Arch Virol (1994) 139 327-336
O Springer-Verlag 1994
Printed in Austria
The complete
nucleotide
sequencesof
the coat protein cistron
and the
3'
non-coding
region
of
a newly-identified
potyyirus
infecting
sweetpotato, as
compared
to
those
of
sweetpotato
feathery
mottle
virus
D. Colinet, J. Kummert, and P. Lepoivre
Faculté des Sciences Agronomiques, Laboratoire de Pathologie Végétale,
Gembloux, Belgium
Accepted August 10, 1994
Summary. Complementary
DNA
representing 728 nucleotides of the 3' endof
the genomic
RNA
of sweetpotato virus G (SPV-G), a newly-identified potyvirus infecting sweetpotato, was cloned and sequenced. This sequence was combinedwith that
previously determinedfor the 5'
terminal
part of
the coat
protein cistron of the virus. The whole sequence contained a single open reading frame(ORF)
of
1065 nucleotides,with
the capacityto
encode a coatprotein
of
355amino acids, significantly larger than
that
ol
other potyviruses. TheORF
waslollowed
by
an untranslated regionof
222 nucleotides and apoly
(A)tail.
The coat protein ol SPV-G was only distantly relatedto
that of known potyviruses,with the
exceptionof
sweetpotato featherymottle virus
(SPFMV).
Indeed, sequenceidentity in
the C-terminal
three quartersof
the coat protein
(morethan 80){)
and
in
the
3'
untranslatedregion (more
Than 70"/.)indicate
thatSPV-G should be considered as closely related to, though distinct from SPFMV.
This
subset relationship issimilar
to
that
previously reportedfor
membersof
the
beanyellow
mosaicvirus
subgroupor
the
beancommon
mosaic virussubgroup.
Introduction
Sweetpotato
may
be
affectedby
several potyviruses
[12],
among
which sweetpotatofeathery
mottle
virus (SPFMV) and
sweetpotatolatent
virus (SPLV) were recently identifiedin
sweetpotato clones originatingfrom
China, using a combined assay of reversetranscription
and polymerase chain reaction(RT-PCR)
with
degenerateprimers derived
from
conserved regionsin
the genome of the potyviruses[5,6].
RT-PCR
and sequence analysis of the variableN-terminal
part of
the coat protein allowed a
third
distinct potyvirus
to
beidentified
in
sweetpotatoesfrom China
[6].
The
name sweetpotatovirus
G (SPV-G) has been tentatively assignedto
this
virus.Vîrology
. ::j:.:,,::, i..
:,:,,j:: . ::.:,
The
useof
sequenceinformation
for
the
classificationof
potyviruses isgenerally
more
reliable
than
schemes basedon
biological and
serologicalproperties,
which
reveal apparent relationshipsthat do not
always correlatewith
one another[15,17],
Comparisons of coatprotein
amino acid sequencesand
of
3'
non-coding nucleotide
sequenceshave
been used
to
establishtaxonomic relationships among potyvirus members L7, 13, 15, 17, 1 8]. Sequence
comparisons show
a
bimodal
distribution of coat protein
sequenceidentity
[15]. Known
distinct
potyvirusesvary
in
coat
protein
amino acid
sequencesimilarity
between 38/,to
7I/",
while known strains of viruses range from 90/"to
997.[15].
Strains of the same virus share83/"
to 99/"
nucleotide sequenceidentity
in the 3' untranslated region, while distinct viruses share generally only39/" to 53/"
identity
in
this
region[7].
In
order
to
further
examinethe
relationship
betweenSPV-G and
other potyviruses we determined the nucleotide sequencesol
the 3' terminalpart of
the coat protein cistron and
of
the
3'
untranslatedregion
of
SPV-G. Thesesequences were combined
with
that
previously determinedfor
the 5' terminalpart
of the SPV-G coat protein cistron[6].
The complete SPV-G coat proteinamino acid
sequenceand
3'
non-coding
region
nucleotide
sequence were compared with those of other potyviruses, and more particularly with the RussetCrack (RC) and the Common (C) strains ol
SPFMV
[1],
and the Taiwan isolateof SPLV (SPLV-T)
(D. Colinet, unpubl. results).Materials
and methodsPlant material
Sweetpotato (I pomoea batatas L.) clone GN58 originated from China, Guangdong province (Dr. Fing ZuXia,Upland Crops Research Institute, Guangzhou) and was maintained in
Gembloux under greenhouse conditions.
RNA extraction and cDNA sYnthesis
Total RNA was extracted from symptomatic ieaves by the method of Chirgwin et al. [4]. Single stranded cDNA was synthesised lrom 5 pg of totai RNA using the Amersham cDNA Synthesis Kit, with the following modifications:
-
oligo(dT) primer was replaced by 0.5pg of hybrid dT,r-adapter primer (s'-GACTCG-AGTCGACAGCGATTTTTTTTTTTTTTTTT- 3J [9],AGTCGACAGCGATTTTTTTTI
-
the reverse transcription reaction 30min. cDNA was diluted 10-foldwas incubated at 42'C for t h, and then at 52'C for
with sterile water.
Amplifcation of cDNA 3' end
Amplification of the cDNA was performed in a voiume
of
100pl of PCR buffer (10mM Tris-HClpH
9.0,2.5mMMgClr,50mM
KCl,0.l7;
Triton x-100) containing 200pM each of dATP. dCTP, dGTP, dTTP, 0.1 nmol each of adapter primer (5'-GACTCGAGT-CGACAGCG-3')t9l
and specific primer, and 2.5 unitsof
Triq DNA polymerase. Alter denaturation at94"C for 5 min, annealing at 50'C for 5min and elongationatT2oC forat94"C for 30sec, annealing at 50"C for
lmin
and DNA synthesisat72'C
for 3 min. A final 15 min elongation step at72'C was performed at the end of the 40 cycles. Amplificationproducts were analysed by electrophoresis of 10 pl of the reaction mixtures in a
l/,
agarose gel, in trisacetate-EDTA buffer[l4].
Bands were visualized by ethidium bromide staining.Cloning and sequencing of the amplifed fragment
After electrophoresis in 1/o agarose gel, the amplified fragment was excised and eluted with
the QIAEX Gel Extraction
Kit
from Qiagen. After digestionwith
BamHI and SalI, the DNA fragment was directionally cioned into the Bluescript plasmid.Nucleotide sequence was obtained by subcloning the amplified fragment after cleavage
with
restriction enzymes. Double-strandedDNA
sequencingby
the
dideoxy chain termination method was performed using T7 DNA polymerase (Pharmacia) according to manufacturer's instructions.Results
Amplifcation
of the3'
terminal region of SPV-G genomeRACE
(RapidAmplification
ofcDNA
Ends) method[9]
was usedto
amplilya
cDNA
fragment correspondingto
the 3'terminal
region of SPV-G genome'The
RACE protocol
allows the 3' and 5' endsof
acDNA to
be amplifiedif
ashort
stretchof
sequence located betrveen the endsis known.
For
the 3' end,RNA
is reverse transcribed using a"hybrid"
primer consisting of oligo(dT) (17residues)
linked
to a
unique
17-baseoligonucleotide
("adapter"
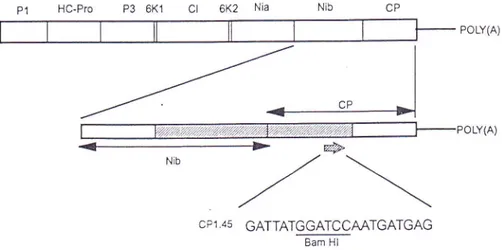
primer).P1 HC-Pro P3 6K1 6K2 Nia
POLY(A)
POLY(A)
,/\
cPl .4s GATTATG_G_ôI_qCAATGATGAG
Fig.
l.
Genetic map of the potyvirus genome showing the position and sequence of thespecific primer (CP 1.45) designed from the known sequence of the 5' terminal part of the coat protein cistron. The primer corresponds to codons 187-193 of the coat protein. The hatched box represents the part of the SPV-G genome previously amplified by RT-PCR
with degenerate primers 16). P1 First protein; HC-Pro helper component-protease; P3 third protein; 6K1 first 6K peptide;
CI
cytoplasmic inclusion protein; 6K2 second 6K peptide; Nic nuclear inclusion a protein; Nib nuclear inclusion b protein (RNA polymerase),CP coat Protein
1.37
-
0.830.56
-Fig. 2. Agarose gel stained with ethidium bromide, showing the result ol amplification ol
the 3' terminal region of SPV-G genome.
I
DNA
size markers, lragment sizes (in kb) indicated on the ieft.2 Product amplified from SPV-G infected Ipontoea setoscr2
Amplification
is subsequently performed using the adapterpt'ir"ç.,
which bindsto
eachcDNA
at
its
3' end, and aprimer
specificto
the geneof
interest.Total RNA
was isolatedfrom
leavesof
Ipontoea setosagrflted with
the sweetpotato cloneGN58
and shownto
be inlectedwith SPV-q
but
not
withSPFMV
and SPLV, using PCR andhybridization
assays(unpu[I.
res.).cDNA
was amplified using oligo(dT, ,)-adapter and amplification was caf ried out usingadapter
primer and
a
specificprimer (CP
1.45) designediro[n the
knownsequenceolthe5'terminal
partolthecoatproteincistronolSP\-Gt6l(Fig'
l).The
three nucleotides 10to
12ol
the
specificprimer
CP
1.45 J'vere modified(GTG
to
TCC)in
orderto
generate aBamHI
site(Fig.
1).A 7ry
bp lragment was amplified (Fig.2)
and clonedinto
the Bluescriptplasmid.
I I
Sequence analysis of the
SPV-G
PCRproduct
I
Because
of
the
low fidelity ol the
Taq
DNA
polymerase, whicfrmay
lead tomisincorporations, the sequence ol the SPV-G amplified fragmer[t was deduced
from
three
independent clones.
This
sequencewas
combir[ed
with
that previously determinedfor
the 5'-terminal
part of
the coat
profieincistron
ol
Spv-c
tot
IThe
complete nucleotide sequenceof
the coat protein cist{on and the
3'untranslated region of SPV-G is presented
in
Fig.
3. There is ar| open readingframe
(ORF)
extendingfor
1065 nucleotides,with
the capacitYlto encode 355amino acids. The ORF is followed by an untranslated region o1122 nucleotides
and a
poly(A)
tail. The
DAG
box
involved
in
aphid
transmlssion[2]
waslocated
at
position 7 of
the coatprotein amino
acid sequenceloi
SPV-G,lor
which aphid-transmissibility has been demonstrated (unpubl. ref.).The 3'
non-coding region
of
SPV-G
contains
the
consensus sequence{AGTGAGG
(position
1238to
1244) andCCTC (position
1250to
1253) se{aratedby
five nucleotides, whichcould form a
step-loop structure possiblyhfvine
arole
inreplication
[3].
L_
TCT GCT GAA GAG ATA TAC GAT GCA GGA AAA ACA GGA AAC ACA GGA AGG GGA A6A GCA CGA GGT 63
S A E E I Y D A G K T G H T G R G R G R G 21
ACT GTG CCT CCG CCG CCG CCA CCC CCT GGG CCA CCA AGA ACA GGT GAC CTG CCT CCA GCA GTG 1?6
T VP PPP PP PGÀPR T GD L PPAU 12
CAG ACÂ GGA CCA TTA CCA CCA GGT GCA GÇC TCA AAÂ CCA CCT ATC ATT GAG GAA ATT CTG CAG 189
AT GPLP PGAASKPP I I EE I LO 63
CCA GA6 TCA CCG AGA ACG AAG GCA TTG CG6 6AA GCG AGA GGG MÂ GCT CCA GCA ACA ATT CCA 252
P E S P R T KA L R EAR GKAPAT I P 84
GAT AGT AGA GGG GTT GAT ACA TCA CAA ATA CCG AGT TTC ACA CCA GGT AGA 6AC CM ACA ATG 315
D S R G V D T S A I P S F T P G R D O T t't 105 ACA CCA ACC CCT CAA AGA ACA AGC ACT GAG GlG AGA GAT AGA GAT GTG MT GCT GGT ACT GTT 378
T P T P A R T S T E V R D R D V N A G T V 126 G6T ACT TTC ATA GTG CCA CGG CCC CAG ATA ACA CAT AGT ÀAG AAA ACA GCA CCA ATG GCA AAT I'11
G T F I V P R P O I T H S K K T A P I'{ A N 147 GGA AGA ATA GTA GTC AAT CTT GAC CAC TTG ACÂ ATC TAC GAC CCT GAA CM ACA AGT CTT TCA 501
G R I V V N L D H L T I Y D P E O T S L S 168
AAT ACT C6A GCA ACA CAG Gfu\ Cfuq TTT AAT GCT TG6 TAC GAG CGT GTC AGG GM GAT TAT GGA 567
N T R A T O E A F N A IJ Y E G V R E D Y G 189 GTG fudT GAT GAG CA,/\ ATG GGG ATA TTG CTC AAT GGG TTA ATG GTT TGG TGC ATC GAG fu\T GGA 630
V N D E O H G I L L H G L H V I'J C t E N G 210 ACA TCC CCA ,ÙqT ATT A,/\T GGA ATG TGG GTC ATG ATG GAT GGT GAT CAA CM GTT ACA TAT CCA 693
T S P N I N 6 It H V I.t I't D G D E O V T Y P ?31
ATA AAA CCT CTA TTG GAT CAT GCT GTC CCC ACA TTT AGG CAG ATA ATG ACA CAC TlT AGC GAC 756
I K P L L D H A V P T F R O I t"I T H F S O 25?
ATC GCT GAÂ CCG TAT ATT 6AA AAG CGC MT AGG ATC AAG GCA TAC ATG CCG AGG TAT GGT CTA 819
t A E A Y I E K R I,I R I K A Y t'I. P R Y G L 273 CÀA CGA AAC TTG ACG GAT ATG AGT ATT GCG CGA TAT GCG lTC GAT TTC TAT GAG CTG CAC TCG 882
O R II L T D H S I A R Y A F D F Y E L H S 294 AAC ACT CCT GTT C6G GCC AGA GAG GCA CAT ATG CAA AlG AAA GCA GCA 6CA CTT MG AÂC GCA 915
II T P V R A R E A H I.I O H K A A A L K H A 315 CÀÀ AAT CGG TTG TTT 6GT TTG GAC GGA AAC GTC TCC AC6 CAG GAA GAA GAT ACG GAG AGG CAC 1OO8
O }I R L F G L D G H V S T O E E D T E R H 336
ACA ACG ACT GAT GTT ACA AGG A,qT ATA CAT MC CTC TTG GGT ATG AGG GGT GTG CAG TAAACAA 1072
TTTDVTRH I HIILLGHRGVA 355
TATATTGGCTCGTACCTTTTAATTTCAGTTTCGCCTTCAGTGTAATTTAAATTCGTATCTTTCAGTCCCGAAGAACCTGGTTT 1155
GGT GCAGAGCT TAAT GAGGT GT TACCT CTAT CT T T GCATT GGAGAAGGGATCT T T CTAT TACGTAT CAlAAGG6ACT CT TAÀA 1 238
GTGAGGTTTTACCTCGTAAGAÂAÂGCCTTTTTGGTTCGTGATCGAGCC POIY (A) 128é
Fig.3. Sequence of the SPV-G coat protein and 3' untranslated region' The predicted
u-ino
acià sequenceof
the single ORF is presented above the nucleotide sequence' Underlined nucleotides indicate ihe region from which the specific primer was derived.comparison of the coat protein sequence of
sPV-G with
sPFMV
(-RC
and-C),
SPLV-T
and other potyuirusesThe SPV-G coat
protein
shared66/.
and65/"
identitywith
SPFMV-RC
and-C,
respectively, a1the amino acid
level(Fig.
a).The
sequenceidentity
with
SPLV-T and other
potyviruseswas
lessthan 60/". The
C-terminal
three-quarters (residues 116to
355)ofthe
SPV-G coat protein sequence shared 83/"ind
S2/"
identity with
SPFMV-RC and -C,
respectively,and
lessthan 70/"
with
SPLV-T
and other potyvirusesat
the amino acid level'SPV. G SP FI'IV - RC sPFtlV- C SPLV-T PPV PVY SCMV TEV SPV. G SPFI"IV-RC SP FMV- C SPLV- T PPV PVY SCMV TEV SPV. G SP FMV. RC SP FMV. C SPLV - T PPV PVY scl"tv TEV SPV-G SPFI{V-RC sPFilV-C SPtV. T PPV PVY scHV TEV SPV. G SP FMV. RC SP Ft'IV. C SPLV. T PPV PVY scMv TEV
lslnlile----rY
l.lrlrl*
r E-
-
F K ttulslr
c o P E- -
F K L_l ADET...-IL ADEREDEEEV AID---Tl AG---ÏV sG---TV KTGNTGRGRGRGTVPPPPPPPGAPRT GD AD -.. AN-. KV-.-KP--_ ----svvT GS- - -AG--- GGGGNAGT AD.-OPPATGAAAOGGAO... PPATGAAA..' ilA DT DA DA OA DAr- [p-F nl v o T G p L [p-Fl c e n s rlF p] r [-le [Ël r L o
ffi
sll
nr
rlr
L-ni rFT
o l*l-1"
ol'
K p KN'lo
ol--
-'
-lf'l'lrlrlrlv
r ole elolclr o nlri
nlrln nlnlrl -le e elc K p K G nr
r
-
- -
-
- ol4t[_jtL!ut
cle elo[lx 0 All!
lloll
Rlol!;;;.;;;; .
..;;;;;;;;;;';;':lx:i:?l:il
R K K R R K K R nlp nr iFlofilnlclvlolSo I
P s F T P G R D o T r'l T P T P a n r[s]r eolro,
'
tltltltlol*lot
tl*E----
- KEsI
vcnsplxc
olrylvTflebl'Ld*lo r
slx c-
-
-
- E Ks MRsvs
Pa RPPOTNTPTO-. - -..P6PSGKGKEVI SAKGGE I I RTS
::ll
::::::::::
::l
::l::l:li::::i:l:;ll
O P P T T O G S O.L P - - . O G G A T G G G G A O T G A G G T G S V T G G .-.A6KKKDOKDDKVAEOArrM
. rltl
l;l;l
l:l;l
t rltl
.-ol!
IN VD VN VO v t'l T TL SV YC TT TS TF TF TH KI TF TA RO RO RL SL Rl! RL OY ALN II{H VLN VLH VVN A T A A A A I 0 T T T s F D u o D N E D N POITHS VKMNAN VKLHTG LRAIPT TKATITS IKAIÏS LKAI'{SK INATIAT IKCGA VKGKA SKGAT AKGOD 11 RGEY R R R K R H I L H H L L K T K R H H H t H D E E K s K K A s s A A A A A N 0 R N o D D o ï T R A A D A E A TSLS FEVA HNIG LD I T VDLS IDIS ODIS IDLS TRSO P0sc TOSA TREE THEQ YEGV YEGV YEAV YEAI HOAV AK TR ÏR TR AR RE KG KG KN KR R l,l KK MT xlôle o ololt ott
olEA G DHOQ TDDE GETE DDTO NEEO I t K H R K R K E D D E G slLl.tN sll.tll pTvltN TVVMS KILLIILHVIJCIEHGTSPIIING L I.IVIJ C I E N G T S P II I }I G LMVIJCIEII6TSPNI}IG v rJ c
t
E |l GlTls P Nll xlG v !r cI
E lt GlTls P Nll NIG v rJ cI
E N CITIS P Nlv Nlc v u cr
E N Glcls P Nll slG v rJ ct
E H GITIS P NIL Nlc SPV. G SP F HV. RC sP Fl'rv - c SPLV- T PPV PVY scr,rv TEV SPV- G SP FItV. RC SPFMV. C SPLV. T PPV PVY SCMV ÏEV SPV. G SPFMV. RC SP Ft',lv - C SPLV. T PPV PVY SCMV TEV SPV. G SP FI,IV. RC SP FI.IV. C SPLV. T PPV PVY SCMV TEV VSYPI VEHPI VÊYPL TVFPL VSYPL LIOF LLDH IVEN VIEN MVEN LV Lll LH LH Flt lll I}l VN IS LH DT ET NE Uts ED T T T L T ,[ùlu vlulr ulul' olulv,lr
lu ululurlult
'[]'
 K K c o F F L F L o R o o o K tl H M K tl Y H I T T E E K T E Y K S R KH YIA CVE YIE YIE YIE OP KA EP ER RP LO LI LO LQ LT LT LR LT IT lts YS MG YS MS r lnl LIAI'[!
LA LA LA LA LS XVS }IVG GIS NVG NVG VAR I.IAH I',I H H MT H TEEN OKOD AEEN OEEN AEED DRN I NRNI.{ SPSH SRNI.{ NRHI.I RGI H VT HT VT I't N LT N T T R ï R R K A A I A R K K K R R K R o K H H o o N G H N P T T AT DG ÏE AG AH tt TT TL cl TL VRGV VKNI'I VOOHH VROFig. 4. Alignment of the deduced amino acid sequence of the coat protein ol SPV-G with
those of SPFMV-RC, -C, SPLV-T, and other potyviruses. Dashes indicate the gaps for
optimum alignment. Only identical residues of SPV-G and of both strains of SPFMV and those which are identical in the five other potyviruses are boxed
Comparison of the SPV-G and
SPFMV (-RC
and-C)
3' non-coding sequences
The 3'
non-codingportion of
the SPV-G
genome hadthe
samelength
(222nucleotides) and sharedT3%identity
with
those ofSPFMV-RC
and -C(Fig.5)'
The sequence
identity
between SPV-G and other potyvirusin
this region wasless
than
55l".I.IMD GD E OVT Y P I KP L LD HAVP T
I.{MD GD E O VT Y P I K P L L D H AVP T
I.ID GD E OV T Y P I K P L L D H AV P T
r,r n o olo
tlolv
s Y P llK PILI
0 FlAlÂlP TN u o cle rlolv E H P I IK PIL L D HlAlKlP T
u x o
cll
e lolv E Y P LIK Pll v E NlAlKlP Trq u o clo
rlolr
v F P tlK PlvI
E NIAlslP Tr{ H D CIE DlolV S Y P LIK P
FROIMTHFSD FROIl'ITHFSD FROII'ITHFSD RIOIIIt.I A HIF S D
rlol'lt,
',l*
o K A Y t.{ P R Y G L O R N L T D I'I S KAYI'IPRYGLORNLTDHS KAYMPRYGTARNLTD}'IS Y l,{ P R v clt- olnxlt
TIDY I.I P R Y GII OIR NIL TID
Y tl P R Y GIL I IR NIL RID
Y M P R Y GIL olR NIL TID
Y r.t P R Y GIL olR Hll TID
RYAFDFYETHS RYAFDFYELHS RYATDFYELHS R Y A F D F Y EIV TIS R Y A F D F Y EIH TIS R Y A F D F Y EIV TIS R Y A F D F Y EII'I NIS R Y A F D F Y EIL TIS EAHIlOMKAAA
r
nnlo
H K A A A EAHMOI{KAAA E AIY rla M K A A A LKNA LKNA LKNA LTNT LRNV LKSA VRGS VRNS E AIH I IO M K A A A ealr
rlo l.r K A A A E AJH IqJO M K A A A E AIH MIO M K A A A NRLFGLDG NRLFGLDG NRLFGLDG RLFGLDG RLFGLDG RLFGLDG RLFGLDG RLFGLDGNVSTOEEDT ERHT TTDVTRN I HNL LGI'IRGVO ItVS T O E ED T E R H T T TD VT RN I H}I L L GI{R GVO
N V S T O E E D T E R H T T T D V T R II I +I N L L G I'I R G VO T E R H TIA TID V
T E R H TID GID V
rlo e e
xlr
e n nrlr
elo vls p s HIHIT LIL Grlo
r r
xlr
r
n H TIA clD vls R N r,{lHlS LIL GSPV.G TAÂÂCAATATATTGGClCGTACCTT-...T]AATTTCAGTTTC...'6CCTTCAGTGTAATTTAÂATÏCGTATClTTCAGT
SPFI.IV.RC GGAC CCTC CAGT GA ATAC G ATC A6TAT ' G
SPFI{V.C GGAC CCTC CAGT G ATAC T6 ATC AGTAT ' G
SPV. G CCCGAAGAÀCCT 6GT T T GGT GCA6AGCT TAAT GA6GT GT TACCT C TAT C T T T GCAT T GGAGAA6GGAT CÏ T Ï CTAT TACGT
SPFI.IV.RC .GA GA TAAGG GATA T GGTA T TCGC
SPFI.,IV.C .GA GAA lAAGG GATA T GGTA T TCGC
SPV.G ATCATAAGGGACTCTTAA-AAGTGAGG..TTTTACCTCGTAAGAAAAGCCTTTTTTG6TTCGTGATCGAGCC POtY (A)
sPFr.rv-Rc - --AG PotY (A)
sPFMv-c - - -AG PolY (A)
Fig. 5. Alignment of the nucleotide sequence ol the 3' untranslated region ol the SPV-G
genome with those oISPFMV-RC and -C. Dashes indicate the gaps lor optimum alignment
Discussion
The deduced coat protein amino acid sequence ol the newly identified potyvirus inlecting sweetpotato
lrom
China
(SPV-G) rvas significantly longerthan
thatof otheidescribed potyviruses (Fig. a). Moreover, coat protein sequence identity
between
SPV-G and other
potyviruses wasin
the
rangeoi
distinct
viruses.However, sequence
identity
above
80/"
in
the
conservedcoat
protein
coreregion revealèd relatively close relationships
[1,8]
and allowed the establish-ment of subgroupingswithin
the aphid-transmitted potyvirusesll3, l'lf
. indeed,il
theN-terminal
divergent sequences are omitted, the overallidentity
betweenthe C-terminal three quarters
ol
the coatprotein
ol
SPV-G i5 morethan
80/"with both
strains
of
SPFMV,
but
lessthan
70\
with
SPLV-T and
other potyviruses,thus
suggestingthat
SPV-G is
more closely relatedto
SPFMV
than
to
other
potyviruses.Thus SPV-G and
SPFMV form
a
closely related subgroup similar to those previously reportedlor
the bean yellow mosaic virussubgroup
[16,
18] and the bean common mosaic virus subgroup[10,
ll,
16].For comparison, the two strains
oISPFMV
shared 84l" identity ior the completecoat protein and
90.5%identity
in
the C-terminal
three quartersol
the
coatprotein at
theamino
acid level[1].
The relationship between SPV-G and both strains of
SPFMV
was confirmedby the
73ii
sequenceidentity
observedin
the
3' non-coding region,which
isUltow the
â37;to
99\
identity found
for
accepted strainsof
viruses[7],
butabove the
maximum identity
ol
50% generally givenfor
distinct
potyviruses.Such intermediate
level
ol
sequenceidentity
is
sometimes observedwithin
subgroups such as the bean yellow mosaic virus subgroup [16, 18].
In
contrast, the 3' non-coding regions ofSPFMV
strains RC and C were98/"
homologoustll
In
conclusion, coat protein and 3' non-coding region Sequence data indicatethat
SPV-G is closely relatedto
SPFMV,
thoughit
should be considered as adistinct potyvirus.
Indeed,the
lengthol
the SPV-G coat protein
(355 aminoacids) is longer
than
that of SPFMV
(316amino
acids). Moreover, the levelsboth
strainsof SPFMV
are much higherthan
betweenSPFMV
and SPV-G.It
remainsto
be determined whether the sequence identities described above between SPV-G andSPFMV
arelinked
to
similaritiesin
termsof
biologicalor
serological properties.Acknowledgements
We thank Dr. Semal for helplul discussions and critical reading ol this manuscript and Dr. Burny in whose laboratory part of this rvork was carried out' This work rvas financially supported by the EEC (Project STD3 n'TS3-CT91-0013).
l.
References
Abad JA, Conkling NfA, Moyer JW (1992) Comparison of the capsid protein cistron from serologically àistinct strains ol sweetpotato feathery mottle virus (SPFMV) Arch
Virol 126: 141-151
Atreya PL, Atreya CD, Pirone TP (199 1)Amino acids substitutions in the coat protein
results in loss of transmissibility ola plant virus. Proc Natl Acad Sci USA 88: 7887 7891
Bryan GT, Gardner RC, Forster RLS (1992) Nucleotide sequence of the coat protein gene
oi
a strain of clover yellorv vein virus from New Zealand. conservation of astem-loop structure in the 3' region of potyvirLrses. Arch
virol
124: 133-146chirgwin J. Przybyia A, MacDonalcl R, Rutter
w
(1979) Isolation ol biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry l8: 5294 5299 Colinet D, Kummert J (1993) Identificationol
a sweetpotato leathery mottle virus isolate from China (SPFMV-CH) by the polymerase chain reation with degenerateprimers. J Virol Methods 45 149-159
bolinet D, Kummert J, Lepoivre P, Semal J (1993) Identification ol distinct potyviruses rn mixedly-infected sweetpotato by the polymerase chain reaction with degenerate
primers. Phytopathology 84: 65 69
Èrenkel MJ, Ward
cw,
snutta
DD
(1989) The useol
3'non-coding nucleotidesequences in the taxonomy of potyviruses: application to watermelon mosaic virus 2
and soybean mosacid virus-N. J Gen Virol 10 2115 2783
FTCNKCI MJ, JiIKA JM, MCKETN NM, StTiKC PM, CIATK JM JT, ShUKIA DD, WATd CW
(199 1) Unexpected diversity in the amino-terminal ends of the coat protein of strains of
7.
sugarcane mosaic virus. J Gen Virol 12:231 -242
Frohman MA (1990)RACE: Rapid amplification olcDNA DH, Sninsky JJ, White TJ (eds) PCR Protocols: a guide Academic Press, San Diego, PP 28 38
ends. In: Innis MA, Gelland
to method and aPPlications.
4.
6.
I l.
t2.
10. McKern
NM,
Edskes HK, Ward CW, Strike Piv{, Barnett OW, Shukla Coat protein of potyviruses. 7. Amino acid sequence of peanut stripe virus.l19:25-35
McKern NM, Mink GA, Barnett OW, Mishra A, Whittaker LA, Silbernagel MJ, Ward CW, Shukla DD (1992) Isolates of bean common mosaic virus comprising two distinct potyviruses. Phytopathology 82: 355 361
Moy.. JW, Salazar, LF (1t89) Virus and virus-like diseases olsweetpotato. Plant Dis 73: 451-455
Rybicki E, Shukla DD (1992) Coat protein phylogeny and systematics of potyviruses' In: Barnett oW (ed) Poryvirus taxonomy. Springer, wien New York, pp 139- 170 (Arch Virol [Suppl] 5)
DD
(1991)Arch Virol
14. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual'
Cold Spring Harbor Laboratory Press, New York
15. Shukla DD-, Ward
cw
(1989) Identification and classification of potyviruses on the basis of coat protein sequence data and serology. Archvirol
106: 171-20016. Uyeda
I
(1gg2) Bean yellow mosaic virus subgroup; searchfor
the group specific,.qu.n..,
in
the 3' terminal regionol
the genome. In: Barnettow
(ed) Potyvirus taxonomy. Springer, Wien New York' pp 371-385 (Arch Virol lSuppl] 5)17, ward
cw,
strutta DD (1991)Taxonomy of potyviruses: current problems and somesolutions. Intervirology 32: 269-296
18. Ward CW, McKern
ffu,
Frenkel MJ, Shukla DD (1992)Sequence data as the major criterion lor potyvirus classification. In: Barnett OW (ed) Potyvirus taxonomy' Springer' Wien New York, pp 283-291 (Arch Virol [Suppl] 5)Authors' address:
D.
Colinet, Faculté des Sciences Agronomiques' Laboratoire dePathologie Végétale, 13, Avenue Marechal Juin, 8-5030 Gembioux, Belgium. Received MaY 19' 1994