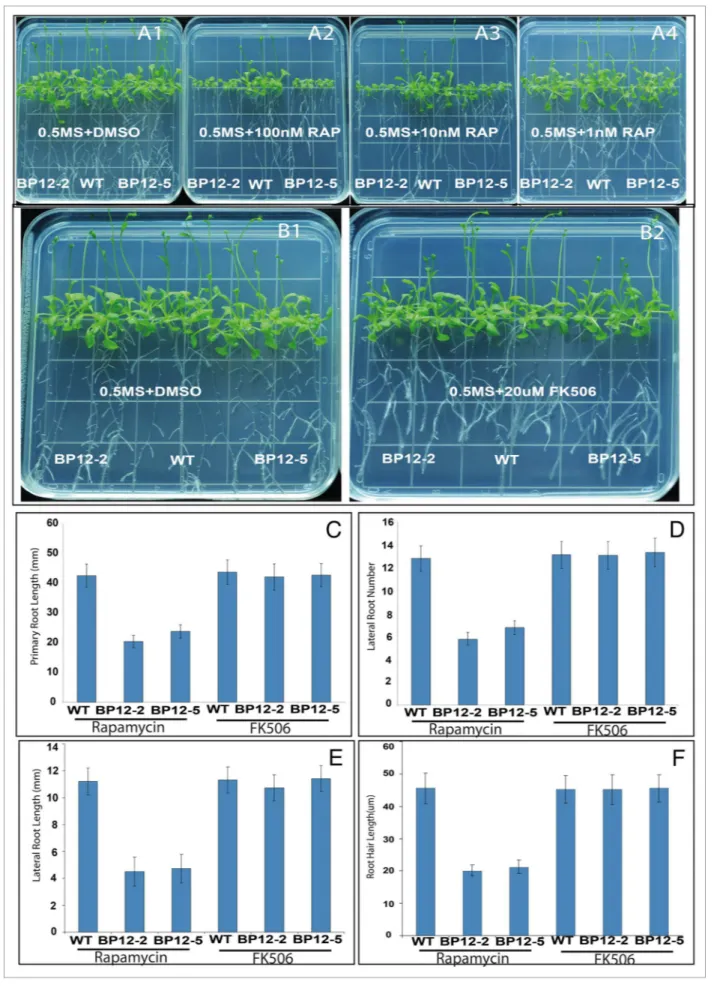
further reduced the rapamycin concentration to the same dosage as that used in yeast and animal cells (10–100nM). As indicated in Figure 1, rapamycin is able to significantly inhibit the pri-mary roots, lateral roots, and root hair growth of BP12 plants at 10–100nM concentration in contrast to WT (Fig. 1A, C–F), whereas the inhibition is undetectable when the concentration is reduced to 1nM. The effects we saw were specific to TOR
inhibition since WT plants with or without rapamycin treatment and BP12 plants without rapamycin treatment were essentially indistinguishable from each other for their growth, development and differentiation. This rapamycin inducible TOR suppression BP12 plants, especially the BP12-2 containing a single copy of ScFKBP12, will contribute to the further dissection of TOR sig-naling network in plants.
FKBP12 acts as a cellular target not only for rapamycin but also for FK506, a macrolide sharing structure similar to rapamy-cin.10,20 FKBP12-FK506 complex further binds to calcineurin
in vivo and inhibits its phosphatase activities.20 FK506
there-fore acts to block CaN-dependent signaling and inhibit the cell growth at 10nM concentration in yeast and animals.20 It should
be very interesting to know whether WT and BP12 Arabidopsis plants can also mediate the action of FK506 by AtFKBP12 or ScFKBP12 protein. We therefore examined the plants with the increasing FK506 concentration: 50, 100, 500, 1,000, 10,000 and 20,000 nM, respectively. Interestingly, the root number, lateral root, root hair, leaf number, and size of WT and BP12 plants didn’t exhibit obviously growth defects at 20μM FK506 concentration in contrast to 0.5xMS media (Fig. 1B–F). These observations suggest that FK506 is unable to inhibit cell growth in Arabidopsis even in the presence of yeast FKBP12. In yeast and animals, FK506 can target a family of proteins that possess the activity of peptidyl-prolyl cis-trans isomerase (PPIase). FKBP12 belongs to the FK506 Binding Protein superfamily and there-fore has dual function to bind rapamycin and FK506. In yeast, the absence of FKBP12 results in rapamycin resistant phenotype and human FKBP12 is able to restore the mutant strain to gain the rapamycin sensitivity, suggesting a functional equivalence between human and yeast.20 However, rapamycin-resistant yeast
strain remains sensitive to FK506, revealing that the other pro-teins involved in the FK506 signal transduction in yeast. These results indicated that ScFKBP12 is the specific target for rapamy-cin, whereas FKBP12 is not essential for FK506 signal transduc-tion. The unique feature of FK506-resistant was observed in plants revealed the different molecular mechanism for FK506 signaling transduction exists in plants.
Although a TOR ortholog was identified in Arabidopsis sev-eral years ago,24,28 the progress in the understanding of TOR
sig-naling in plants has been very limited so far. This is partly due to the lack of a well-established rapamycin sensitive system in
Arabidopsis. In this study, we have developed rapamycin
hyper-sensitive BP12 plants in Arabidopsis by using yeast FKBP12 gene. Rapamycin can effectively inhibit the growth and development of BP12 plants at the concentration as low as that in yeast and animals. Thus the in vitro high efficiency rapamycin sensitive system for suppression of TOR activity has been well established with BP12 plants in our study. TOR signaling rhythmically controls cell growth and development in response to growth fac-tor such as insulin in heterotroph eukaryotic model systems.14,15
However, no equivalents of insulin have been found in plants, but auxin, brassinosteriold, cytokinin are the major growth fac-tor to affect growth and cell proliferation in plants. Thus, it will be important to examine the crosstalk and interplays among TOR signaling pathway, nutrition, light, and hormones signals.
Figure 1. (See previous page). Characterization of rapamycin and FK506 sensitivity in Wt, BP12–2, and BP12–5 plants. (A) Growth performance of Wt, BP12–2, and BP12–5 plants grown on 0.5mS+DmSo, 100nm, 10nm, and 1nm rapamycin media for 25 d. Wt plants showed insensitivity to rapamycin on any concentration of rapamycin media whereas BP12–2 and BP12–5 plants were not obviously inhibited by rapamycin until the concentration was reduced to 1nm as indicated in Figure a4. (B) FK506 does not inhibit the growth of Wt and BP12 plants containing the exog-enous yeast FKBP12. (C–F) the primary root length (C), lateral root num-ber (D), lateral root length, (E) and root hair length (F) of 25 DaG Wt and BP12 plants grown on rapamycin (10nm) or FK506 (20μm) medium were measured, respectively, indicating that rapamycin can specifically inhibit BP12 plants, but FK506 is unable to suppress plant growth in the absence or presence of exogenous yeast FKBP12. Error bars indicate SD for triplicates (n = 20).
Further analyses of the BP12 lines may facilitates our understand-ing of TOR signalunderstand-ing network and contribute to reveal some key biological process and components of TOR signaling in plants.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grant number 2013AA102601 (Functional genomic research and application in cotton) from National 863 Projects.
References
1. Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009; 460:392-5; PMID:19587680
2. Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science 2010; 328:321-6; PMID:20395504; http://dx.doi. org/10.1126/science.1172539
3. Vézina CKA, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975; 28:721-6; PMID:1102508; http://dx.doi.org/10.7164/ antibiotics.28.721
4. Sehgal SN, Baker H, Vézina C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J Antibiot (Tokyo) 1975; 28:727-32; PMID:1102509; http://dx.doi.org/10.7164/antibiotics.28.727 5. Martel RR, Klicius J, Galet S. Inhibition of the
immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol 1977; 55:48-51; PMID:843990; http://dx.doi.org/10.1139/y77-007 6. Calne RY, Collier DS, Lim S, Pollard SG, Samaan
A, White DJ, Thiru S. Rapamycin for immuno-suppression in organ allografting. Lancet 1989; 2:227; PMID:2568561; http://dx.doi.org/10.1016/ S0140-6736(89)90417-0
7. Morris RE, Wu J, Shorthouse R. A study of the contrasting effects of cyclosporine, FK 506, and rapamycin on the suppression of allograft rejection. Transplant Proc 1990; 22:1638-41; PMID:1697111 8. Stepkowski SM, Chen H, Daloze P, Kahan BD.
Rapamycin, a potent immunosuppressive drug for vascularized heart, kidney, and small bowel transplantation in the rat. Transplantation 1991; 51:22-6; PMID:1987691; http://dx.doi. org/10.1097/00007890-199101000-00002 9. Fiebig HH, Berger DP, Winterhalter BR, Plowman
J. In vitro and in vivo evaluation of US-NCI com-pounds in human tumor xenografts. Cancer Treat Rev 1990; 17:109-17; PMID:2272027; http://dx.doi. org/10.1016/0305-7372(90)90034-D
10. Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991; 253:905-9; PMID:1715094; http://dx.doi.org/10.1126/science.1715094
11. Bjornsti MA, Houghton PJ. The TOR pathway: a tar-get for cancer therapy. Nat Rev Cancer 2004; 4:335-48; PMID:15122205; http://dx.doi.org/10.1038/ nrc1362
12. Vignot S, Faivre S, Aguirre D, Raymond E. mTOR-targeted therapy of cancer with rapamycin deriva-tives. Ann Oncol 2005; 16:525-37; PMID:15728109; http://dx.doi.org/10.1093/annonc/mdi113 13. Hidalgo M, Rowinsky EK. The rapamycin-sensitive
signal transduction pathway as a target for cancer ther-apy. Oncogene 2000; 19:6680-6; PMID:11426655; http://dx.doi.org/10.1038/sj.onc.1204091 14. Wullschleger S, Loewith R, Hall MN. TOR
signal-ing in growth and metabolism. Cell 2006; 124:471-84; PMID:16469695; http://dx.doi.org/10.1016/j. cell.2006.01.016
15. Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science 2010; 328:321-6; PMID:20395504; http://dx.doi. org/10.1126/science.1172539
16. Zheng XF, Florentino D, Chen J, Crabtree GR, Schreiber SL. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell 1995; 82:121-30; PMID:7606777; http://dx.doi.org/10.1016/0092-8674(95)90058-6 17. Sabatini DM, Erdjument-Bromage H, Lui M,
Tempst P, Snyder SH. RAFT1: a mammalian pro-tein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994; 78:35-43; PMID:7518356; http://dx.doi. org/10.1016/0092-8674(94)90570-3
18. Madeira da Silva L, Beverley SM. Expansion of the target of rapamycin (TOR) kinase family and func-tion in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infec-tivity. Proc Natl Acad Sci U S A 2010; 107:11965-70; PMID:20551225; http://dx.doi.org/10.1073/ pnas.1004599107
19. Stan R, McLaughlin MM, Cafferkey R, Johnson RK, Rosenberg M, Livi GP. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem 1994; 269:32027-30; PMID:7528205
20. Koltin Y, Faucette L, Bergsma DJ, Levy MA, Cafferkey R, Koser PL, Johnson RK, Livi GP. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol Cell Biol 1991; 11:1718-23; PMID:1996117
21. Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 2002; 10:457-68; PMID:12408816; http://dx.doi.org/10.1016/ S1097-2765(02)00636-6
22. Xu Q, Liang S, Kudla J, Luan S. Molecular char-acterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J 1998; 15:511-9; PMID:9753776; http://dx.doi. org/10.1046/j.1365-313X.1998.00232.x
23. Sormani R, Yao L, Menand B, Ennar N, Lecampion C, Meyer C, Robaglia C. Saccharomyces cerevisiae FKBP12 binds Arabidopsis thaliana TOR and its expression in plants leads to rapamycin susceptibility. BMC Plant Biol 2007; 7:26-33; PMID:17543119; http://dx.doi.org/10.1186/1471-2229-7-26
24. Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. Expression and disruption of the Arabidopsis TOR (target of rapamy-cin) gene. Proc Natl Acad Sci U S A 2002; 99:6422-7; PMID:11983923; http://dx.doi.org/10.1073/ pnas.092141899
25. Mahfouz MM, Kim S, Delauney AJ, Verma DP.
Arabidopsis TARGET OF RAPAMYCIN interacts
with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 2006; 18:477-90; PMID:16377759; http://dx.doi. org/10.1105/tpc.105.035931
26. Leiber RM, John F, Verhertbruggen Y, Diet A, Knox JP, Ringli C. The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 2010; 22:1898-908; PMID:20530756; http://dx.doi. org/10.1105/tpc.109.073007
27. Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, et al. Target of rapamycin signaling regulates metabo-lism, growth, and life span in Arabidopsis. Plant Cell 2012; 24:4850-74; PMID:23275579; http://dx.doi. org/10.1105/tpc.112.107144
28. Ren M, Qiu S, Venglat P, Xiang D, Feng L, Selvaraj G, Datla R. Target of rapamycin regulates develop-ment and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol 2011; 155:1367-82; PMID:21266656; http://dx.doi.org/10.1104/ pp.110.169045