HAL Id: hal-03162773
https://hal.archives-ouvertes.fr/hal-03162773
Submitted on 8 Mar 2021
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Distributed under a Creative Commons Attribution - NonCommercial - NoDerivatives| 4.0
International License
Seed maturation: Simplification of control networks in
plants
Martine Devic, Thomas Roscoe
To cite this version:
Martine Devic, Thomas Roscoe. Seed maturation: Simplification of control networks in plants. Plant
Science, Elsevier, 2016, 252, pp.335-346. �10.1016/j.plantsci.2016.08.012�. �hal-03162773�
ContentslistsavailableatScienceDirect
Plant
Science
jo u r n al h om ep ag e :w w w . e l s e v i e r . c o m / l o c a t e / p l a n t s c i
Review
article
Seed
maturation:
Simplification
of
control
networks
in
plants
Martine
Devic
∗,
Thomas
Roscoe
RégulationsEpigénétiquesetDéveloppementdelaGraine,ERL3500CNRS-IRDUMRDIADE,CentreIRDdeMontpellier,911avenueAgropolisBP64501, 34394,Montpellier,France
a
r
t
i
c
l
e
i
n
f
o
Articlehistory:Received25March2016
Receivedinrevisedform5August2016 Accepted21August2016
Availableonline22August2016 Keywords:
Seedmaturation AFLmultigenefamily Redundancy Simplification
Minimalcontrolnetwork
a
b
s
t
r
a
c
t
Networkscontrollingdevelopmentalormetabolicprocessesinplantsareoftencomplexasaconsequence oftheduplicationandspecialisationoftheregulatorygenesaswellasthenumerouslevelsof tran-scriptionalandpost-transcriptionalcontrolsaddedduringevolution.Networksservetoaccommodate multicellularcomplexityandincreaserobustnesstoenvironmentalchanges.Mathematicalsimplification byregroupinggenesorpathwaysinalimitednumberofhubshasfacilitatedtheconstructionof mod-elsforcomplextraits.Inacomplementaryapproach,abiologicalsimplificationcanbeachievedbyusing geneticmodificationtounderstandthecoreandsingularancestralfunctionofthenetwork,whichislikely tobemoreprevalentwithintheplantkingdomratherthanspecifictoaspecies.Withthisviewpoint,we reviewexamplesofsimplificationsuccessfullyundertakeninyeastandotherorganisms.Astrategyof progressivecomplementationofsingle,doubleandtriplemutantsofseedmaturationconfirmedthe fun-damentalroleoftheAFLsub-familyofB3transcriptionfactorsasmasterregulatorsofseedmaturation, illustratingthatbiologicalsimplificationofcomplexnetworkscouldbemorewidelyappliedinplants. Definingminimalcontrolnetworkswillfacilitateevolutionarycomparisonsofregulatoryprocessesand theidentificationofanessentialgenesetforsyntheticbiology.
©2016TheAuthors.PublishedbyElsevierIrelandLtd.ThisisanopenaccessarticleundertheCC BY-NC-NDlicense(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Contents
1. Introduction...336
2. Seeddevelopmentandmaturation...336
3. AFLproteinsintheplantkingdom...336
4. TheAFLfamilyofregulators−redundancy,specificityandregulation...338
5. Approachestosimplifygeneregulatorynetworks...340
5.1. Simplificationofabiologicalnetworkbymathematicalmodelling...340
5.1.1. Floweringtime...340
5.1.2. Seedmaturation ... 341
5.2. Simplificationofacontrolnetworkbybiologicalapproaches...341
5.2.1. Dissectionoftheprocessinsimpleorganisms...341
5.2.2. Simplificationbyfunctionalcharacterisationinaheterologoussystem...341
Theexampleofhumanp53regulationinS.cerevisiae...341
TheexampleofOLEOSIN1inPhyscomitrellapatens...341
5.2.3. Simplificationoftheprocessinmorecomplexorganisms...342
LearningfromtheexampleofthecellcyclecontrolinS.pombe...342
6. TowardstheelucidationofthefundamentalrolesofAFLproteinsduringseedmaturation...342
7. Benefitsofnetworksimplification...343
8. Generalisationoftheconceptof“minimalcontrolnetwork”inplantsandevolutionaryconsiderations...343
9. Conclusions...344
Acknowledgements...344
∗ Correspondingauthor.
E-mailaddress:martine.devic@ird.fr(M.Devic).
http://dx.doi.org/10.1016/j.plantsci.2016.08.012
0168-9452/©2016TheAuthors.PublishedbyElsevierIrelandLtd.ThisisanopenaccessarticleundertheCCBY-NC-NDlicense( http://creativecommons.org/licenses/by-nc-nd/4.0/).
AppendixA.Supplementarydata...344 References...344
1. Introduction
Seedsareanimportantcomponentofhumanandanimal nutri-tionsupplyingcaloriesintheformofstarch,sugar,andoiltogether withaminoacids,vitaminsandmicroelements.Thesynthesisof theseedreservesoccursafterpatternformationoftheembryo dur-ingmaturation,thesecondphaseofseeddevelopment.Inthefinal phaseofembryogenesis,desiccationtoleranceisacquiredand dor-mancyisestablished.Anunderstandingofthecontroloftheseed maturationphaseisessentialtoimproveseedqualitytraits.The biochemicalpathwaysleadingtotheproductionofseedreserves inArabidopsishavebeengeneticallyanalysed.Severalmaster regu-latorsofseedmaturationhavebeenidentified.Despitethisinsight, thereisnosimplemodelofthecontrolofinitiationandthe pro-gressionthroughmaturation.Thisispartlyduetotheredundancy amongthemasterregulatorsandthemultiplelevelsofregulation addedduringevolution.Itisnecessarytodefinethecore compo-nentsofthesystemratherthantodescribeitscomplexityinorder tounderstandthegeneregulatorynetworkcontrollingseed mat-uration.Aminimalcontrolnetworkmaythereforerepresentthe extantequivalentofanancestralregulatorygene.Here,we high-lightthenecessityforsimplificationandtheapproachestodefine minimalcontrolnetworks.
2. Seeddevelopmentandmaturation
Seedsarecomplexstructures,whichhaveariseninthe Sper-matophytes(seedbearingplants)morethan300millionyearsago followingwholegenomeduplication[1].Gymnospermseedsare composedofadiploidembryo(onematernalhaploidsetof chro-mosomes=1m and one paternal=1p) and a nourishing female gametophyte(2m).Angiospermseedsaretheproductsofadouble fertilisationandarecomposedoftheseedcoat,adiploidmaternal tissue(2m),thenutritiveendosperm(oftentriploid2m+1p)and thediploidembryo(1m+1p),reviewedin[2].Theemergenceof theseedtraithasconferredadvantagesofprotectionofthe repro-ductivestructuresandofdisseminationasdryquiescentmaterial. “Orthodox”Seedsaredefinedbytheirtolerancetodesiccation(they canloseupto90% ofwaterduring seedripening).Theembryo entersintoaquiescentstateandfallsintodormancyconcomitantly withtheacquisitionofdesiccationtolerance.“Recalcitrant”seeds arelesstolerantorareintoleranttodesiccation,arelessquiescent andcannotbewellconserved,buttheyhavetheadvantageofrapid germination.Allseedsaccumulatecarbohydrate,lipidandprotein reservespredominantlyintheembryoofGymnospermsandof ex-albuminousAngiospermseedsorintheendospermofalbuminous Angiospermseeds.Theformationoftheembryopattern,the accu-mulationofreserves,theacquisitionoftolerancetodesiccationand theentryintodormancyarethemainprogrammesofseed devel-opmentandareimportantagronomictraitsdefiningseedquality. Collectively,thesedevelopmentalprocessescompriseseed matu-ration.Irrespectiveofthequalitativedifferenceslistedabove,all seedsessentiallyfollowacommonphaseofseedmaturation.
GeneticanalysesinArabidopsishaveledtotheidentificationof fourmasterregulatorsofseedmaturation:ABSCISSICACID INSEN-SITIVE3(ABI3)[3],FUSCA3(FUS3)[4],LEAFYCOTYLEDON2(LEC2)
[5]andLEAFYCOTYLEDON1(LEC1)[6].WhilstLEC1isasubunitof theCCAATbindingcomplex(HAP3orNF-YB9),ageneraleukaryotic transcriptionalregulatorycomplex,ABI3,FUS3andLEC2 (collec-tivelynamedAFL)aretranscriptionfactorswithaplantspecificB3
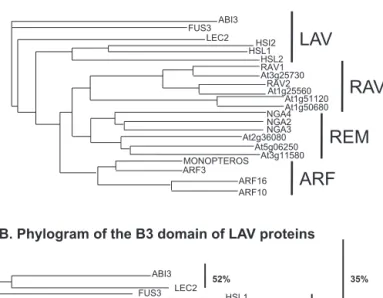
A. Ph
ylogram of
proteins
with B3
domains
ABI3 FUS3 LEC2 HSI2 HSL1 HSL2 At3g25730 RAV1 At1g25560 RAV2 At1g51120 At1g50680 NGA4 NGA2 NGA3 At2g36080 At5g06250 At3g11580 MONOPTEROS ARF3 ARF16 ARF10 LEC2 ABI3 FUS3 HSL1 HSL2 HSI2B. Ph
ylogram
of
the B3
domain of
LAV proteins
52% 68% 35%
LAV
RAV
REM
ARF
Fig.1. ProteinspossessingaB3DNAbindingdomaininArabidopsis.
A.SubsetofB3domainproteins.TheArabidopsisgenomecontains118genes encod-ingproteinspossessingaB3DNAbindingdomain.Theaminoacidsequencesofa selectionof22proteinscontainingaB3DNAbindingdomainwereusedto con-structaphylogramusingClustalWandClustalTree(ebi.ac.uk).Theseproteinscanbe dividedintofourmajorfamilies.TheLAV(LEC-ABI3-VAL)familycontainsthethree AFLandthethreeHSI/VALwiththeadditionalEAR(ERF-associatedamphiphilic repression)domain.TheRAV(RelatedtoABI3/VP1)familyhas13members,first identifiedbytheirhomologytoVP1.CertainRAVproteinspossessanadditional AP2domain.TheARF(AuxinResponseFactor)familyhas23members.ARF1,the foundermember,wasidentifiedasbindingupstreamseveralauxinresponsegenes. TheREMfamily(REproductiveMeristem)has76membersthatcanbesubdivided into6subgroups.MostoftheREMproteinscontainmorethanoneB3DNAbinding domain.ThegraphicillustratesthatthenearestneighbourproteinstotheAFLare theHSI/VALproteins.Membersofthethreeotherfamilies,ARF,RAVandREM, con-stituteseparateclades.
B.RelatednessoftheaminoacidsequencesoftheB3domainofAFLandHSI/VAL transcriptionfactors.Thenumbersindicatethepercentageofaminoacididentityof theDNAbindingdomainonly.
DNAbindingdomain(Fig.1).TheAFLregulatorsinfluencemost aspectsofseedmaturation.Forexample,theabi3andfus3mutants areintoleranttodesiccation,fus3isnotdormant(viviparous:can germinateonthemotherplant)andabi3,fus3andlec2seedscontain lessstorageproteinandlipidreservesandmorestarch.
HomologuesofAFLproteinshavebeenidentifiedinthegenomes ofallseedplantssequencedtodateandinsomemossandalgae.It ishypothesisedthattheirfunctioninseedmaturationisconserved amongSpermatophytes[7–9].DefiningthecorerolesofAFLinthe modelplantArabidopsiswillprovideinsightastoregulationofthe maturationphaseofembryogenesisincultivatedcrops.
3. AFLproteinsintheplantkingdom
Amongthe118proteinsinArabidopsisthatcontainaB3DNA bindingdomain,ABI3,FUS3andLEC2havethemostconservedB3 domainandaremoresimilartoeachotherthantoanyotherB3 protein[10](Fig.1).AnalysisofindividualAFLlossoffunction phe-notypeshasdemonstratedtheexistenceofastrongredundancy intheirmodeofactiontogetherwithalimitedspecificity,thatis, specialisationoffunction[11].
Table1
NumberofAFLorthologuesintheplantkingdom.
Species ABI3 FUS3 LEC2 ZmAFL4 ZmAFL5,6 Other Total References
Chlorophytes
Chlamydomonasreinhardtii 1ancestralAFL/VAL 1 [18]
Volvoxcarteri 1ancestralAFL/VAL 1 [18]
Bryophytes Physcomitrellapatens 3 0 0 0 0 2 5 [15,18,21] Lycopodiophytes Selaginellamoellendorffii 2 0 0 0 0 2 4 [15] Amborellales Amborellatrichopoda 1 0 0 0 0 2 3 Gymnosperms piceaabies 1 ND ND ND ND ND ND [15] Chamaecyparisnootkatensis 1 ND ND ND ND ND ND [91] Monocots Brachypodiumdistachyon 1 1 0 1 1 0 4 Brachypodiumstacei 1 1 0 1 1 1 5 Hordeumvulgare 1 1 0 1 1 0 4 [41,42] sorghumbicolor 1 1 0 1 1 0 4 [15,92] Oryzasativa 1 1 0 2 1 0 5 [10,15,16,40] Zeamays 1 1 0 1 2 0 5 [15,17] Dicotyledons Arabidopsisthaliana 1 1 1 0 0 0 3 [15] Glycinemax 2 2 1 0 0 0 5 [15] Medicagotruncatula 1 1 1 0 0 0 3 [93,94] Populustrichocarpa 1 2 2 0 0 0 5 [15] Manihotesculenta 2 2 1 0 0 0 5 [15] Solanumlycopersicum 2 1 0 0 0 0 3 [95] Solanumtuberosum 2 1 0 0 0 0 3 Theobromacacao 1 1 1 0 0 0 3 Vitisvinifera 1 1 0 0 0 1 3 ND:notdetermined.
AlthoughhomologuesofAFLgenesarefoundinthegenomesof seedplants,theirpresenceandnumbervariesamongspecies.The resourcesofGRAMENE[12],Phytozome[13],Inparanoid[14],data from[15]andadditionalsearchesandphylogenetictreeanalyses
(Figs.2and3,SupplementaryFig.1)wereusedtoproducealistof
AFLorthologuesinselectedplantspecieswhosegenomehasbeen entirelysequenced(Table1).Sincefewgenomeshavebeen specifi-callyexaminedforAFLorthologues,thesedatarepresenttheresult ofaglobalsearchfororthologousgenes.Furthermore,functional characterisationhasbeenlimited(referencedinTable1)and fre-quentlyrestrictedtoanindividualAFLinasinglespecies.Despite thiscaveat,thedatainTable1revealtheubiquityofAFLinseed plantsandoftheirpresenceinlowerplantsaswell.Homologues ofABI3,FUS3andLEC2arefoundinsoybean,wheretherearetwo ABI3,twoFUS3andoneLEC2.OrthologuesofLEC2havenotbeen unambiguouslyidentifiedintomato(Solanumesculentum),potato (Solanumtuberosum)andgrape(Vitisvinifera).HomologuesofABI3 andFUS3havebeenidentifiedinmonocotsspeciesbutnotofLEC2. Instead,additionalclassesofB3transcription factors,for exam-pleIDEF1(orthologueofZmAFL5)initiallyidentifiedinrice[16]
andZmAFL4inmaize,arepresent.ThetotalnumberofAFL-like isfourinbarley,fiveinbothriceandmaize.Furthermore,since Arabidopsisandmostdicotyledonousseedsdonotaccumulatea largeamountofstarchincontrasttocereals,itisofinterestthat themaizeZmAFL4protein,associatedwithstarchaccumulationin theendosperm[17],hasorthologuesinallmonocotsexamined.To datethreegenomesofgymnospermshasbeensequencedbutnone havebeenintegratedyetintoGramene,PhytozomeandInparanoid databases.Basedontheworkof[15]andsearchesonESTdatabases, itwaspossibletoidentifyABI3homologuesinpinespeciesbutno otherAFL.ThegenomeofAmborella,abasalangiosperm,contains oneABI3geneandtwoadditionalAFLgenes,oneofwhich resem-blesLEC2.Inconclusion,homologuesofABI3areunambiguously
recognisedandidentifiedin allfloweringplants butorthologies withLEC2andtoalesserextentwithFUS3arelessevidentbetween monocotsanddicots.
NoAFLwasidentifiedinthegenomeofthepicoalga Ostreococ-cuslucimarinus,butsomeAFLhomologueswerefoundinnon-seed plants(Table1.Chlamydomonas,volox,Physcomitrellaand Selagi-nalla).ABI3/HSI,presentasasinglecopyingreenalga,isthought tobethefoundermemberoftheentireB3DNAbindingdomain superfamily(Fig.1)[18].
ThephylogenetictreeoftheABI3/VP1proteins(Fig.2)groups closelytheAmborellaandgymnospermsproteins,awayfromthe dicotandmonocotclades.Thenonseed-plantproteinsare posi-tionedclosertothemonocotsthantoAmborellaproteins.
ABI3hastwomainrolesduringseeddevelopment:seed mat-urationinvolvingthebindingto theRYcis-element (afunction commontoeachAFL)andactivationofLEA(LateEmbryogenesis Abundant)genestoacquiredesiccationtolerance.Thislatterroleis morespecifictoABI3.RYelementsareover-representedinthe pro-motersofseedreservebiosyntheticgenes(maturation),butnotin thepromotersofLEAgenes[19].ThisexplainswhyasingleAFLcan compensatetheabsenceoftheothermembersforreserve accu-mulationbutisinefficientforlateembryogenesistraits[11].The functionalityofABI3orthologueshasbeentestedinPhyscomitrella
[20,21].ThethreeABI3/VP1-like(PpABI3A,PpABI3B,PpABI3C)genes
inthemosshavebeenshowntoactivateLEAgenesinpresenceof abscisicacid(ABA)andtobenecessaryfordroughttolerance.When expressedinabi3seeds, PpABI3Acanactivatetheexpressionof storageproteinsandoleosin2genes,inducechlorophyllbreakdown (seeddegreening),andpartiallyrestoreABAsensitivityat germi-nation.Unexpectedly,thetransformedseedsarestilldesiccation intolerant[21].Anexplanationforanincompletecomplementation wasthattheinteractionofABI5(bZIP)isweakerwithPpABI3Athan
Fig.2.PhylogeneticrelationshipsofABI3proteins.
Theproteinsequences(supplementaryFig.1)wereanalysedbyClustalOMEGA (http://www.ebi.ac.uk/Tools/msa/clustalo/). The multiple alignment was used tobuildaphylogenetictree atClustalphylogeny(http://www.ebi.ac.uk/Tools/ phylogeny/clustalw2phylogeny/).Thecladogram ispresentedasa Neighbour-joiningtreewithoutdistancecorrections.Themonocotsanddicotsproteinswere clearlyseparated,withthenon-seedplantproteinsclosetomonocots.Amborella ABI3wasgroupedwiththetwoexamplesofgymnospermABI3.
Chlamy:Chlamydomonasreinhardtii;Volvox:Volvoxcarteri;Physco:Physcomitrella patens;Sel:Selaginellamoellendorffii;Ambo:Amborellatrichopoda;Picea:piceaabies; Cyp:Chamaecyparisnootkatensis;Brdi:Brachypodiumdistachyon;Brast: Brachy-podiumstacei;Hv:Hordeumvulgare;Sorbi:sorghumbicolor;Os:Oryzasativa;Zm: Zeamays;At:Arabidopsisthaliana;Soybn:Glycinemax;Medtr:Medicagotruncatula; POPTR:Populustrichocarpa;Manes:Manihotesculenta;Solyc:Solanumlycopersicum; Soltub:Solanumtuberosum;Thecc:Theobromacacao;Vitis:Vitisvinifera.
withAtABI3.Atpresent,thefunctionalidentitiesoftheadditional AFLinPhyscomitrellaandSelaginellaremaininconclusive.
4. TheAFLfamilyofregulators−redundancy,specificity andregulation
CommonandspecificfunctionsofindividualAFLwereassessed inArabidopsisinthecontextofreserveaccumulation[11]. Speci-ficityclearly existsas shown by their distinct gene expression profilesduringseeddevelopment,differencesinthelevelof con-trolexertedoverfattyacidmodification,triacylglycerolandstorage protein accumulation, mediated in part through their binding affinitiesforpromotersofseed reserve-relatedgenes.However, strongredundancyisevidentsincesingleAFLmutantscontinueto accumulatesignificantlipidandproteinreserves.Storageprotein andoleosingenesarecommontargetsofABI3andFUS3as estab-lishedbychromatinimmunoprecipitation[22,23]andofLEC2[24]. Inaddition,whenanindividualAFLisectopicallyover-expressed, theminorspecificitiesarenolongerevidentandtherefore,theAFL exhibitanequivalentfunction[11].Inthiscontext,theunique func-tionalAFLhaslostmostofitsregulationandinteractionwiththe twootherAFLandadditionalregulators.Thislastpointraisesthe questionoftheimportanceoftheAFLnetworkanditsregulationin reserveaccumulation.Furthermore,sincethenumberofAFLisnot constantamongtheplantgenomes,thearchitectureofthenetwork maybevariableaccordingtothespecies.
TheAFLnetworkcontrolsreservesynthesiseitherdirectlyby activatinggenesencodingenzymesoffattyacidandstorageprotein synthesisorindirectlythroughactivationofsecondary
transcrip-Fig.3. RelationshipsamongFUS3/LEC2/ZmAFL2/ZmAFL4-6proteins.
TheB3domainproteinsthatwerenotABI3-likewereanalysedandrepresentedas inFig.2.Globally,theFUS3/LEC2proteinsofdicotsareseparatedfromthedistinct ZmAFL2,4–6proteinsofmonocots.Theproteinsfromnon-seedplantsaregrouped togetherandclosetothedicotproteins.
tionfactorssuchasWRI1.RegulationoftheAFLnetwork inthe developing Arabidopsis seed is currentlyunderstood asa tran-scriptionalactivationatthetransitionfrompatternformationto maturationandprincipallyanepigeneticrepressionatthe transi-tionfromseedtogerminationandseedlingdevelopment[25,26
andreferencestherein].RobustcontroloftheAFLnetworkisbased
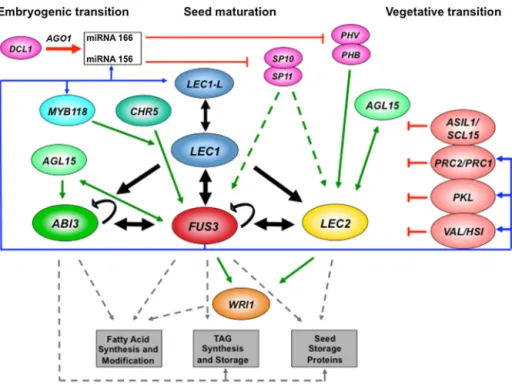
onauto-andmutualregulationandFUS3isprominentwithinthe networkmediatingactivationandrepressionduringseed develop-mentviafeedbackloops(Fig.4).
Activationof the AFLnetwork is effected by diversefactors actingearlyinseeddevelopment.OverexpressionofAGAMOUS15 (AGL15)up-regulatesLEC1andLEC2andsubsequentlyABI3,FUS3 and LEC2have been confirmed asdirect targets of AGL15 [27]. MYB118hasbeenidentifiedasanactivatorofAFLatthe vegeta-tivetoembryonictransitionwhereover-expressionresultsinan increasedexpressionofLEC1andaccumulationofstoragelipidin vegetativetissues[28].ExpressionofCHR5duringearlyembryo developmentleadstoactivationofAFL.Byassociating withthe promotersofABI3andFUS3,CHR5establishesanactivechromatin state[29].The timingofactivation ofmaturation-relatedgenes is critical. Mutationsin DCL1confirma role for miRNAs inthe inductionofthematurationprogram.Indirectevidencesuggests thatduringembryopatterning,miR156actingonitstargetgenes
Fig.4. RegulationoftheAFLnetworkduringembryodevelopmentandtransitiontovegetativegrowthinArabidopsis.
Thefigureillustratestranscriptionalandpost-transcriptionalcontroloverAFLactivationatthetransitionfromembryomorphogenesistomaturationandepigeneticrepression ofthenetworkatgermination.Blackarrowsindicateautoandmutualcontrolandthegraphicalrepresentationoffactorsdoesnotimplyahierarchyofgeneticinteractions. Greenarrowsindicateactivationofthenetworkcomponentsorpotentialactivation(brokengreenarrows),bluearrowsindicatefeedbackcontrolandbrokengreyarrows indicateactivationofreserve-relatedmaturationgenes.T-barsindicaterepressionorsilencing.WRI1isatissuespecificenhanceroffattyacidsynthesis.Targetgenesofthe AFLnetworkareboxed.=WRINKLED1,TAG=Triacylglycerol.
SPL10andSPL11,preventtheprecociousexpressionofmaturation genesincludingLEC2andFUS3[30,31].Atthe morphogenesis-to-maturationtransitionSPL10andSPL11transcriptionisenhanced and repression is overcome. The morphogenesis-to-maturation transitionmayalsobecontrolledbymiR166sinceamongitstargets PHAVOLUTA(PHV)andPHABULOSA(PHB),thelatterwasconfirmed asa direct activatorof LEC2[32].FUS3 isinvolved in feedback regulation of the activation network since AGL15, MYB118 and miRNA156wereshowntobedirecttargets[22].
The transition to vegetative development requires that the AFLnetworkisefficientlyrepressedduringgerminationto facil-itate the transcriptional reprogramming necessary for seedling development.TheAFLnetworkissubjecttoastrongepigenetic controlmediatedbyH3K27me3transcriptionalsilencing.VAL/HSI B3factorsare closelyrelated totheAFLfactors andbind com-petitivelyto theRY cis-element. Interaction with co-repressors isproposedtolead totherecruitmentofa histonedeacetylase complexthattargetsgenescontainingRYmotifsandactive chro-matinmarkstotranscriptionallysilencechromatin[25].Repression of the AFL network in vegetative tissues is also controlled by PolycombRepressiveComplexes(PRC)actinginconcertwith chro-matinre-modelingproteins.PRC2trimethylatesHistoneH3such thattargetgenes,includingtheAFLareH3K27me3modifiedand hencerepressed.PRC1interactswithproteinsthateffectaswitch inchromatinstatewhereerasureofactiveH3K4me3marksand replacementbyrepressiveH3K27me3marksrepressseed devel-opmentalgenesatthevegetativetransition[33].PRC1alsoactsin concertwithPRC2,recognisingH3K27me3markstoinducehistone 2Amonoubiquitinationtofurtherstabiliserepressionoftargetloci includingAFL.ThechromatinremodellerPICKLEisalsoinvolvedin therepressionoftheAFLnetworkmediatedbycooperationwith PRC2atlocibearingrepressivemarks[34].Additionalproteins con-tributetorepression,forexampleASIL1,whichobstructsaccessof AFLtotheRY-element[35]andSCL15requiredforregulationof achromatinremodellingfactortorepressembryonictraitsduring
vegetativegrowth[36].FUS3alsohasaroleincontrolling repres-sionoftheAFLnetworkbybindingtoVAL1,toPICKLEand the RING1bsub-unitofPRC1innegativefeedbackloops.
Despite the elaborate epigenetic, transcriptional and post-transcriptionalcontroloftheAFLnetworkduringseed develop-ment,itisremarkablethatthiscomplexregulationcanbeovercome bytheectopicexpressionofanyindividualAFL. Theabilityofa singleAFLtoinducereservesynthesis,(LEC2,[37])supportsour argumentthatredundancyexistsamongtheAFLandisbasedon thepresenceofaB3DNAbindingdomaincommontoeachfactor. ThattheproductionofreservelipidsinArabidopsisseedlings fol-lowinginductionofFUS3expressionisindependentofinductionof LEC1,LEC1-L,LEC2orABI3[38]clearlyillustratesthispoint.
Seed development between monocot and dicot species dif-ferswithrespecttotheformationofstorageorgans(scutellum, cotyledon, endosperm) and the partitioning of starch, protein andlipidreserves.InformationconcerningAFLcontrolofreserve accumulationinmonocotsisfragmentedandtheirintra-and inter-regulation, largelyunknown. Studieshave focussedon thelate functionsofVP1(VIVIPAROUS1,orthologoustoAt-ABI3)inrelation withABA,dormancy,germination,LEAactivationinmaize[39],rice
[40]andbarley[41].WeexaminedevidenceforAFLredundancyon seedreservesinmonocots.Co-expressionanalyseshaveshowna relativeconservationoftheABI3,FUS3andLEC1networks (includ-ingstorageproteinandcarbohydratemetabolism),butnotofLEC2 betweenmonocotsanddicots[7].Thiscomparisonwasbasedon systemsbiology studiesmainlyinwildtype seeddevelopment. Functionalstudiesoneachmemberofthenetworkanddetailed descriptionoftheirmodeofactionarenecessary.Inmaize,a com-prehensivestudy[17]hasdescribed thestructure oftheZmAFL family.TheZmAFL3(VP1)andZmAFL4factorseachtransactivatea maizeoleosin2promoterinamossheterologoussystemandmay actin synergy.However,theactivationbyZmAFL4 maynot be biologicallyrelevantsinceZmAFL4isexpressedintheendosperm andoleosin2intheembryo.Thus,certainZmAFLmembersretain
acommonabilitytoactivatetheaccumulationofdistincttypes ofseedreservesduringthematurationphase,buttheyhavealso gainedspecialisationbytheirspatio-temporalexpressionpattern andsequencedivergence,analogoustotheArabidopsisAFL.Totest thefullextentoftheirredundancyinvivowillrequiremultiple mutantcombinationsorRNAiwithlimitedspecificitysinceseveral membersofthemaizeAFLfamilyareexpressedinthesametissues. Remarkably,monocotsoftenhaveoneortwoextraAFLbelonging totheFUS3/LEC2typeincomparisontodicots(Table1,Fig.3).Since ZmAFL4isexpressedintheendospermandfoundinallthe mono-cotsexamined,theadditionalmember(s)mayfunctiontoregulate reserveaccumulationinthisstarchaccumulatingtissue.
AFLproteinsinmonocotshaveretainedasimilarmodeofaction withrespecttothecontrolofseedreserve synthesis.The pres-enceoftheRYcis-elementhasbeendescribedinthepromoters ofgenesencodingreserveproductsindifferentmonocots(in bar-ley,-hordeinstorageproteinandtrypsininhibitorBTI-CMe[42], inSetariapF128,aseedspecificpromoterforstorageprotein[43]). HvFUS3hasbeenshowntobeinvolvedseedstorageprotein accu-mulationintheendospermandintheembryo[42].Furthermore, ABI3hasbeenshowntoactivatestorageproteingenesin gym-nosperms[44].
ExaminationoftheAFLwithintheplantkingdomleadsusto proposethatthecorefunctionoftheAFLisconservedamong Sper-matophytespecies,butthattheirgenenetworks(thewaytheglobal functionisdistributedamongtheAFLmembersandtheir regula-tion)isnotconserved.Forexample,amutationinVP1inmaize inducesprecociousgermination,whereasinArabidopsis,alesion inFUS3resultsinmoresevereviviparythanisevidentinabi3seeds
[45].TheredundancyinherentwithintheAFLobscuresthe funda-mentalanduniversalroleoftheAFLfamilyinthecontrolofseed reservesandthereforetheregulatorynetworkshouldbesimplified toitsminimalcomponent:asingleproteinpossessingacommon AFLfunctionality(Fig.5).Wereturntothisconceptinthefollowing section.
5. Approachestosimplifygeneregulatorynetworks 5.1. Simplificationofabiologicalnetworkbymathematical modelling
A simplification of the system components is necessary to understandthebasicmechanismthat governsa developmental andmetabolicpathway.Thismaybeachievedbymathematical modellingusingsimplificationorreduction,byregroupinggenes orproteinsorenzymaticreactionsintokernelswhilepreserving thecorestructure[46].Weillustratethisprinciplebythefollowing examples.
5.1.1. Floweringtime
Plantspeciesusedifferentpathwaysinresponseto environmen-talcuesallowingthemtoflowerinthecorrectseason.Keytraits suchasirreversibilityandrobustnesstofluctuatingsignalshave beenconservedandinvolvethekeyregulatorsofthefloral transi-tion(perceptionofsignalsandfloralmeristemspecification).These includetheFLOWERINGLOCUS T(FT) functionandthe interac-tionswithFLOWERINGLOCUSD,APETALA1,LEAFYandTERMINAL FLOWER1,which are allconserved in diversespecies including tomato,riceandArabidopsis.Themajordynamicpropertiesofthe floraltransitionwerecapturedinminimalregulatorynetworksof corecomponentsusingmathematicalmodellingandsimplification
[47,48].Sincemanyhundredsofgenesaffectfloweringtime,this
wasachievedbydescribingonlythemajoractivitiesofgroupsof genes,referredtoas“regulatoryhubs”thatrepresentoneormore genesandproteins.Simplifiedmodelslackthefinerdetailsofthe
Fig.5. GeneticsimplificationleadingtoMinimalControlNetworkconstruction. Seedmaturation
A:Duringnormalseeddevelopment,acomplexauto-andinter-transcriptional net-workbetweenthethreeAFL(ABI3,FUS3andLEC2)controlstheinitiationandthe progressionofthedifferentprocessesoftheseedmaturationphase[96].This net-workcontrolsthesynthesisandtheaccumulationofproteinandlipidreservesin theseed.
B:InplantsharbouringmutationsinAFLgenes,transformationwithasingle consti-tutivelyexpressedAFLtransgenerestoresreservesynthesis[11].Thisactivationis independentoftheidentityoftheAFLmemberandofitsregulationandismediated throughtheactionoftheconservedB3DNAbindingdomain.Thatreserves accu-mulateinadoublemutanttransformedwithasingleAFLindicatestheexistenceof athresholdlevelforAFLfunctionnecessaryforactivation.This‘UniqueAFL’system constitutesaPlantMinimalNetworkforseedreserveaccumulation.
Mitoticcellcycle(basedon[69])
C:Inanormalcell,themitoticcellcycleoffissionyeastreliesontheassociationof aCdk(Cdc2)withvariouscyclins(Cdc13,Puc1,Cig1,Cig2).Thelevelofactivityof aCdk-cyclincouple(redgradient)regulatedbyseveralfactorstriggersthephase transitionG1-S,G2-M.
D:InyeastcellwheretheCdkandthe4cyclinshavebeendeletedandreplaced byaproteinfusionbetweenCdc13andCdc2,theoscillationofCdkactivityisstill occurringandissufficientforthenormalprogressionandthedirectionofthecore cellcycle.TheinteractionofCdkwithspecificcyclinsateachcellcyclephase tran-sitionisnotessential.ThissystemrepresentstheMinimalControlNetworkofthe cellcycleindependentofthemainregulators.
biologicalsystem,yettheyprovideanunderstandingoftheoverall systembehaviourandhencerepresentthecorestructure underly-ingthefloraltransitionbysimplefeed-forwardloops.Molecular geneticstudiesof floweringtime in responseto environmental clueshavealsobeenperformedinrice,ashort-dayplant. Compar-isonsoftheregulatorynetworkinArabidopsis,along-dayplant, andinrice,haveshownthatthecorefloweringpathway (mini-malcontrolnetwork)isconservedandthatotherpathwayshave evolvedfor novelfunctions, increasingthe diversity of flower-ingbehaviours[49–51].Inaddition,Arabidopsisandricedonot usethesamenumberof genestoperformthesecorefunctions
(e.g.oneflorigeninArabidopsisFT,[52],twoinriceHd3aandRFT1
[53,54]).Althoughthecorenetworkintegratingtheenvironmental
cuesisconserved,itsconsequentmechanismshavediverged dur-ingevolutiontoproducesometimesoppositefloweringresponses (long-day for Arabidopsisand short-dayfor rice [49]).This has allowedricetoflower evenduring long-dayconditionsusing a uniqueregulatorypathway[55].Furthermore,floweringin Ara-bidopsisis controlled by a small number of large-effectgenes, whereasinmaize,itiscontrolledbymanyadditivesmall-effect quantitativetraitloci[56].Interestingly,butasyetinsufficiently documented,someAFLplayaroleinfloweringtimeinArabidopsis (forAtABI3[57])andinricefor(OsLFL1=FUS3[58]).
Thus,themannerinwhichthecorefunctionisdistributedwith redundancy and/or specificity among regulatorygenes and the presenceorabsenceofgenefamilies,isvariableandprobably rep-resentsthebasisforthefine-tuningofthecorenetworkandpermits robustnessandadaptationforvariousgrowthconditions.
5.1.2. Seedmaturation
Similarly,itisimportanttodeterminethecoreandconserved functionsoftheAFLduringseedmaturationandmorespecifically, ofeachsubsetoffunctionsincludingembryonicidentity,reserve accumulation,dormancy,desiccationtoleranceandgermination. Severalrepresentationsof OMICsdata forseed developmentin individual species and for cross-speciescomparisons are avail-able(PaNethttp://aranet.mpimp-golm.mpg.de/[59]).Theyallow thecombinedrepresentationsofmetabolicpathwaysandfluxes and gene expression data. PlaNet was appliedto compare the co-expressionnetworksduringseedmaturationinmonocotsand dicots species [7] and showedthe relative conservationof the ABI3/FUS3andLEC1networksbutnotofLEC2asdiscussedabove. These representations demonstratethe complexity of the gene regulatorynetworksbutdonotfacilitatetheextractionthecore functions.Suchrepresentationshavehowever,facilitatedmultiple speciescomparisonsandledtotheidentificationofyieldrelated genesforcropimprovement[7].
Simplifiedmodels,suchastheonesdevelopedfortheregulation ofthefloweringtime,separatingthedistinctfunctionsoftheAFL masterregulators,arenecessary.
RIMAS (Regulatory Interaction Maps of Arabidopsis Seed Development, http://rimas.ipk-gatersleben.de/ [60]) proposes a simplifiedrepresentationbasedonthedesignofelectroniccircuits SystemBiologyGraphicalNotation(SBGN).TheadvantageofRIMAS istohavedissectedthedifferentfunctionsoftheAFLintoseparate networks(maturationgenecontrol,hormonemetabolism, epige-neticcontrol).Integratingtheredundancyandthresholdlevelof thethreeAFLfortheinitiationofseedreservesandupdatingthe actualdatabase,mayconstituteabasistorepresenttheminimal controlnetworkofAFL.
5.2. Simplificationofacontrolnetworkbybiologicalapproaches Keygenesareuniqueinasimplifiednetwork,althoughtheymay representseveralgenesandproteinsthathavebeencombinedinto an“activityhub”.Empiricalobservationisthefoundationforthe constructionofthemathematicalmodelsandtheirsimplification ingroupsormodules.Intuitively,it iseasiertobuildand testa modelinplantsinwhichthekeyregulatorygenesareencodedby asinglegeneratherthanbyagenefamily.Thissimplificationmay beachievedinthreeways:
5.2.1. Dissectionoftheprocessinsimpleorganisms
TheeffectsofauxinintheliverwortMarchantia[61]andthe effectsofabscisicacidinthebryophytemossPhyscomitrellapatens
[21]areexamples.Howeverthisapproachmaybeprecludedorof limitedvalueforprocessesspecifictoplantssuchasfloweringtime
whereonlysubsetsofthepathwaysmaybestudied.Inaddition, simpleorganismsmayhaveunanticipatedcomplexity,byexample PhyscomitrellapatenshasthreeABI3-likegenes(Table1).
5.2.2. Simplificationbyfunctionalcharacterisationina heterologoussystem
Thepresenceofgenefamilies,themultiplefunctionsofcertain regulatoryproteinsandthemultiplelevelsofcontrol(epigenetic, transcriptionalandpost-transcriptional)togetherexacerbatethe difficultyof determining thecorecomponentsand structure of anetwork.Analternativeand/oradditionalapproachtosimplify a network would be to isolate partof its componentsinto an heterologoussystem,whichcontainssomeoftheorthologous com-ponents,butnotall.Thisapproachallowsthestudyofonepartof thefunctionalityofapathwayoroftheregulationofakeygene, independentlyoftheothercomponents.
Theexampleofhumanp53regulationinS.cerevisiae
p53 is conserved from worms to humans. It functions as a sensorofDNAdamageandtriggerscellcyclearrest(life)or apo-ptosis (death). A complex protein network, mostly involved in post-translationalmodifications,tightlyregulatesitsactivity.Asa consequence,itisdifficulttounderstandtheregulatoryprinciples inthep53signallingnetwork.Theminimalrequirementsfor func-tionallyrelevantp53post-translationalmodificationswerestudied byexpressingthehumanp53togetherwithitsbest-characterised modifierMdm2inbuddingyeasttocircumventthiscomplexity
[62].Buddingyeastdoesnotcontainp53norMdm2homologues andthusisanappropriatetooltostudyp53-Mdm2interactionin isolationfromitsotherregulators,stillwithinacellularcontext. Thisp53-Mdm2modulewassufficienttofaithfullyrecapitulatekey aspectsofp53regulationofhighereukaryotes.Asimilarapproach couldbetakenwiththeAFLbyisolatingpartofthenetwork, par-ticularlytoaddresspost-translationalcontrols.
TheexampleofOLEOSIN1inPhyscomitrellapatens
Whilst some higher plant-specific developmental processes cannot be directly transposed to moss, Physcomitrella patens remainsaninteresting modelsystemtoexploreplantfunctions
[63,64]and todissect cellularprocesses includingtranscription
[65], aswell ashormonaland signalling pathways[21,66].The developmentofa heterologoussysteminPhyscomitrellafor the analysisof transcription factor–DNAinteractions hasbeen vali-datedwiththeBANYULSpromoter(BANYULSencodesanenzyme involvedinproanthocyanidinsynthesis)anditsmainregulators
[65].ThissystemhasbeenusedtodissecttheAFLnetworksand transcription complexes activatingthepromoter of anoilbody proteingene(OLEOSIN1)[67].ABI3,LEC2andLEC1actinsynergy and LEC2,LEC1 (NF-YB9)and NF-YC2 canforma ternary com-plex.FUS3wasnotfoundtoparticipateinthissynergy,althoughin isolation,itactivatestheOLEOSIN1promoter.SincePhyscomitrella hasthreeABI3-likegenesandtwoadditionalAFL-likegenes,itis notanentirelyneutralsystemforcharacterisingAFLfactors.The Physcomitrellasystemdiffersfromayeastone-hybridsystemsince itrequiresadditionalplantproteinsforactivationofthereporter gene. The yeast systemonly requires the binding of the plant transcriptionfactortoitscognateciselement,thecomplete tran-scriptionmachinery isindependent oftheidentity of theplant protein.Incontrast,inPhyscomitrella,OLEOSIN1activation proba-blyutilisesamossbZIPtointeractwithArabidopsisAFLraisingthe questionastowhethersequencedivergenceinfluencesthe effi-ciencyofactivationasin[21].However,thePhyscomitrellasystem hastheadvantageover yeastthatagreaternumber ofproteins canbeaddedsimultaneouslyandArabidopsisbZIPfactorssimilar
Fig.6.Benefitsofbiologicalsimplifications.
Approachestodeterminethecoreandessentialfunctionsofacelland/oramulticellularorganism.Theconceptofmathematicallygroupingofgenesunderasingleoneis similartothebiologicalsimplificationoftheAFLorcyclin-CDKminimalcontrolnetwork.Isolatingpartofthenetworkintoanheterologoussystemfacilitatestheidentification ofessentialinteractions.Takentogether,thecorearchitecturesdeducedfrommathematicalmodelling,thecorefunctionsfromthebiologicalsimplification,theessential interactionsofthenetworkandtheminimalgenesetwillaidminimalcelldesignwithoptimisedmetabolism.
tothosedescribedin[68]maybeaddedtofurthercustomisethe Physcomitrellasystem.
5.2.3. Simplificationoftheprocessinmorecomplexorganisms Afirstapproximationwouldbetoreducethegenefamilytoa singleregulatoryentitytoconstructthemodel.Themodelthen couldbeappliedtomorecomplexspecies.Aprerequisitewouldbe todeterminetheextentoftheredundancywithinthegenefamilyin plantabythestudyofthephenotypeofsingleandmultiplemutants. LearningfromtheexampleofthecellcyclecontrolinS.pombe
Thecontrolofthecellcycleinfissionyeast[69]servesasa con-ceptualframeworktodevelopasimplifiedplantcontrolnetwork forthedissectionofseedmaturationinArabidopsis.The eukary-oticcellcycleusesthefunctionofenzymesofthecyclin-dependent proteinkinase(Cdk)family,which interactwithspecific regula-torysubunits,thecyclins,toinitiatetheSphase(DNAreplication) andtheMphase(mitosis).Fissionyeastpossessessixcyclinsanda singleCdk.Researchhasprovidedanaccuratedescriptionofthe molecularmechanismscontrollinggenome duplicationand cell division[70].Stillknowledgeofthecomplexityofcellcycle reg-ulationhasmadeitdifficulttoelucidateitsbasicdeterminants. Asyntheticapproachthatgeneratedaminimalcellcyclecontrol
systemwasusedtoinvestigatethecoreofcellcycleregulation. Pre-viousresults[71]demonstratedthatasingleBcyclincansubstitute forG1cyclinsandregulateS-phaseandmitosisbytheoscillation ofCdkactivity.AfusionproteinbetweenasingleCdkanda sin-glecyclinissufficienttocontrolthetwomaintransitionsofthe cellcycle (S/G1, G2/M)duringmitosis ormeiosisof yeast cells depletedofalltheotherendogenouscyclins[69,72].Furthermore, cells operatingwiththis minimal module(lackingmuch ofthe knownregulation)havenoabnormalphenotype.Thus, composi-tionallydistinctcomplexesarenotabsolutelyrequiredformitotic andmeioticcellcycleprogression.Thissurprisingsimplicityofthe corecellcyclenetworkchallengesanumberofparadigmsincell cyclecontrol(Fig.5).
6. TowardstheelucidationofthefundamentalrolesofAFL proteinsduringseedmaturation
Anobjectiveofresearchintoseeddevelopmentistoidentify theessentialfunctionsandrequirementsoftheAFLin establish-ingembryonic identityandreserveaccumulation.Conceptually, ourworkontheAFL genes[11] is a firststep tothebiological simplificationof theseedmaturationnetwork (Fig.5).We sys-tematicallyanalysed combinationsof aflmutant phenotypesby
complementationandconfirmedtheextentoffunctional redun-dancyamongtheAFLregulators.Auniqueectopicallyexpressed AFLessentiallycomplementstheseedmaturationdefectsofreserve accumulationand morphologyofaflembryos(the bendingand theshapeof thecotyledonsand oftheaxis),but notthe toler-ance todesiccationanddormancy. Thisstudyfurtherindicated theexistenceofathresholdnecessaryforfunctionwithintheAFL pool.Furthermore,sincenoindividualAFLwasabletosuppress thetolerancetodesiccation,mid-andlate-maturationprograms wereuncoupled.Such aminimalregulatoryarchitectureshould bevalidforthecontrolofreserveaccumulationinmostflowering plants.Fromthissimplebasicmodel,itispossibletoincrementally increasethecomplexity.TheArabidopsisgenomepossessesthree HSI/VALgenesinadditiontothethreeAFLgenes[73,74].Theyalso havehighlyredundantfunctions.TheHSI/VALproteinscontainaB3 DNAbindingdomainverysimilartothatoftheAFL(Fig.1),which recognisesimilartargetpromoters.However,incontrasttotheAFL, theyactmainlyasrepressorsmediatedthroughtheirEAR (ERF-associatedamphiphilicrepression)domain[75].Thesesixgenes arederivedfromacommonancestralgene.HSI/VALproteinsclearly represstheactionofAFLduringgerminationbuttheirconflictual rolesareunknownduringseeddevelopment[76].Reductiontoa simplifiedAFLandHSI/VALnetworkcomprisingasingleAFLand asingleHSI/VALwouldfacilitateunderstandingofthemechanism underlyingtheAFL-HIS/VALantagonismduringseeddevelopment. OrthologuesofHIS/VALgenesalsoexistinthegenomeofotherplant species.However,ricehasfiveAFLandtwoHSI/VALgenes[10],and soagain,thenetworkanditsmembersarenotconserved.An under-standingoftheinteractionbetweena“genericAFL”anda“generic HSI/VAL”willbeapplicabletomostplantspecies.
AbiologicalreductiontoasingleAFLisalsoconceptually sim-ilar to mathematical modular (top-down) control analysis: the complexityof thesystemis reducedbygroupinggenesor pro-teinsorreactionsandreactantsintolargemodulesconnectedbya smallnumberofintermediates(Fig.6).Webelievethatbiological reductionwillleadtoasimplifiedmathematicalmodellingforthe phasetransitionfromembryogenesistomaturation.This biologi-calreductionalsoshowedthat,despitethedegreeofredundancy thatexistsamongtheAFL,theircommonsequence(essentiallythe B3DNAbindingdomain),issufficientfortheactivationoflipid and proteinreserve accumulation,but not for processes occur-ringduringthelatematurationprogramme.Thusthesimplification obtainedwithasingleAFLonlyappliestomid-maturationphase. In theabsence of experimentation,this wassuspected but not demonstrated.Throughthisapproach,weobtainedexperimental evidencefortheseparationofseedmaturationintodistinct mod-ules.Combiningthestudyofsimplifiedbiologicalmaterialwiththe establishmentofmathematicalmodelsisexpectedtofacilitatethe identificationofthecoremechanismofametabolicor develop-mentalprocessorpathway.Forexample,acomputationalmodel ofthemolecularinteractionsoftheminimalcontrolnetworkofcell cycleinfissionyeastbasedontheCdk-cyclinfusionproteinandon thenotionofquantitativeCdKactivitywasbuiltandchallenged byexperimentation[77].Themodelsupportsthe“quantitative” regulationofthecellcycleincontrasttothe ¨qualitative ¨modelof theinteractionsofCdkwithspecificcyclins.Italsoprovidednew insightsintotheregulatoryeffectoftheinhibitoryphosphorylation ofCdk.
7. Benefitsofnetworksimplification
Theuseofthesimplifiedfissionyeastpossessingtheminimal controlnetwork forcellcyclehasallowedtheestablishmentof thequantitativemodelofCdk.Theoscillationofactivitywithtwo thresholds(lowatG1/SandhighatG2/M)issufficientto
inde-pendentlyregulatethetwocellcyclephasesanddoesnotrequire specificcyclins.Inaddition,mostcanonicalregulationsbyWee1, Mik1andCdc25havedispensablerolesfortheminimalcellcycle. Thisminimalsystemdescribedinfissionyeast,revealsthecore con-trolofthe“generic”eukaryoticcellcycle[72].Thediscoverythat theoscillationofCdkactivityactsastheprimaryorganiserofthe cellcyclewouldnothavebeenpossible(oratleast,greatlydifficult) ifthediversityandcomplexityofthenumerouscyclinswouldhave beentakenintoaccount.Theconstructionoftheminimalcontrol networkhasrevealedtheessentialcomponentsofthecellcycle control.
Thevalidatedreductionistyeastmodelofp53-Mdm2 interac-tionallowsfurtherdissectionofnetworksandthestudyoftheir dynamics,bytheprogressiveadditionofcomponentstothis min-imalnetwork.Thisnovelapproachmaybeexpectedtoleadtothe elucidationofthecoremechanismsofp53regulationandallow testingofthestrategiestocounteractp53malfunctions.
8. Generalisationoftheconceptof“minimalcontrol network”inplantsandevolutionaryconsiderations
Plantsareexcellentcandidatesforapplyingbiological reduc-tionismtounderstandtheregulationofcomplexprocessessince theirgenomesencodelargeandnumerousgenefamilies[78].The availabilityofseveralcollectionsofT-DNAinsertionalmutantsin Arabidopsis hasinitially facilitatedthefunctionalstudy of indi-vidual members of these families, evidencing their degree of redundancy.Subsequently,doubleandmultiplecombinationsof mutantshavehelped todiscovertheroleof afamily orclosely relatedmembersactinginspecificpathways.Forexample,the com-pletemutantsetoftheFLOWERINGLOCUST/TERMINALFLOWER 1family hasbeenstudied(sextuplemutant,[79]).Tointroduce inthesecomplexmutantbackgroundsasinglefunctional mem-berofthefamily,ubiquitouslyandover-expressedwillalsohelp toidentifyspecificandredundant functions.For example,actin is a highlyconserved ubiquitousprotein, which isessential for cellularprocesses.Arabidopsiscontainseightactingenesthatare groupedintovegetativeandreproductiveclasses,functionally dis-tinct.Mutationsinanyofthethreevegetativeactinsrevealedonly mildphenotypes.Whenauniquevegetativeactinisover-expressed inadoublemutant,normalplantswererecovered[80].Webelieve thatthistypeofapproachcouldbemorewidelyusedtoidentifythe minimalgenesetand/orminimalcontrolnetworks.Reducingthe complexityofanetworktoasinglegenewillfacilitatethestudyof itsinteractionwithotherpartiallyredundantnetworks,whichin turn,canbesimplified.Onceaminimalcontrolnetworkis estab-lishedinamodelplant,itmaybemoreeasilyextendedtomore complexspecies.
Theminimalcontrolnetworkintuitivelyrepresentsthe ances-tralnetwork.Thegrowthphenotypeoftheyeastlackingallthe cyclinsand genesbutexpressingthesingleCdk-cyclinfusionis comparable tothatof thewildtype yeastwithanefficiencyof 80–90%.Inaway,thismodelarguesagainstthedogmathatspecific cyclinsarerequiredforspecificcellcyclephasetransition. How-ever,thissimpleproteinfusionisprobablynotsufficientlyrobust for cellsurvival in naturein competitionwith wildtype yeast. TheancestraleukaryotemayhavepossessedasingleCdk-cyclin complexwithoneCdkandonecyclin.ExtraCdksandcyclinsare necessarytofine-tunetheregulationandtheprogressionthrough thecellcycle.Selectionforadditionalregulatorycomponentsin the modern yeast cell hasestablished a requirement for these factors[72]suchthatthecoreprocessandmechanismarenow obscured.Byanalogy,itcanbehypothesisedthatauniqueAFLgene inanancestralplantgaverisetomultiplegenesinmodernplants. Arabidopsis expressing a single functional AFL represents this
progenitorandallowsaccesstothecorefunctionsofAFLinthe controloftheseedmaturationphase.
AnotherimportantaspectoftheidentificationofPlant Mini-malControlNetworksisthepossibilitytoextendthemtocomplex organismsinordertofacilitateevolutionary-developmental bio-logical comparisons. Gene regulatory networks can be used as evolutionarycharacterstocompareorganisms[81].Resolvingthe GeneRegulatoryNetworksthatunderliedevelopmentalprocesses in severalspecies would enable comparison of these networks andidentifysimilaritiesanddifferencesintheircomponentsand interactions.We anticipatethat minimalcontrol networkcould constitutea“buildingblock”forevolutionarydevelopmental com-parisonsofregulatoryprocessesamongdiverseplantspeciesto determineancestralrelationshipsandprovideinsight astohow regulatoryprocessesevolve.
Defining plant minimal control network can constitute an approachtoattainingoneofthegoalsincellbiology:toidentifythe universalminimalgeneset(essentialgenes)requiredtosustainlife. Searchesforessentialgeneshavebeenperformedinprokaryotes andinmulticellulareukaryotes[82],includingArabidopsis[83,84]. This has facilitated the creation of synthetic prokaryotic cells containingaminimalgenome[85].Initiallythesesyntheticcells survivedonlyunderdefinedgrowthconditions.Whenchallenged bytheenvironmentand incompetitionwithotherprokaryotes, therequirementfor additional genesfor fitnessisevident. The fine-tuning and the robustness of the networks require many moregenes,conservedinmostlivingorganisms,ratherthanthe bareessential [86]. Thisis nolongerthecase for thesynthetic prokaryotes.Severalsyntheticprokaryoteshavebeenconstructed withgrowthcomparabletotheiroriginalstrains.Manysynthetic pathwayshavebeenintroducedintomicroorganisms.Frequently therate-limitingstepsinthepathwaysarestrengthenedandthe unnecessarypathways,whichuseintermediatemetabolitesor pro-ducecompetingby-products,areremovedtoincreaseyield[87]. Theachievementofaminimalyeastcelliswithinreach.Canthis beachievedinplantcellsandeventuallyinmulticellularplants? Microalgaeareusedascellfactoriestoproducehigh-value prod-ucts(e.g.terpenoids,[88]).Syntheticbiologyisalsousedinplant biologywithambitiousgoalssuchasengineeringC4rice plants
[89]orintroducingnitrogenfixationinnon-leguminousplants[90]. Informationonminimalgene setand minimalcontrol network willfacilitatethecreationofmodifiedorsyntheticplantcellsby optimisinggenomereductionorediting.
9. Conclusions
TheArabidopsisAFLcanbestudiedasauniqueentityina mini-malcontrolnetworktounderstandthecoreprocess.Thisbiological reduction(a)revealedthatindividualAFLsharepartially redun-dantfunctionsand(b)permitedthedissociationandsubsequent independentstudyofdistinctdevelopmentalphasesoccurring dur-ingseedmaturation.Bycombiningtheunderstandingoftheplant minimalcontrolnetworkdeterminedinamodelplant,withthe specialisedfunctionsandinteractingnetworksofeachcomponent ofthenetworkinagivenspecies,itwillbepossibletoreconstitute thecoreprocessatitsoriginwithitsfine-tuninginthemodern plants.
Acknowledgements
Thisperspectivewasinspiredbythepresentation “Understand-ingCellCycleControl”ofSirPaulNurse.Thisworkwassupported inpartbytheFrenchAgenceNationaldelaRecherche,Grant ANR-10-BLAN-1238(CERES)andGrantANR-11-ISV7-0002(SYNERGY).
AppendixA. Supplementarydata
Supplementarydataassociatedwiththisarticlecanbefound,in theonlineversion,athttp://dx.doi.org/10.1016/j.plantsci.2016.08.
012.
References
[1]Y.Jiao,etal.,Ancestralpolyploidyinseedplantsandangiosperms,Nature473 (2011)97–100.
[2]C.Baroux,C.Spillane,U.Grossniklaus,Evolutionaryoriginsoftheendosperm infloweringplants,GenomeBiol.3(2002)(reviews1026).
[3]J.Giraudat,B.M.Hauge,C.Valon,J.Smalle,F.Parcy,H.M.Goodman,Isolation oftheArabidopsisABI3genebypositionalcloning,PlantCell4(1992) 1251–1261.
[4]H.Luerssen,V.Kirik,P.Herrmann,S.Miséra,FUSCA3encodesaproteinwitha conservedVP1/AB13-likeB3domainwhichisoffunctionalimportancefor theregulationofseedmaturationinArabidopsisthaliana,PlantJ.15(1998) 755–764.
[5]S.L.Stone,etal.,LEAFYCOTYLEDON2encodesaB3domaintranscription factorthatinducesembryodevelopment,Proc.Natl.Acad.Sci.U.S.A.98 (2001)11806–11811.
[6]T.Lotan,etal.,ArabidopsisLEAFYCOTYLEDON1issufficienttoinduceembryo developmentinvegetativecells,Cell93(1998)1195–1205.
[7]N.Sreenivasulu,U.Wobus,Seed-developmentprograms:asystems biology-basedcomparisonbetweendicotsandmonocots,Annu.Rev.Plant Biol.64(2013)189–217.
[8]P.Agarwal,S.Kapoor,A.K.Tyagi,Transcriptionfactorsregulatingthe progressionofmonocotanddicotseeddevelopment,Bioessays33(2011) 189–202.
[9]Schallau,etal.,Phylogeneticfootprintsinfernspore-andseed-specificgene promoters,PlantJ.53(2008)414–424.
[10]K.Swaminathan,K.Peterson,T.Jack,TheplantB3superfamily,TrendsPlant Sci.13(2008)647–655.
[11]T.J.Roscoe,etal.,Complementationofseedmaturationphenotypesbyectopic expressionofABSCISICACIDINSENSITIVE3,FUSCA3andLEAFYCOTYLEDON2 inArabidopsis,PlantCellPhysiol.54(2015)1215–1228.
[12]M.K.Tello-Ruiz,etal.,Gramene2016:comparativeplantgenomicsand pathwayresources,NucleicAcidsRes.44(2016)D1133–D1140.
[13]D.M.Goodstein,etal.,Phytozome:acomparativeplatformforgreenplant genomics,NucleicAcidsRes.40(2012)D1178–D1186.
[14]E.L.Sonnhammer,G.Östlund,InParanoid8:orthologyanalysisbetween273 proteomesmostlyeukaryotic,NucleicAcidsRes.43(2015)D234–D239.
[15]Y.Li,etal.,StepwiseoriginandfunctionaldiversificationoftheAFLsubfamily B3genesduringlandplantevolution,J.Bioinform.Comput.Biol.8(2010) 33–45.
[16]T.Kobayashi,etal.,ThetranscriptionfactorIDEF1regulatestheresponseto andtoleranceofirondeficiencyinplants,Proc.Natl.Acad.Sci.U.S.A.104(48) (2007)19150–19155.
[17]Grimault,etal.,RoleofB3domaintranscriptionfactorsoftheAFLfamilyin maizekernelfilling,PlantSci.236(2015)116–125.
[18]E.A.Romanel,etal.,EvolutionoftheB3DNAbindingsuperfamily:new insightsintoREMfamilygenediversification,PLoSOne4(2009)e5791.
[19]G.Guerriero,etal.,TheRY/Sphelementmediatestranscriptionalrepression ofmaturationgenesfromlatematurationtoearlyseedlinggrowth,New Phytol.184(2009)552–565.
[20]Yotsui,etal.,ABSCISICACIDINSENSITIVE3regulatesabscisicacid-responsive geneexpressionwiththenuclearfactorYcomplexthroughtheACTT-core elementinPhyscomitrellapatens,NewPhytol.199(2013)101–109.
[21]H.H.Marella,Y.Sakata,R.S.Quatrano,Characterizationandfunctional analysisofABSCISICACIDINSENSITIVE3-likegenesfromPhyscomitrella patens,PlantJ.46(2006)1032–1044.
[22]F.Wang,S.E.Perry,IdentificationofdirecttargetsofFUSCA3,akeyregulator ofArabidopsisseeddevelopment,PlantPhysiol.161(2013)1251–1264.
[23]G.Mönke,etal.,TowardtheidentificationandregulationoftheArabidopsis thalianaABI3regulon,NucleicAcidsRes.40(2012)8240–8254.
[24]S.A.Braybrook,etal.,GenesdirectlyregulatedbyLEAFYCOTYLEDON2 provideinsightintothecontrolofembryomaturationandsomatic embryogenesis,Proc.Natl.Acad.Sci.U.S.A.103(2006)3468–3473.
[25]H.Jia,H.M.Suzuki,D.R.McCarty,Regulationoftheseedtoseedling developmentalphasetransitionbytheLAFLandVALtranscriptionfactor networks,Rev.Dev.Biol.3(2014)135–145.
[26]C.Fatihi,D.Boulard,S.Bouyer,B.Baud,L.Dubreucq,DecipheringLepiniecand modifyingLAFLtranscriptionalregulatorynetworkinseedforimproving yieldandqualityofstoragecompounds,PlantSci.250(2016)198–204.
[27]Y.Zheng,N.Ren,H.Wang,A.J.Stromberg,S.E.Perry,Globalidentificationof targetsoftheArabidopsisMADSdomainproteinAGAMOUS-Like15,PlantCell 21(2009)2563–2577.
[28]X.Wang,Q.W.Niu,C.Teng,C.Li,J.Mu,N.H.Chua,J.Zuo,Overexpressionof PGA37/MYB118andMYB115promotesvegetative-to-embryonictransitionin Arabidopsis,CellRes.19(2009)224–235.
[29]Y.Shen,M.Devic,L.Lepiniec,D.X.Zhou,Chromodomain,Helicaseand DNA-bindingCHD1proteinCHR5,areinvolvedinestablishingactive
chromatinstateofseedmaturationgenes,PlantBiotechnol.J.13(2015) 811–820.
[30]M.D.Nodine,D.P.Bartel,MicroRNAspreventprecociousgeneexpressionand enablepatternformationduringplantembryogenesis,GenesDev.24(2010) 2678–2692.
[31]M.R.Willmann,A.J.Mehalick,R.L.Packer,P.D.Jenik,MicroRNAsregulatethe timingofembryomaturationinArabidopsis,PlantPhysiol.155(2011) 1871–1884.
[32]X.Tang,etal.,MicroRNA-mediatedrepressionoftheseedmaturation programduringvegetativedevelopmentinArabidopsis,PLoSGenet.8(2012) e1003091.
[33]A.M.Molitor,Z.Bu,Y.Yu,W.H.Shen,ArabidopsisALPHD-PRC1complexes promoteseedgerminationthroughH3K4me3-to-H3K27me3chromatinstate switchinrepressionofseeddevelopmentalgenes,PLoSGenet.10(2014) e1004091.
[34]H.Zhang,B.Bishop,W.Ringenberg,W.M.Muir,J.Ogas,TheCHD3remodeler PICKLEassociateswithgenesenrichedfortrimethylationofhistoneH3lysine 27,PlantPhysiol.159(2012)418–432.
[35]M.J.Gao,etal.,Repressionofseedmaturationgenesbyatrihelix
transcriptionalrepressorinArabidopsisseedlings,PlantCell.21(2009)54–71.
[36]M.J.Gao,etal.,SCARECROW-LIKE15interactswithHISTONEDEACETYLASE19 andisessentialforrepressingtheseedmaturationprogramme,Nat. Commun.6(2015)7243.
[37]M.SantosMendoza,B.Dubreucq,M.Miquel,M.Caboche,L.Lepiniec,LEAFY COTYLEDON2activationissufficienttotriggertheaccumulationofoiland seedspecificmRNAsinArabidopsisleaves,FEBSLett.29(2005)4666–4670.
[38]M.Zhang,X.Cao,Q.Jia,J.Ohlrogge,FUSCA3activatestriacylglycerol accumulationinArabidopsisseedlingsandtobaccoBY2cells,PlantJ.(2016),
http://dx.doi.org/10.1111/tpj.13233.
[39]D.R.McCarty,T.Hattori,C.B.Carson,V.Vasil,M.Lazar,I.K.Vasil,The Viviparous-1developmentalgeneofmaizeencodesanoveltranscriptional activator,Cell66(1991)895–905.
[40]T.Hattori,T.Terada,S.T.Hamasuna,Sequenceandfunctionalanalysesofthe ricegenehomologoustothemaizeVp1,PlantMol.Biol.24(1994)805–810.
[41]Z.Abraham,etal.,Adevelopmentalswitchofgeneexpressioninthebarley seedmediatedbyHvVP1(Viviparous-1)andHvGAMYBinteractions,Plant Physiol.170(2016)2146–2158.
[42]M.A.Moreno-Risueno,N.González,I.Díaz,F.Parcy,P.Carbonero,J. Vicente-Carbajosa,FUSCA3frombarleyunveilsacommontranscriptional regulationofseed-specificgenesbetweencerealsandArabidopsis,PlantJ.53 (2008)882–894.
[43]Y.Pan,X.Ma,H.Liang,Q.Zhao,D.Zhu,J.Yu,Spatialandtemporalactivityof thefoxtailmillet(Setariaitalica)seed-specificpromoterpF128,Planta241 (2015)57–67.
[44]Y.Zeng,N.Raimondi,A.R.Kermode,RoleofanABI3homologueindormancy maintenanceofyellow-cedarseedsandintheactivationofstorageprotein andEmgenepromoters,PlantMol.Biol.51(2003)39–49.
[45]M.Keith,N.G.Kraml,P.Dengler,McCourt,fusca3:aheterochronicmutation affectinglateembryodevelopmentinArabidopsis,PlantCell6(1994) 589–600.
[46]J.R.Kim,etal.,Reductionofcomplexsignalingnetworkstoarepresentative kernel,Sci.Signal.4(2011)ra35.
[47]K.E.Jaeger,etal.,Interlockingfeedbackloopsgovernthedynamicbehaviorof thefloraltransitioninArabidopsis,PlantCell25(2013)820–833.
[48]N.Pullen,etal.,Simplenetworkmotifscancapturekeycharacteristicsofthe floraltransitioninArabidopsis,PlantSignal.Behav.8(2013)e26149.
[49]Z.Milec,M.Valárik,J.Bartoˇs,J.Safáˇr,Canalatebloomerbecomeanearlybird? Toolsforfloweringtimeadjustment,Biotechnol.Adv.32(2014)200–214.
[50]R.Shrestha,J.Gómez-Ariza,V.Brambilla,F.Fornara,Molecularcontrolof seasonalfloweringinrice,arabidopsisandtemperatecereals,Ann.Bot.114 (2014)1445–1458.
[51]C.Sun,D.Chen,J.Fang,P.Wang,X.Deng,C.Chu,Understandingthegenetic andepigeneticarchitectureincomplexnetworkofricefloweringpathways, ProteinCell12(2014)889–898.
[52]Corbesier,etal.,FTproteinmovementcontributestolong-distancesignaling infloralinductionofArabidopsis,Science316(2007)1030–1033.
[53]R.Komiya,A.Ikegami,S.Tamaki,S.Yokoi,K.Shimamoto,Hd3aandRFT1are essentialforfloweringinrice,Development135(2008)767–774.
[54]H.Tsuji,K.Taoka,K.Shimamoto,Regulationoffloweringinrice:twoflorigen genes,acomplexgenenetwork,andnaturalvariation,Curr.Opin.PlantBiol. 14(2011)45–52.
[55]H.Tsuji,K.Taoka,K.Shimamoto,Florigeninrice:complexgenenetworkfor florigentranscription,florigenactivationcomplex,andmultiplefunctions, Curr.Opin.PlantBiol.16(2013)228–235.
[56]E.S.Buckler,etal.,Thegeneticarchitectureofmaizefloweringtime,Science 325(2009)714–718.
[57]S.Kurup,H.D.Jones,M.J.Holdsworth,Interactionsofthedevelopmental regulatorABI3withproteinsidentifiedfromdevelopingArabidopsisseeds, PlantJ.21(2000)143–155.
[58]L.T.Peng,etal.,OverexpressionoftranscriptionfactorOsLFL1delays floweringtimeinOryzasativa,J.PlantPhysiol.165(2008)876–885.
[59]Mutwil,etal.,PlaNet:combinedsequenceandexpressioncomparisonsacross plantnetworksderivedfromsevenspecies,PlantCell23(2011)895–910.
[60]A.Junker,A.Hartmann,F.Schreiber,H.Bäumlein,Anengineer’sviewon regulationofseeddevelopment,TrendsPlantSci.15(6)(2010)303–307.
[61]E.Flores-Sandoval,D.M.Eklund,J.L.Bowman,ASimpleauxintranscriptional responsesystemregulatesmultiplemorphogeneticprocessesinthe LiverwortMarchantiapolymorpha,PLoSGenet.11(2015)e1005207.
[62]B.DiVentura,etal.,ReconstitutionofMdm2-dependentpost-translational modificationsofp53inyeast,PLoSOne3(2008)e1507.
[63]D.J.Cove,etal.,ThemossPhyscomitrellapatens:anovelmodelsystemfor plantdevelopmentandgenomicstudies,ColdSpringHarb.Protoc.2(2009) (pdbemo115).
[64]M.J.Prigge,M.Bezanilla,Evolutionarycrossroadsindevelopmentalbiology: Physcomitrellapatens,Development137(2010)3535–3543.
[65]J.Thevenin,etal.,Anewsystemforfastandquantitativeanalysisof heterologousgeneexpressioninplants,NewPhytol.193(2012)504–512.
[66]P.F.Perroud,etal.,DefectiveKernel1(DEK1)isrequiredfor
three-dimensionalgrowthinPhyscomitrellapatens,NewPhytol.203(2014) 794–804.
[67]S.Baud,etal.,Decipheringthemolecularmechanismsunderpinningthe transcriptionalcontrolofgeneexpressionbyL-AFLproteinsinArabidopsis seed,PlantPhysiol.(2016),http://dx.doi.org/10.1104/pp.16.00034. [68]A.Yamamoto,Y.Kagaya,R.Toyoshima,M.Kagaya,S.Takeda,T.Hattori,
ArabidopsisNF-YBsubunitsLEC1andLEC1-LIKEactivatetranscriptionby interactingwithseed-specificABRE-bindingfactors,PlantJ.58(2009) 843–856.
[69]D.Coudreuse,P.Nurse,DrivingthecellcyclewithaminimalCDKcontrol network,Nature468(2010)1074–1079.
[70]C.J.McInerny,Cellcycleregulatedgeneexpressioninyeasts,Adv.Genet.73 (2011)51–85.
[71]D.L.Fisher,P.Nurse,AsinglefissionyeastmitoticcyclinBp34cdc2kinase promotesbothS-phaseandmitosisintheabsenceofG1cyclins,EMBOJ.15 (1996)850–860.
[72]P.Gutierrez-Escribano,P.Nurse,Asinglecyclin-CDKcomplexissufficientfor bothmitoticandmeioticprogressioninfissionyeast,Nat.Commun.6(2015) 6871.
[73]H.Tsukagoshi,A.Morikami,K.Nakamura,TwoB3domaintranscriptional repressorspreventsugar-inducibleexpressionofseedmaturationgenesin Arabidopsisseedlings,Proc.Natl.Acad.Sci.U.S.A.104(2007)2543–2547.
[74]M.Suzuki,H.H.Wang,D.R.McCarty,RepressionoftheLEAFYCOTYLEDON 1/B3regulatorynetworkinplantembryodevelopmentbyVP1/ABSCISICACID INSENSITIVE3-LIKEB3genes,PlantPhysiol.143(2007)902–911.
[75]Ohta,etal.,RepressiondomainsofclassIIERFtranscriptionalrepressors shareanessentialmotifforactiverepression,PlantCell13(2001)1959–1968.
[76]H.Jia,D.R.McCarty,M.Suzuki,DistinctrolesofLAFLnetworkgenesin promotingtheembryonicseedlingfateintheabsenceofVALrepression, PlantPhysiol.163(2013)1293–1305.
[77]C.Gerard,etal.,CellcyclecontrolbyaminimalCdknetwork,PLoSComput. Biol.11(2015)e1004056.
[78]Y.L.Guo,Genefamilyevolutioningreenplantswithemphasisonthe originationandevolutionofArabidopsisthalianagenes,PlantJ.73(2013) 941–951.
[79]W.Kim,etal.,Generationandanalysisofacompletemutantsetforthe ArabidopsisFT/TFL1familyshowsspecificeffectsonthermo-sensitive floweringregulation,J.Exp.Bot.64(2013)1715–1729.
[80]M.K.Kandasamy,E.C.McKinney,R.B.Meagher,Asinglevegetativeactin isovariantoverexpressedunderthecontrolofmultipleregulatorysequences issufficientfornormalArabidopsisdevelopment,PlantCell21(2009) 701–718.
[81]A.H.Fischer,J.Smith,Evo-devointheeraofgeneregulatorynetworks,Integr. Comp.Biol.52(2012)842–849.
[82]R.Zhang,Y.Lin,DEG5.0:adatabaseofessentialgenesinbothprokaryotes andeukaryotes,NucleicAcidsRes.37(2009)D455–458.
[83]M.Devic,Theimportanceofbeingessential:EMBRYO-DEFECTIVEgenesin Arabidopsis,C.R.Biol.331(2008)726–736.
[84]D.Meinke,etal.,IdentifyingessentialgenesinArabidopsisthaliana,Trends PlantSci.13(2008)483–491.
[85]M.Juhas,L.Eberl,J.I.Glass,Essenceoflife:essentialgenesofminimal genomes,TrendsCellBiol.21(2011)562–568.
[86]A.K.Ramani,etal.,Themajorityofanimalgenesarerequiredforwild-type fitness,Cell148(2012)792–802.
[87]D.Choe,etal.,Minimalgenome:worthwhileorworthlesseffortstoward beingsmaller?Biotechnol.J.11(2016)199–211.
[88]D.Gangl,etal.,Biotechnologicalexploitationofmicroalgae,J.Exp.Bot.66 (2015)6975–6990.
[89]R.T.Furbank,WalkingtheC4pathway:past,present,andfuture,J.Exp.Bot. (2016)(pii:erw161).
[90]F.Mus,etal.,Symbioticnitrogenfixationandchallengestoextendingitto non-legumes,Appl.Environ.Microbiol.(2016)01055–01056(pii:AEM.).
[91]Y.Zeng,Y.A.R.Kermode,AgymnospermABI3genefunctionsinasevere abscisicacid-insensitivemutantofArabidopsis(abi3-6)torestorethe wild-typephenotypeanddemonstratesastrongsynergisticeffectwithsugar intheinhibitionofpost-germinativegrowth,PlantMol.Biol.56(5)(2004) 31–46.
[92]F.Carrari,etal.,Cloningandexpressionofasorghumgenewithhomologyto maizevp1:Itspotentialinvolvementinpre-harvestsproutingresistance, PlantMol.Biol.45(6)(2001)631–640.
[93]J.Verdier,etal.,Aregulatorynetwork-basedapproachdissectslate maturationprocessesrelatedtotheacquisitionofdesiccationtoleranceand longevityofMedicagotruncatulaseeds,PlantPhysiol.163(2)(2013)57–74.
[94]J.Verdier,etal.,GeneexpressionprofilingofM.truncatulatranscription factorsidentifiesputativeregulatorsofgrainlegumeseedfilling,PlantMol. Biol67(6)(2008)567–580.
[95]G.W.Bassel,R.T.Mullen,J.D.Bewley,ABI3expressionceasesfollowing,but notduring,germinationoftomatoandArabidopsisseeds,J.Exp.Bot.57(6) (2006)1291–1297.
[96]To,etal.,Anetworkoflocalandredundantgeneregulationgoverns Arabidopsisseedmaturation,PlantCell18(2006)1642–1651.