HAL Id: pasteur-01457825
https://hal-pasteur.archives-ouvertes.fr/pasteur-01457825
Submitted on 6 Feb 2017
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Distributed under a Creative Commons Attribution| 4.0 International License
epidemic arthropod-borne virus.
Marie Vazeille, Karima Zouache, Anubis Vega-Rúa, Jean-Michel Thiberge,
Valérie Caro, André Yébakima, Laurence Mousson, Géraldine Piorkowski,
Catherine Dauga, Marie-Christine Vaney, et al.
To cite this version:
Marie Vazeille, Karima Zouache, Anubis Vega-Rúa, Jean-Michel Thiberge, Valérie Caro, et al..
Impor-tance of mosquito ”quasispecies” in selecting an epidemic arthropod-borne virus.. Scientific Reports,
Nature Publishing Group, 2016, 6, pp.29564. �10.1038/srep29564�. �pasteur-01457825�
Importance of mosquito
“quasispecies” in selecting an
epidemic arthropod-borne virus
Marie Vazeille
1, Karima Zouache
1, Anubis Vega-Rúa
1,2, Jean-Michel Thiberge
3, Valérie Caro
3,
André Yébakima
4, Laurence Mousson
1, Géraldine Piorkowski
5,6, Catherine Dauga
7,
Marie-Christine Vaney
8, Mosè Manni
9, Giuliano Gasperi
9, Xavier de Lamballerie
5,6&
Anna-Bella Failloux
1Most arthropod-borne viruses (arboviruses), perpetuated by alternation between a vertebrate host and an insect vector, are likely to emerge through minor genetic changes enabling the virus to adapt to new hosts. In the past decade, chikungunya virus (CHIKV; Alphavirus, Togaviridae) has emerged on La Réunion Island following the selection of a unique substitution in the CHIKV E1 envelope glycoprotein (E1-A226V) of an East-Central-South African (ECSA) genotype conferring a higher transmission rate by the mosquito Aedes albopictus. Assumed to have occurred independently on at least four separate occasions, this evolutionary convergence was suspected to be responsible for CHIKV worldwide expansion. However, assumptions on CHIKV emergence were mainly based on viral genetic changes and the role of the mosquito population quasispecies remained unexplored. Here we show that the nature of the vector population is pivotal in selecting the epidemic CHIKV. We demonstrate using microsatellites mosquito genotyping that Ae. albopictus populations are genetically differentiated, contributing to explain their differential ability to select the E1-226V mutation. Aedes albopictus, newly introduced in Congo coinciding with the first CHIKV outbreak, was not able to select the substitution E1-A226V nor to preferentially transmit a CHIKV clone harboring the E1-226V as did Ae. albopictus from La Réunion.
Arboviruses have succeeded in escaping from their sylvatic cycles by changing their host range to increase infec-tion rates in humans1–4. Subsequent epidemics may be facilitated by minor changes in the viral genome that
enable the virus to adapt to new vectors, including invasive species. Competitive displacement of an indigenous mosquito species by an invasive competitor can have a significant impact on vector-borne diseases5,6. The
mos-quito species Aedes albopictus has attracted great interest as an invasive vector and potential competitor of resi-dent mosquitoes, principally Aedes aegypti5–8. In the past decade, chikungunya virus (CHIKV; genus Alphavirus,
family Togaviridae) has emerged in regions where Ae. albopictus has succeeded in occupying vacant ecological niches6.
CHIKV strains are distributed among three phylogroups: East/Central/South African (ECSA), West-African, and Asian. The CHIKV Indian Ocean lineage (IOL) emerged from the ECSA phylogroup and has spread
1Department of Virology, Institut Pasteur, Arboviruses and Insect Vectors, 75724 Paris cedex 15, France. 2Institut Pasteur of Guadeloupe, Laboratory of Medical Entomology, Environment and Health, Guadeloupe, BP 484, Morne Jolivière, 97183 Abymes, Guadeloupe. 3Department of Infection and Epidemiology, Institut Pasteur, Environment and Infectious Risks/Laboratory for Urgent Response to Biological Threats (CIBU), Pole of Pathogen Genotyping, 75724 Paris cedex 15, France. 4Centre de Démoustication/Conseil général de La Martinique, 97234 Fort-de-France, Martinique. 5Aix Marseille Université, IRD French Institute of Research for Development, EHESP French School of Public Health, EPV UMR_D 190 ‘Emergence des Pathologies Virales’, 13005 Marseille cedex 05, France. 6IHU Méditerranée Infection, APHM Public Hospitals of Marseille, 13385 Marseille cedex 05,France. 7Center for Bioinformatics, BioStatistics and Integrative Biology, International Group for Data Analysis, 75724 Paris cedex 15, France. 8Department of Virology, Institut Pasteur, Structural Virology, 75724 Paris cedex 15, France. 9Department of Biology and Biotechnology “Lazzaro Spallanzani”, Laboratorio degli insetti di interesse agrario e sanitario,Università degli Studi di Pavia, Viale Ferrata 9, 27100, Pavia, Italy. Correspondence and requests for materials should be addressed to A-B.F. (email: anna-bella.failloux@pasteur.fr)
Received: 23 February 2016 accepted: 16 June 2016 Published: 07 July 2016
OPEN
throughout many tropical regions9. The IOL lineage was also introduced in Europe where autochthonous
trans-mission was documented in Italy (2007)10 and in France (2010)11,12. It predominates among the currently
circu-lating CHIKV strains in areas where the vector Ae. albopictus is present/dominant, in part due to selection of an Ae. albopictus-adaptive substitution in the CHIKV E1 envelope glycoprotein (E1-A226V). This substitution results in more efficient infection and dissemination in Ae. albopictus13,14. This region in E1 has been shown to
play a role in viral entry via fusion of viral and endosomal membranes15. The adaptation of the ECSA genotype to
Ae. albopictus is assumed to have occurred independently on at least four separate occasions13,16: on La Réunion
Island (2004–2005)4 and India (2008)17 for the IOL lineage18, and in Cameroon (2006) and Gabon (2007) for the
Central African lineage19.
In Africa, CHIKV was first isolated in Tanzania in 1952–195320, the Democratic Republic of Congo (DRC)
in 195821 and again in 196022. After almost 40 years of silence, two large CHIKV outbreaks were reported in
Kinshasa, DRC in 1999 and 200023 caused by ECSA CHIKV strains24. From 2004 to 2010, sequential epidemics
were reported in Cameroon (2006) and Gabon (2007 and 2010) with CHIKV isolates harboring the E1-A226V substitution16 coinciding with the first detection of Ae. albopictus in the region25,26. In 2011, a CHIKV outbreak
was reported in Congo with both Ae. aegypti and Ae. albopictus acting as vectors of CHIKV isolates presenting the
Ae. albopictus-adaptive substitution E1-A226V27,28. Recent findings stressed the role of small genetic changes in
the viral genome as the main factor leading to adaptation of new vectors for CHIKV emergence13,14,29. By contrast,
thus far, there has been no evidence for a role of the genetic differentiation within mosquito populations (hereaf-ter named quasipecies) on viral emergence.
Here, we performed 10 alternate passages mimicking host alternation between indigenous mosquitoes and a CHIKV presenting the original ECSA E1-226A isolated from Central Africa (DRC). We found that Ae. albopictus Congo behave differently from Ae. albopictus La Réunion; it was unable to (i) select the substitution E1-A226V and (ii) preferentially transmit CHIKV E1-226V when exposed to a blood-meal containing equal proportion of viral clones differing by a single mutation A or V at E1-226. Assuming that these differences can be attrib-uted to mosquito genetics, we demonstrated that Ae. albopictus Congo were clearly different from Ae. albopictus La Réunion using mosquito genotyping with microsatellites, stressing the importance of the nature of the vector population in arbovirus emergence.
Results
Ae. albopictus transmit CHIKV Congo_2011 less efficiently than Ae. aegypti.
Both Ae. aegypti and Ae. albopictus from Congo were orally infected with DRC_2000 or Congo_2011 via an infectious blood-meal (Fig. 1). When analyzing viral dissemination at 7 days post-infection (dpi), disseminated infection rates (DIR) were higher in Ae. aegypti than in Ae. albopictus regardless of the viral strain (P < 0.05, Fig. 1a). Transmission rates (TR), as determined by the presence of virus in saliva, were similar for both mosquito species infected with DRC_2000 and higher for Ae. aegypti infected with Congo_2011 (P < 0.05, Fig. 1b). However, the viral titer pres-ent in saliva was higher in Ae. albopictus regardless of the viral genotype (P < 0.05, Fig. 1c). Finally, in contrast to expectations, Ae. albopictus from Congo was not more efficient in transmitting CHIKV with the Ae.albopic-tus-adaptive mutation E1-226V.
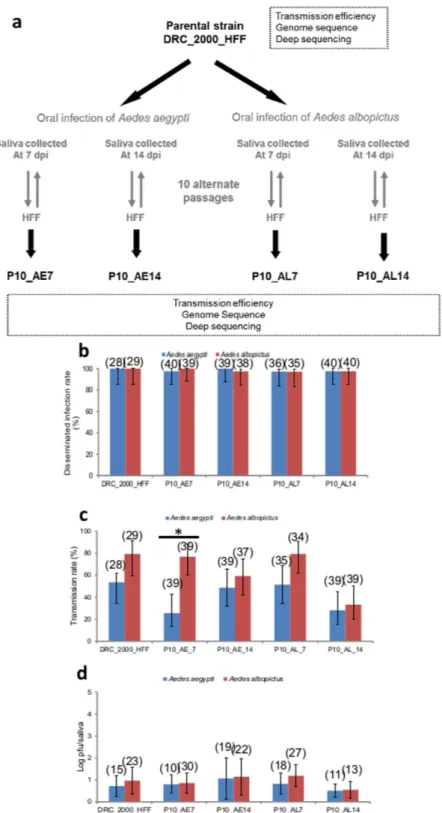
Alternate passages of CHIKV in human cells and mosquitoes do not impact viral dissemination
and transmission in mosquitoes.
Alternate passages between mosquitoes and human HFF cells were per-formed to experimentally attempt to select the mutation E1-226V from CHIKV DRC_2000 that has the E1-226A genotype. HFF cells were infected with CHIKV DRC_2000 followed by 10 alternate passages between mosquitoes and HFF cells and mosquitoes were analyzed for virus at 7 and 14 dpi (Fig. 2a). The resulting viral strains were orally provided to mosquitoes to determine their ability to disseminate and be transmitted. The first passage of CHIKV DRC_2000 on human HFF cells (DRC_2000_HFF) generated a viral strain causing an enhanced dis-semination in both mosquito species ( < 0.05, Fig. 2b), which did not however lead to a significant increase in transmission (P > 0.05, Fig. 2c) or increased viral load in saliva (P > 0.05, Fig. 2d). Overall, alternate passages of CHIKV did not improve the transmission potential of the virus when provided to the same mosquito species used for passages and in fact, reduced the viral titer in mosquito saliva in all cases except CHIKV P10_AE14.Substitutions are located in both non-structural and structural regions.
To identify whether mutations generated by passages can be specific to one mosquito species, the viral genomes were sequenced. All viruses were grouped in an independent cluster within the ECSA phylogroup (Supplementary Fig. 1). When compared to the S27 strain isolated in 1952 in Tanzania30, the parental strain DRC_2000 and the viral populationobtained after the 10th alternate passage differed in 66 amino-acid substitutions: 40 in the non-structural proteins
and 26 in the structural proteins. Of the 66 amino-acid substitutions, eight were found to be unique to one strain (Supplementary Table 1).
Genetic diversity is increased in Ae. aegypti passaged strains.
Genetic diversity was examined by estimating mutation frequencies at each position of the viral genome by comparison to the DRC_2000_HFF ref-erence sequence. Viral strains resulting from alternate passages harbored mainly fixed mutations with 13 detected in non-structural genes along with three mutations in the E2 gene (Table 1). The Ae. aegypti passaged strains: P10_ AE7 and P10_AE14 contained a higher number of variants with fixed mutations than did Ae. albopictus passaged strains: P10_AL7 and P10_AL14 (Table 1). With the exception of the nsP2 A3210C (K510T) polymorphism, these fixed mutations segregated according to the mosquito species used for the alternate passages (Table 1). The three fixed mutations in the E2 glycoprotein detected in Ae. aegypti (I217V and G249R) and Ae. albopictus (G82R) were located on the surface of the virion without any obvious interaction with the position E1-226 (Supplementary Fig. 2). Unexpectedly, an amino-acid change C-> Y at the position 483 of the nsP4 polymerase gene was identified inthe CHIKV P10_AE7 strain (Table 1) coinciding with the substitution identified by31. All clones of the CHIKV
P10_AE7 presented this mutation and none in the P10_AE14 strain (Extented Data Fig. 3a). The nsP4-C483Y substitution did not induce a replicative cost in either Ae. aegypti Aag2 or Ae. albopictus U4.4 cells (Extented Data Fig. 3b–e). The non-persistence of this mutation in passages initiated using late-collected saliva coincided with a higher viral load detected in saliva of mosquitoes infected with the P10_AE14 (Fig. 2d).
The biological clone E1-V IOL is better transmitted by Ae. albopictus La Réunion but not by
Ae. albopictus Congo.
Competition assays were performed to assess transmission efficiencies of two bio-logical clones E1-A (with E1-226A) or E1-V (with E1-226V) IOL in Ae. albopictus and Ae. aegypti from Congo in comparison with Ae. albopictus from La Réunion; saliva of 15–20 mosquitoes orally infected were examined at 7 dpi (Fig. 3). When provided alone (Fig. 3a), the E1-V biological clone presenting the Ae. albopictus-adaptiveFigure 1. Disseminated infection rates, transmission rates and viral loads of saliva for mosquitoes infected with the two parental strains Congo_2011 and DRC_2000. (a) Disseminated infection rates, (b) transmission
rates and (c) viral loads in saliva detected at 7 dpi for Aedes aegypti and Aedes albopictus from the Congo orally infected with the parental strains Congo_2011 and DRC_2000 provided at a titer of 106.5 pfu/mL in the
Figure 2. Experimental design for selecting CHIKV strains by alternate passages and vector competence analysis of samples. (a) The DRC_2000_HFF was alternatively passaged 10 times between mammalian HFF
cells and mosquitoes (Ae. aegypti or Ae. albopictus) to mimic host alternation. Transmission efficiency was estimated after oral infection of mosquitoes. Genetic changes were identified by comparing the consensus sequence obtained with the reference CHIKV S27. Genetic diversity was determined by estimating variants in the viral population by deep sequencing. (b) Disseminated infection rates, (c) transmission rates and
(d) viral loads of saliva at 7 dpi for mosquitoes orally infected with DRC_2000_HFF, and the 10 alternate
passages between HFF cells and mosquitoes, provided at a titer of 106.5 pfu/mL in the blood-meal. Error bars
show the confidence intervals (95%). Significant p values are indicated by an asterix. In brackets, the number of mosquitoes tested.
substitution E1-A226V was two-fold better transmitted by Ae. albopictus La Réunion as expected, whereas
Ae. albopictus Congo did not exhibit any marked differences. On the other hand, Ae. aegypti Congo better
trans-mitted the E1-A clone harboring the E1-226A substitution. In individuals exposed to a blood-meal containing an equal proportion of the two biological clones (Fig. 3b), the mean proportion of E1-V in saliva was 3 fold lower for
Ae. albopictus Congo compared to Ae. albopictus La Réunion (P < 0.05). The biological clone E1-V IOL was better
transmitted by Ae. albopictus La Réunion but not by Ae. albopictus Congo.
Ae. albopictus Congo and Ae. albopictus La Réunion are genetically distinct.
The location of the four E1-A226V substitution emergence events are shown on the map (Fig. 4a): La Réunion Island (2004–2005)4and India (2008)17 for the IOL lineage18, and Cameroon (2006) and Gabon (2007) for the Central African lineage19.
Date of introduction or first description of Ae. albopictus is indicated for each concerned country. Genetic relat-edness between 16 Ae. albopictus populations was assessed by genotyping mosquitoes using 11 microsatellites. All loci were polymorphic with 4–21 alleles scored (Supplementary Table 2). The neighbor-joining tree based on Cavalli-Sforza and Edwards’s chord distances showed three distinct groups with high bootstrap support at nodes separating them (Fig. 4b). The two most distant groups included the two populations from La Réunion (STANDRE and ALPROV) and the two populations from Congo (MFILOU and CONG). Mosquitoes from La Réunion were grouped with Ae. albopictus from France (BL), whereas mosquitoes from Congo were grouped with Ae. albopictus from India (CAL) suggesting different sources of introduction. A third group comprising nine populations from America was subdivided into two subgroups: a subgroup associating mosquitoes from South America and another subgroup comprised of mosquitoes from Central/North America.
Discussion
A key feature of many invasive species is their ability to displace and eventually replace the indigenous species that occupies the same ecological niche. Once it arrives in a region, Ae. albopictus tends to displace the indigenous
Ae. aegypti through its larval competitive advantage for resources25,32,33 and which may alter vector
compe-tence34,35. Cameroon36, Gabon37, or the Central African Republic38 were the main source of the Ae. albopictus
invasion into Congo. Based on microsatellite genotyping, we showed that Ae. albopictus from Congo are genet-ically different from Ae. albopictus from La Réunion where the Ae. albopictus-adaptive substitution E1-A226V has been selected. Furthermore, when exposed to an equal proportion of CHIKV clones differing by a single substitution A or V at E1-226, Ae. albopictus from Congo did not preferentially transmit the clone harboring the E1-226V mutation as expected.
CHIKV has recently spread throughout many tropical regions due in part to an amino-acid substitution in the E1 glycoprotein (E1-A226V) that enhances infectivity for Ae. albopictus. It is assumed that this mutation has been selected for on at least four separate occasions including Central Africa, and appears to coincide with the occurrence of Ae. albopictus as a CHIKV vector13,16. Cases of CHIK disease were first reported in Brazzaville
nt position nt change Gene aa change
Frequency of mutations (%) Ae. aegypti Ae. albopictus P10_AE7 P10_AE14 P10_AL7 P10_AL14
562 A -> G nsp1 162 (L)# 99.9 978 C -> T nsp1 T301I 99.8 1017 C -> T nsp1 T314M 100 1851 G -> A nsp2 A57T 99.9 2635 C -> T nsp2 318 (T)# 99.8 3210 A -> C nsp2 K510T 99.6 99.3 99.7 3734 A -> G nsp2 I685D 99.9 4424 G -> A nsp3 G117R 99.9 4897 C -> T nsp3 274 (T)# 99.8 5942 C -> T nsp4 P93S 97.2 93 6157 C -> T nsp4 164 (F)# 98.7 6724 C -> T nsp4 353 (F)# 100 7113 G -> A nsp4 C483Y 99.9 8785 G -> A E2 G82R 12.2* 9190 A -> G E2 I217V 99.9 9286 G -> A E2 G249R 98.8
Table 1. Population genetic diversity in the four selected CHIKV strains. nt, nucleotide; aa, amino-acid; #no
amino-acid change, synonymous mutation; * minority variant not found in the consensus sequence; P10_AE7, 10th passage between HFF cells and Ae. aegypti initiated with saliva collected at 7 dpi; P10_AE14, 10th passage
between HFF cells and Ae. aegypti initiated with saliva collected at 14 dpi; P10_AL7, 10th passage between
HFF cells and with Ae. albopictus initiated with saliva collected at 7 dpi; P10_AL14, 10th passage between HFF
cells and Ae. albopictus initiated with saliva collected at 14 dpi. The DRC_2000_HFF was used as the reference genome.
(Congo) in June 2011. This emergence was assumed to be related to a host switching event from Ae. aegypti to the Asian Ae. albopictus as both mosquito species were found naturally infected in the field28 in a situation similar
to that observed in Cameroon and Gabon26,39. We compared the native Ae. aegypti and the recently introduced
Ae. albopictus collected from the same breeding site and showed that the Congo Ae. albopictus was less able to
transmit the most recent epidemic CHIKV strain, Congo_2011, which harbours the E1-226V mutation. This was surprising, as it has been shown that under some circumstances, this mutation enhances CHIKV transmission by Ae. albopictus13,14.
The CHIKV strain Congo_2011 differs mainly from DRC_2000 at two amino-acid positions. CHIKV DRC_2000 possesses the prototypic E1-226A and E2-337V residues, while CHIKV Congo_2011 contains the E1-226V and E2-377I mutations (Supplementary Table 3). We attempted to experimentally enhance the poten-tial of the DRC_2000 strain to be transmitted by Ae. albopictus by alternately passaging the virus between either
Ae. aegypti or Ae. albopictus mosquitoes from Congo and human cells. After the 10th passage, we conducted
genome sequence analysis and showed that genetic changes in the consensus sequence were located in both non-structural and structural regions (Table 1). We then analyzed the genetic diversity of these viral popula-tions using deep sequencing and showed that the proportion of fixed mutapopula-tions (frequency > 80%) was lower in sequences derived from virus passaged in Ae. albopictus. These findings suggest that the environment within
Figure 3. The biological clone E1-V IOL lineage is better transmitted by Ae. albopictus from La Réunion but not by Ae. albopictus from Congo. Both strains of Ae. albopictus (Congo and La Réunion) and Ae. aegypti
from Congo were exposed to an infectious blood-meal containing an individual clone or both clones provided at equal titers. Saliva collected at 7 dpi were inoculated on Vero cells. (a) For mosquitoes infected with one viral clone, the percentage of infectious saliva was determined. (b) For mosquitoes infected with both viral clones, saliva were collected and inoculated on Vero cells. Then, lytic plaques were collected for RNA extraction and sequencing to define the identity of the amino-acid at the position E1-226. Proportion of E1-V among clones examined in saliva was estimated at 7 dpi for mosquitoes blood-fed with E1-A and E1-V provided at equal proportions. Error bars show the confidence intervals (95%).
Ae. albopictus exerts a stronger selective pressure on the virus and acts to bottleneck the viral genetic diversity
with consequences on CHIKV adaptive evolution40.
Under the selective pressure of Ae. aegypti however, the frequency of mutations within the viral population was higher. In addition, we observed a higher number of mutations in virus collected at 14 dpi when compared to 7 dpi suggesting that there may be a temporal effect on population diversity. Of particular interest, we iso-lated a fixed mutation in CHIKV P10_AE7, which results in an amino-acid substitution (nsP4-C483Y), pre-viously described by31. Located within the viral RNA dependent RNA polymerase (RdRp), this mutation was
shown to cause an increase in replication fidelity. This results in the generation of a viral population with reduced genetic diversity, potentially leading to reduced viral fitness both in Ae. aegypti mosquitoes and in vertebrate hosts31. Unsurprisingly, this mutation was not recovered from any other sample populations further supporting
this hypothesis. Nevertheless, the CHIKV nsp4 C483Y mutation did not appear to impact viral replication as P10_AE7 replicates well in both Ae. aegypti Aag2 and Ae. albopictus U4.4 cells (Supplementary Fig. 3b–e). In
Ae. aegypti mosquitoes, virus recovered from passages using late-collected saliva at 14 dpi showed both increased
genetic diversity (Table 1) and increased viral load in saliva (Fig. 2d). These data suggest that genetic diversity of CHIKV is important in maximizing viral transmission. Two amino-acid substitutions in the E2 glycoprotein at positions 217 and 249 (Table 1) were also identified. Neither of these residues have been shown to interact with E1 position 226 and in fact, are located on the outside of the trimeric spike where they could potentially play a role in the induction of the host protective immune response41 or interaction of the virion with host receptor proteins42
(Supplementary Fig. 2).
Ae. albopictus from Congo are different from those on La Réunion when comparing their transmission
potential for CHIKV IOL. When exposed to an equal proportion of viral clones differing by a single substi-tution A or V at E1-226, Ae. albopictus from Congo did not preferentially transmit the clone harboring the
Ae. albopictus-adaptive substitution E1-A226V as expected (Fig. 3b). One explanation for this is that Ae. albopictus
from Congo are genetically different from Ae. albopictus from La Réunion; geographically-distant populations
Figure 4. Genetic differentiation of Aedes albopictus populations. (a) Map showing the four E1-A226V
substitution emergence events in La Réunion Island33, India55, Cameroon36 and Gabon37 and spreading of
CHIKV ECSA lineage correlated with the presence of Ae. albopictus. Date of introduction or first description of Ae. albopictus in the country is indicated. (b) Neighbor-joining cluster analysis (unrooted) based on Cavalli-Sforza and Edwards’s chord distance. Apparent root (CAL from India) is for visual purposes only. Numbers indicate bootstrap values above 65%. Genotyping included DNA extraction from 30 mosquitoes (15 males and 15 females), PCR amplification of 11 microsatellites and sequencing of fragments for scoring haplotypes. A dendrogram based on microsatellite Cavalli-Sforza & Edwards’s genetic distance clustering by the NJ method was constructed using 16 Ae. albopictus populations: ALPROV, Saint-Denis, La Réunion; BL, Bar-sur-Loup, France; CAL, Calcutta, India; CONG, Brazzaville, Congo; JRB, Jurujuba, Brazil; MAN, Manaus, Brazil; MFILOU, Brazzaville, Congo; MIA, Misiones, Argentina; MXA, Tapachula, Mexico; PNA, Colon, Panama; PNM, Parnamirim, Brazil; SAN, Santos, Brazil; STANDRE, Saint-André, La Réunion; STR, Santarém, Brazil; TYS, Tyson, United States; VRB, Florida, United states. F1 mosquitoes were used except for the lab colony CAL. The map was modified using PowerPoint from http://www.powerpointslides.net/powerpointgraphics/ powerpointmaps.html using a map previously published in56.
have lower genetic connectedness and higher population genetic differentiation with potential consequences for their transmission of infectious agents. We showed using mosquito genotyping with microsatellites that
Ae. albopictus from Congo and from La Réunion formed divergent groups (Fig. 4b) with probably a different
history of colonisation33,43. The process of ongoing competitive displacements of Ae. aegypti by Ae. albopictus
in Central Africa44 provides a situation different from La Réunion Island where Ae. albopictus was the unique
vector32. The selection of the mutation E1-226V of an ECSA genotype conferring a higher transmission by
Ae. albopictus was unlikely to have been the scenario that occurred in the Congo as it was in La Réunion.
Therefore, CHIKV ECSA E1-226V was most likely introduced into the Congo rather than selected in situ. Thus, attention should be paid to the existence of specific interactions between mosquito and virus genotypes that have differing potential for leading to an emergence event.
Methods
Ethics Statement.
The Institut Pasteur animal facility has received accreditation from the French Ministry of Agriculture to perform experiments on live animals in compliance with the French and European regulations on care and protection of laboratory animals. This study was approved by the Institutional Animal Care and Use Committee (IACUC) at the Institut Pasteur. No specific permits were required for the described field studies in locations, which are not protected in any way and did not involve endangered or protected species.Mosquitoes.
Eggs were collected from ovitraps placed in the city of Brazzaville (Congo) on the ORSTOM campus in august 2011 and sent to the Institut Pasteur in Paris where they were reared in standardized conditions. After morphological identification, Ae. aegypti and Ae. albopictus adults were placed in different cages and reared at 28 ± 1°C, at relative humidity of 80% and a light:dark cycle of 16 h:8 h. A constant supply of 10% sucrose was provided to adults. To produce eggs, females were fed three times a week on anesthetized mice (OF1 mice from Charles River laboratories, France). Oral infection experiments were performed using mosquitoes from the F2-F4 generations. The F12 generation of Ae. albopictus Providence (ALPROV) collected in 2007 on La Réunion Island was used for comparison.Cell cultures.
C6/36 (Ae. albopictus) cells were maintained at 28 °C in Leibovitz L-15 medium supplemented with non-essential amino-acids (1X), 10% fetal bovine serum (FBS), 100 units/mL penicillin and 100 μ g/mL streptomycin. They were used for production of parental viral stocks. Vero (green monkey kidney) cells used for CHIKV titrations were maintained at 37 °C, 5% CO2 in Dulbecco’s Modified Eagle medium (DMEM) with 10%FBS, 100 units/mL penicillin and 100 μ g/mL streptomycin. HFF (Human Foreskin Fibroblast) cells were main-tained at 37 °C, 5% CO2 in Dulbecco’s Modified Eagle medium (DMEM) supplemented with pyruvate, 10% FBS,
100 units/mL penicillin and 100 μ g/mL streptomycin. These cells were used for the alternate passages as a proxy to mimic the human host. Two other types of mosquito cells were used to assess replication kinetics: (i) an Ae.
aegypti-derived Aag2 cells and (ii) an Ae. albopictus-derived U4.4 cells. Aag-2 cells were maintained in Schneider
Drosophila medium, 10% FBS and 1% P/S and U4.4 cells were maintained in L-15 medium supplemented with 10% FBS and 10% tryptose phosphate broth at 28 °C.
Viruses.
The CHIKV DRC_2000 and Congo_2011 strains were isolated from patients during the epidemics of 2000 in the DRC and 2011 in Congo and were both provided by the French National Reference Center for Arboviruses at the Institut Pasteur. Both isolates belong to the ECSA phylogroup and the Central African lineage. The DRC_2000 and Congo_2011 differ in two major substitutions in E1-226 and E2-337: the DRC_2000 strain harbours E1-226A and E2-337V whereas the Congo_2011 strain presents E1-226V and E2-337I (Supplementary Table 3). Viral stocks were produced on C6/36 and viral titers were determined via plaque assay as previously described14. In addition, biological clones E1-A and E1-V of IOL lineage which differ by a single position E1-226from an alanine (A) to a valine (V), produced by plaque purification from two CHIKV isolates from La Réunion Island, respectively CHIKV 06.21 and CHIKV 05.115 were also used14,45.
Mosquito oral infections.
Infection assays were performed with one-week-old females starved 24 hrs prior to infection in a BSL-3 laboratory. Mosquitoes were allowed to feed for 15 min through a piece of pork intestine (for Ae. aegypti) or chicken skin (for Ae. albopictus) covering the base of a Hemotek feeder containing the infec-tious blood-meal maintained at 37 °C. The blood-meal was composed of 1/3 of viral supernatant, 2/3 of washed rabbit erythrocytes isolated from arterial blood, and adenosine triphosphate at a final concentration of 10−3 M46.To assess the vector competence, adult females of F2 generation (for CHIKV DRC_2000 or Congo_2011 strains) and of F3/F4 generation (for the four CHIKV strains resulting from the 10th passage; Fig. 1) were exposed to
infectious blood-meals at a titre of 106.5 pfu (plaque forming units)/mL and engorged females were separated and
incubated under controlled conditions (28 ± 1°C, relative humidity of 80%, light:dark cycle of 16 h:8 h). At 7 dpi, vector competence was assessed based on two phenotypes: (i) viral dissemination from the midgut into mosquito general cavity and (ii) transmission potential with virus detected in mosquito saliva. Disseminated infection rate (DIR) was defined as the percentage of engorged mosquitoes with virus detected in heads suggestive of successful viral dissemination from the midgut. Transmission rate (TR) was calculated as the percentage of mosquitoes with viral particles detected in saliva among mosquitoes that developed a disseminated infection. Saliva was collected as described in Dubrulle et al.47. Briefly, wings and legs were removed from each individual and its proboscis
was inserted into a 20 μ L tip containing 5 μ L of FBS. After 45 min, FBS containing saliva was expelled in 45 μL of Leibovitz L15 medium (Invitrogen) for titration. Experimentally induced salivation is widely used to demonstrate the transmission of pathogens ingested by hematophagous insects48. DIR and TR were calculated by titrating head
Competitive assays between two viral clones E1-V and E1-A from IOL lineage.
Competition assays were performed by providing the two biological clones E1-A (with E1-226A) and E1-V (with E1-226V) from IOL lineage in an equal proportion (1:1) to mosquitoes in blood-meals prepared at a final titre of 106.5 pfu/mL. At 7 dpi, viruses were isolated from saliva. Briefly, viruses were used to infect six-well-plates containing confluent monolayers of Vero cells. Cells were incubated for 3 days at 37 °C and 5% CO2 under an overlay
con-sisting of DMEM (1X) with 2% FBS, 1% L-Glutamine, 1% agarose and 1% penicillin/streptomycin/amphotericin (Invitrogen). For each mosquito species, 30 saliva samples were analyzed and lytic plaques were removed by suc-tion using a pipette. Each agarose plug containing an individual clone was dissolved overnight at + 4°C in 50 μ L of DMEM. RNA was extracted using NucleoSpin RNA kit (Macherey Nagel) according to the manufacturer’s instruction. A one-step RT-PCR reaction targeting a region comprising the position 226 in the E1 gene was performed using the Titan One Tube kit (Roche). The sequencing reaction was conducted using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit version 3.1 (Applied Biosystems)45.
Alternate passages of the parental CHIKV DRC_2000 strain.
Alternate passages are summarised on Fig. 2. Before alternate passages, a stock of DRC_2000 was produced on HFF human cells (DRC_2000_HFF) using the DRC_2000 parental strain at a multiplicity of infection (MOI) of 0.1. For the first passage in mosqui-toes, the F2 generation of both species (Ae. aegypti and Ae. albopictus) were orally infected with the produced DRC_2000_HFF supernatant provided in a blood-meal at a final titer of 106.5 pfu/mL. Engorged mosquitoeswere incubated at 28 °C for either 7 or 14 days and then processed for saliva collection. Saliva was pooled by species and day of collection (4 samples, 20 salivations per sample) and the volume of each sample adjusted to 600 μ L with DMEM prior to filtration through a Millipore H membrane (0.22 μ m). An aliquot of 300 μ L of each sample was used to inoculate a sub-confluent flask (25 cm2) of HFF cells (considered as the first passage of saliva
on human cells). After 1 hour, the inoculum was discarded and cells rinsed once with DMEM medium. 5 mL of DMEM medium complemented with 2% FBS were added and cells were incubated for 3 days at 37 °C. Cell culture supernatants were then collected and stored at − 80 °C until used. For each passage and each condition (P10_AE7, P10_AE14, P10_AL7 and P10_AL14, where AE and AL stand for Ae. aegypti and Ae. albopictus, respectively) (Fig. 2a), saliva were pooled from 10 to 20 individuals depending on the mosquito feeding rate. Passages P2 to P6 were performed with mosquitoes of the F3 generation and passages P7 to P10 with mosquitoes of the F4 generation. HFF supernatants collected at each passage were used undiluted for the next mosquito blood-meal without titration.
Replication kinetics.
To measure viral replicative fitness, growth curves were conducted in Ae. aegypti Aag2 and Ae. albopictus U4.4 cells. Confluent cell monolayers were prepared and inoculated with viruses simulta-neously in duplicates at a MOI of 0.1 pfu/cell. Cells were incubated for 1 hour in appropriate conditions. Viral inoculum was removed and cell monolayers were washed 3 times with PBS to eliminate unbound virus. Two mL of medium supplemented with 5% FBS were then added and cells were incubated at 28 °C. At various times (0, 4, 6, 8, 10 and 24 hrs) post-inoculation, supernatants were collected and titrated by Vero plaque assays. To estimate the number of RNA copies, RNA was extracted from the same samples using Nucleospin RNA II kit (Macherey-Nagel) and a one-step qRT-PCR was performed to detect the number of viral RNA copies accord-ing to protocols described by Vazeille et al.14. Primers have been designed in the E2 structural gene: senseChik⁄E2⁄9018⁄+ (CACCGCCGCAACTACCG) and anti-sense Chik⁄E2⁄9235⁄− (GATTGGTGACCGCGGCA).
Genome sequencing.
Viral RNA was extracted from supernatants of DRC_2000, P10_AE7, P10_AE14, P10_AL7 and P10_AL14. RT-PCR was performed using SuperScript One-Step RT-PCR with platinum Taq (Invitrogen) using 21 sets of primers targeting the complete CHIKV genome as described by Schuffeneckeret al.4. Amplicon sequencing reactions were performed using Big Dye Terminator v1.1 cycle sequencing
kit (Applied Biosystems) and sequencing was performed using an ABI3730XL sequence analyzer (Applied Biosystems). Sequence analysis, contig assembly and sequence alignments were performed using BioNumerics version 6.5 (Applied-Maths, Saint-Martens-Latem, Belgium). For phylogenetic analysis, maximum-likelihood tree was constructed using MEGA version 5 (www.megasoftware.net), based on the Tamura-Nei model. Reliability of nodes was assessed by bootstrap resampling with 1,000 replicates.
Viral diversity.
Whole genome sequences (excluding the first 19 nucleotides of the 5′ UTR and the 41 nucleo-tides upstream the polyA tail) were determined for DRC_2000_HFF and the last passages (P10_AE7, P10_AE14, P10_AL7 and P10_AL14) using the Ion PGM Sequencer (Life Technologies) as described by Rothberg et al.49 andsequence analysis was conducted using CLC Genomics Workbench 6 software. For deep sequencing, a set of four primer pairs (Supplementary Table 4) was used to generate amplicons with 3 μ L of nucleic acid extract and the Superscript III One-Step RT-PCR Platinum TaqHifi kit (Life Technologies) according to manufacturer’s instruc-tions using the following cycling parameters: 50 °C for 30 min, 94 °C for 2 min followed by 45 cycles of 94 °C for 15 sec, 56 °C for 30 sec and 68 °C for 4 min. PCR products were verified by gel electrophoresis and amplicons were purified using Amicon Ultra – 0.5 mL 30 K kit (Millipore) according to the manufacturer’s instructions. For each sample, an equimolar mix of all amplicons was used to build a library and produce the corresponding sequences for the Ion PGM Sequencer according to the manufacturer’s instructions. The reads obtained were trimmed: first using quality score and then by removing the primers used for amplification. Reads were mapped to the genome sequence of CHIKV DRC_2000_HFF, which was used as a reference. Mutation frequencies (proportion of viral genomes with the mutation) at each position were calculated as the number of reads with a mutation compared to the reference divided by the total number of reads at that site. Only substitutions with a mutation frequency ≥ 5% were considered significant for further analysis.
Mosquito genotyping.
DNA was extracted from each mosquito with a commercial purification kit (Nucleospin 96 DNA Tissue kit, Macherey-Nagel) following manufacturer’s instructions for vacuum processing. Briefly, 30 mosquitoes (15 males and 15 females) per population were individually grounded in 180 μ L lysis buffer supplemented with 25 μ L of Proteinase K. The homogenates were then passed through columns allowing binding of total nucleic acids. Silica membranes were further desalted and DNA was finally eluted in 100 μ L of elution buffer. Eleven microsatellites were amplified by PCR using specific primers flanking the repeated region50. PCRreaction mixtures consisted of 50 ng genomic DNA, 1x PCR buffer, 1.5 mM MgCl2, 0.27 mM dNTPs (Invitrogen), 1 U Taq polymerase (Invitrogen), and 10 μ M of each primer, one of which was 5′ labelled with a fluorescent dye, in a final volume of 15 μ L. PCR cycling conditions were 94 °C for 5 min, 30 cycles of 94 °C for 30 s, 59 °C for 30 s, and 72 °C for 30 s, followed by a final extension step of 72 °C for 7 min. Aliquots of PCR products were separated by electrophoresis on 2% agarose gels stained with ethidium bromide, and visualized under UV light. Each PCR product was then diluted 1:10 in ddH2O water and 2 μ L of this dilution was added to 10 μ L of a mixture of deionized formamide and GeneScan-500 ROX size standard (Applied Biosystems). The fragments were then resolved on an ABI3730XL sequence analyzer (Applied Biosystems). Data were analysed using GeneScan and Genemapper software. The program TANDEM51 was used to limit ambiguous genotyping.
Statistical/Phylogenetical analysis.
Statistical analyses were conducted using the STATA software (StataCorp LP, Texas, and USA). Rates/proportions (disseminated infection rate, transmission rate, percentage of infectious saliva, proportion of E1-V clones) were compared using Chi square test. The numbers of infectious particles in saliva were compared using Kruskal-Wallis test. P-values > 0.05 were considered non-significant. Allelic frequencies were obtained using the GENEPOP software (version 4.0) of Raymond & Rousset52. To assessrelatedness among populations, Cavalli-Sforza & Edwards’s53 chord distance for each pair of populations was
cal-culated in PHYLIP 3.69 (GENDIST module) and the resulting distance matrix was used to create a dendrogram. A neighbor-joining tree was constructed with node confidence inferred via 100 bootstrap replicates in PHYLIP 3.69 (modules SEQ- BOOT, GENDIST, NEIGHBOUR and CONSENSE). The neighbor-joining network should not be taken as a true phylogeny since microsatellites are not ideal markers for recovering evolutionary history54.
Rather, the analysis should be considered as a method to visualize discrete genetic clustering of populations.
References
1. Wang, E. et al. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol 74, 3227–3234 (2000). 2. Moncayo, A. C. et al. Dengue emergence and adaptation to peridomestic mosquitoes. Emerg Infect Dis 10, 1790–1796, doi: 10.3201/
eid1010.030846 (2004).
3. Anishchenko, M. et al. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc Natl Acad
Sci USA 103, 4994–4999, doi: 10.1073/pnas.0509961103 (2006).
4. Schuffenecker, I. et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 3, e263, doi: 10.1371/journal.pmed.0030263 (2006).
5. Juliano, S. A. & Lounibos, L. P. Ecology of invasive mosquitoes: effects on resident species and on human health. Ecol Lett 8, 558–574, doi: 10.1111/j.1461-0248.2005.00755 (2005).
6. Juliano, S. A. Species interactions among larval mosquitoes: context dependence across habitat gradients. Annu Rev Entomol 54, 37–56, doi: 10.1146/annurev.ento.54.110807.090611 (2009).
7. Hawley, W. A. The biology of Aedes albopictus. J Am Mosq Control Assoc Suppl 1, 1–39 (1988).
8. Lounibos, L. P. Invasions by insect vectors of human disease. Annu Rev Entomol 47, 233–266, doi: 10.1146/annurev. ento.47.091201.145206 (2002).
9. Powers, A. M. & Logue, C. H. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol 88, 2363–2377, doi: 10.1099/vir.0.82858-0 (2007).
10. Rezza, G. et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370, 1840–1846, doi: 10.1016/ S0140-6736(07)61779-6 (2007).
11. Rezza, G. et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370, 1840–1846, doi: 10.1016/ S0140-6736(07)61779-6 (2007).
12. Grandadam, M. et al. Chikungunya virus, southeastern France. Emerg Infect Dis 17, 910–913, doi: 10.3201/eid1705.101873 (2011). 13. Tsetsarkin, K. A., Vanlandingham, D. L., McGee, C. E. & Higgs, S. A single mutation in chikungunya virus affects vector specificity
and epidemic potential. PLoS Pathog 3, e201, doi: 10.1371/journal.ppat.0030201 (2007).
14. Vazeille, M. et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS One 2, e1168, doi: 10.1371/journal.pone.0001168 (2007).
15. Chatterjee, P. K., Vashishtha, M. & Kielian, M. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J Virol 74, 1623–1631 (2000).
16. de Lamballerie, X. et al. Chikungunya virus adapts to tiger mosquito via evolutionary convergence: a sign of things to come? Virol J
5, 33, doi: 10.1186/1743-422X-5-33 (2008).
17. Santhosh, S. R., Dash, P. K., Parida, M., Khan, M. & Rao, P. V. Appearance of E1: A226V mutant Chikungunya virus in Coastal Karnataka, India during 2008 outbreak. Virol J 6, 172, doi: 10.1186/1743-422X-6-172 (2009).
18. Volk, S. M. et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol 84, 6497–6504, doi: 10.1128/JVI.01603-09 (2010).
19. Peyrefitte, C. N. et al. Circulation of Chikungunya virus in Gabon, 2006–2007. J Med Virol 80, 430–433, doi: 10.1002/jmv.21090 (2008).
20. Robinson, M. C. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. I. Clinical features. Trans R
Soc Trop Med Hyg 49, 28–32 (1955).
21. Osterrieth, P. & Blanes-Ridaura, G. [Research on the Chikungunya virus in the Belgian Congo. I. Isolation of the virus in upper Uele]. Ann Soc Belg Med Trop (1920) 40, 199–203 (1960).
22. Osterrieth, P., Rathe, E. & Deleplanque-Liegeois, P. [Simultaneous isolation of the yellow fever and Chikungunya viruses at Bili (Haut-Uele, Belgian Congo)]. Ann Soc Belg Med Trop (1920) 41, 207–212 (1961).
23. Muyembe-Tamfum, J. J. et al. [Epidemic of Chikungunya virus in 1999 and 200 in the Democratic Republic of the Congo]. Med Trop (Mars) 63, 637–638 (2003).
24. Pastorino, B. et al. Epidemic resurgence of Chikungunya virus in democratic Republic of the Congo: identification of a new central African strain. J Med Virol 74, 277–282, doi: 10.1002/jmv.20168 (2004).
25. Kamgang, B. et al. Geographic and ecological distribution of the dengue and chikungunya virus vectors Aedes aegypti and Aedes albopictus in three major Cameroonian towns. Med Vet Entomol 24, 132–141, doi: 10.1111/j.1365-2915.2010.00869.x (2010).
26. Paupy, C., Kassa Kassa, F., Caron, M., Nkoghe, D. & Leroy, E. M. A chikungunya outbreak associated with the vector Aedes albopictus in remote villages of Gabon. Vector Borne Zoonotic Dis 12, 167–169, doi: 10.1089/vbz.2011.0736 (2012).
27. Kelvin, A. A. Outbreak of Chikungunya in the Republic of Congo and the global picture. J Infect Dev Ctries 5, 441–444 (2011). 28. Mombouli, J. V. et al. Chikungunya virus infection, Brazzaville, Republic of Congo, 2011. Emerg Infect Dis 19, 1542–1543,
doi: 10.3201/eid1909.130451 (2013).
29. Tsetsarkin, K. A. et al. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun 5, 4084, doi :10.1038/ncomms5084 (2014).
30. Ross, R. W. The Newala epidemic. III. The virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond) 54, 177–191 (1956).
31. Coffey, L. L., Beeharry, Y., Borderia, A. V., Blanc, H. & Vignuzzi, M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci USA 108, 16038–16043, doi: 10.1073/pnas.1111650108 (2011).
32. Bagny, L., Delatte, H., Quilici, S. & Fontenille, D. Progressive decrease in Aedes aegypti distribution in Reunion Island since the 1900s. J Med Entomol 46, 1541–1545 (2009).
33. Delatte, H. et al. The invaders: phylogeography of dengue and chikungunya viruses Aedes vectors, on the South West islands of the Indian Ocean. Infect Genet Evol 11, 1769–1781, doi: 10.1016/j.meegid.2011.07.016 (2011).
34. Alto, B. W., Lounibos, L. P., Higgs, S. & Juliano, S. A. Larval Competition Differentially Affects Arbovirus Infection in Aedes Mosquitoes. Ecology 86, 3279–3288 (2005).
35. Alto, B. W., Lounibos, L. P., Mores, C. N. & Reiskind, M. H. Larval competition alters susceptibility of adult Aedes mosquitoes to dengue infection. Proc Biol Sci 275, 463–471, doi: 10.1098/rspb.2007.1497 (2008).
36. Fontenille, D. & Toto, J. C. Aedes (Stegomyia) albopictus (Skuse), a potential new Dengue vector in southern Cameroon. Emerg
Infect Dis 7, 1066–1067, doi: 10.3201/eid0706.010631 (2001).
37. Coffinet, T. et al. First record of Aedes albopictus in Gabon. J Am Mosq Control Assoc 23, 471–472, doi: 10.2987/5636.1 (2007). 38. Diallo, M., Laganier, R. & Nangouma, A. First record of Ae. albopictus (Skuse 1894), in Central African Republic. Trop Med Int
Health 15, 1185–1189, doi: 10.1111/j.1365-3156.2010.02594.x (2010).
39. Paupy, C. et al. Comparative role of Aedes albopictus and Aedes aegypti in the emergence of Dengue and Chikungunya in central Africa. Vector Borne Zoonotic Dis 10, 259–266, doi: 10.1089/vbz.2009.0005 (2010).
40. Forrester, N. L., Guerbois, M., Seymour, R. L., Spratt, H. & Weaver, S. C. Vector-borne transmission imposes a severe bottleneck on an RNA virus population. PLoS Pathog 8, e1002897, doi: 10.1371/journal.ppat.1002897 (2012).
41. Goh, L. Y. et al. Neutralizing monoclonal antibodies to the E2 protein of chikungunya virus protects against disease in a mouse model. Clin Immunol 149, 487–497, doi: 10.1016/j.clim.2013.10.004 (2013).
42. Asnet Mary, J., Paramasivan, R., Tyagi, B. K., Surender, M. & Shenbagarathai, R. Identification of structural motifs in the E2 glycoprotein of Chikungunya involved in virus-host interaction. J Biomol Struct Dyn 31, 1077–1085, doi: 10.1080/07391102.2012. 721496 (2013).
43. Kamgang, B. et al. Genetic structure of the tiger mosquito, Aedes albopictus, in Cameroon (Central Africa). PLoS One 6, e20257, doi: 10.1371/journal.pone.0020257 (2011).
44. Ngoagouni, C., Kamgang, B., Nakoune, E., Paupy, C. & Kazanji, M. Invasion of Aedes albopictus (Diptera: Culicidae) into central Africa: what consequences for emerging diseases? Parasit Vectors 8, 191, doi: 10.1186/s13071-015-0808-3 (2015).
45. Arias-Goeta, C., Mousson, L., Rougeon, F. & Failloux, A. B. Dissemination and transmission of the E1-226V variant of chikungunya virus in Aedes albopictus are controlled at the midgut barrier level. PLoS One 8, e57548, doi: 10.1371/journal.pone.0057548 (2013). 46. Vazeille-Falcoz, M., Mousson, L., Rodhain, F., Chungue, E. & Failloux, A. B. Variation in oral susceptibility to dengue type 2 virus of
populations of Aedes aegypti from the islands of Tahiti and Moorea, French Polynesia. Am J Trop Med Hyg 60, 292–299 (1999). 47. Dubrulle, M., Mousson, L., Moutailler, S., Vazeille, M. & Failloux, A. B. Chikungunya virus and Aedes mosquitoes: saliva is
infectious as soon as two days after oral infection. PLoS One 4, e5895, doi: 10.1371/journal.pone.0005895 (2009).
48. Boorman, J. Induction of salivation in biting midges and mosquitoes, and demonstration of virus in the saliva of infected insects.
Med Vet Entomol 1, 211–214 (1987).
49. Rothberg, J. M. et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352, doi: 10.1038/nature10242 (2011).
50. Manni, M. et al. Molecular markers for analyses of intraspecific genetic diversity in the Asian Tiger mosquito, Aedes albopictus.
Parasit Vectors 8, 188, doi: 10.1186/s13071-015-0794-5 (2015).
51. Matschiner, M. & Salzburger, W. TANDEM: integrating automated allele binning into genetics and genomics workflows.
Bioinformatics 25, 1982–1983, doi: 10.1093/bioinformatics/btp303 (2009).
52. Raymond M. & R. F. Genepop (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86, 248–249 (1995).
53. Cavalli-Sforza, L. L. & Edwards, A. W. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet 19, 233–257 (1967).
54. Takezaki, N. & Nei, M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144, 389–399 (1996). 55. Gratz, N. G. Critical review of the vector status of Aedes albopictus. Med Vet Entomol 18, 215–227, doi: 10.1111/j.0269-283X.2004.
00513.x (2004).
56. Zouache, K. & Failloux, A.-B. Insect–pathogen interactions: contribution of viral adaptation to the emergence of vector-borne diseases, the example of chikungunya. Curr Opin Insect Sci 10, 14–21, doi: http://dx.doi.org/10.1016/j.cois.2015.04.010 (2015).
Acknowledgements
We thank Thérèse Couderc for providing HFF cells, Anna Malacrida and Laure Diancourt for their advices in genotyping mosquitoes. We are grateful to François Rougeon for his critical comments. We warmly thank Richard Paul for correcting the manuscript. This study was funded by the Institut Pasteur and the French Government’s Investissement d’Avenir program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (grant n°. ANR-10-LABX-62-IBEID). KZ was supported by the project “VECTORIE” and the Foundation Inkermann (Fondation de France).
Author Contributions
M.V. contributed to conceive, perform and analyze the experiments. K.Z. contributed in designing and performing the experiments. J.M.T. and V.C. contributed in sequencing viral strains and genome analysis. A.V.R., M.M. and G.G. genotyped mosquitoes with microsatellites. A.Y. was involved in providing mosquito samples. L.M. participated in experiments on mosquitoes. G.P. and X.D.L. were responsible for NGS experiments, analysis and interpretation of data. M.C.V. contributed in drawing the atomic structure of CHIKV. C.D. contributed to microsatellite analysis. A.B.F. participated in conceiving and designing the experiments, analyzing/interpreting the data and the writing of the article. All authors reviewed the paper.
Additional Information
Supplementary information accompanies this paper at http://www.nature.com/srep Competing financial interests: The authors declare no competing financial interests.
How to cite this article: Vazeille, M. et al. Importance of mosquito “quasispecies” in selecting an epidemic
arthropod-borne virus. Sci. Rep. 6, 29564; doi: 10.1038/srep29564 (2016).
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/