Streptomyces albus G serine f-lactamase
Probing of the catalytic
mechanism via
molecular
modelling of
mutant
enzymesJosette LAMOTTE-BRASSEUR, Francoise
JACOB-DUBUISSON,
GeorgesDIVE, Jean-Marie FRERE and Jean-Marie GHUYSENCentred'Ingenierie des Proteines, Universite deLiege, Institut deChimie, B6, B-4000 Sart tilman (Liege 1), Belgium
Inprevious studies, several amino acids of the active site of class A,6-lactamases have been modified by site-directed mutagenesis. On the basis ofthe catalytic mechanism proposed for the Streptomyces albus G ,J-lactamase [Lamotte-Brasseur,Dive, Dideberg,Charlier,Frere &Ghuysen(1991)Biochem. J.279,213-221], the influence that these mutations exert on the hydrogen-bonding network of the active site has been analysed by molecular mechanics. The results
satisfactorily explain the effects of the mutations on the kinetic parameters of the enzyme's activity towards a set of substrates. The present study also shows that, upon binding a properly structured ,J-lactam compound, theimpaired
cavity ofa mutantenzymecanreadopta functionalhydrogen-bonding-network configuration.
INTRODUCTION
Theserine ,-lactamases of classes A, C andDhydrolyse the ,-lactamantibiotics byanacylation-deacylation mechanism.
Sev-eral fl-lactamases of class A have been studied byX-ray crys-tallography (Dideberg et al., 1987; Herzberg & Moult, 1987;
Moews et al., 1990; Herzberg, 1991). They have almost com-pletelysuperimposable three-dimensionalstructures.Their active
siteislocatedatthejunction betweenanall-adomain andan
a/,/
domain which consists ofafive-stranded f-sheetwithadditional helices on bothfaces (Ghuysen, 1991).
Thestructureof theclassAStreptomycesalbus G ,J-lactamase
has been resolved to 0.17nm (1.7 A) and optimized byenergy minimization (Dideberg et al., 1987; Lamotte-Brasseur et al., 1991). The active site is a dense hydrogen-bonding network in
which nineamino acids, namely S70, K73, S130, N132, E166, N170, K234, T235 andA237, andtwowatermolecules, WI and
W2, are involved. The S70VFK73 motif, where S70 is the
nucleophilic serine residue, ona2 ofthe all-adomain, occupies a centralposition inthe cavity;the E166PELN170 motif, on a loop betweena6 and a8 of theall-a domain, isattheentranceof
thecavity;theS130DN1 32 motif, between a4 anda5of the all-adomain, lieson one side ofthecavity; the K234T235GA237 motif,ontheinnermost /33strand of thea/fldomain, forms the
other side.
On the basis of these structural data, molecular-mechanics
studies have allowed us to propose a mechanism of proton
abstraction-donation during hydrolysis ofbenzylpenicillin by
theStreptomyces albusG /3-lactamase (Lamotte-Brasseur etal., 1991). The same procedure has now been used to study the changesintheenzyme-ligand interactions resulting from changes
inaminoacidstowhicharolehas beenassignedincatalysis. As
shown below, the rearrangements undergone by the active-site hydrogen-bonding networkarecompatible with theeffects that
these mutations haveon thesecond order-rate constantfor the
acylation of the essential serine residue, characterized by the kcat./Km ratio.
MATERIALS AND METHODS
The active site of the Streptomyces albus G /,-lactamase,
defined by all the amino acids and water molecules present
0
withina 1.5nm(15 A)radius aroundS7OCa,was optimized by energy minimization (Lamotte-Brasseur et al., 1991) using the
Ambermolecular-mechanicsV3framework (Weineretal., 1984).
As an initial guess for optimization, the torsion angles ofthe
wild-typeactive sitewereretainedasmuchaspossibletosimulate theamino acid changes in themutantenzymes. Minimization of
thestructureswascarriedoutuntiltheroot-mean-squaregradient
was <0.0419kJ/nm (0.1 kcal/A). Water moleculeswere
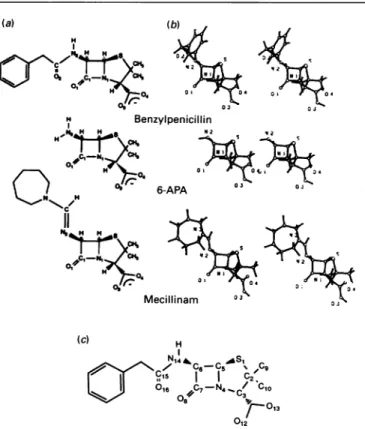
gener-(a) (b) 32 342 0 4 Mecillinam (c) H 14Ak as, <\ I' C6-C5 C,Cg I I 0p°16 3tC, N4-C/ CIO 012
Fig.1.(a) Arbitrary numbering of benzylpenicilHin used in the present study, (b) optimized structures of benzylpenicilin, 6-APA and
mecillinam, and(c)standard chemicalnumberingofpenicillins
Abbreviation used:6-APA,6-aminopenicillanate.
J-J.
Lamotte-Brasseur
and others ated by a Monte Carlo bath (using the Amber procedure) andtheir co-ordinates were minimized together with the
enzyme-mutant co-ordinates.
The structures of benzylpenicillin, 6-aminopenicillanate (6-APA) and mecillinam,usedasligands,wereoptimized (Fig. 1)by the AMI quantum-chemistry method(Dewaretal., 1985). Each optimized moleculewasdocked in the cavity ofthewild-typeand
mutant fi-lactamases. The ,-lactam carbonyl carbon C-i was placed closeto the O-yof S70 and theoxygenatoms 0-3 and 0-4 of the carboxylate were oriented towards the bottom of the
cavity,i.e. K234. Thestructuresof theMichaeliscomplexes and, in one case, ofthe tetrahedral intermediate leading to enzyme acylation, were optimizedasdescribed for the wild-typeenzyme, using CNDO charges on the ligand. The bond lengths, bond angles and ring dihedral angles of the
fl-lactam
ligands were constrained tothe AMI values.RESULTS
Theatom numbering of the/,-lactam ligands is arbitrary (see
Fig.1). Theamino acids numbering of the wild-type Streptomyces
/I-lactamaseand theenzymemutantsis that described by Ambler
et al. (1991).
Selected
fl-lactamase
mutantsThe goal of the present study was to seek an answer to
questions regarding the relationships between the alterations in the activity of the Streptomyces albus G
fi-lactamase
caused bysingle amino acid changes and the modifications that these
aminoacid changesinduce in the configuration of the
hydrogen-bondingnetwork of the enzymecavity.
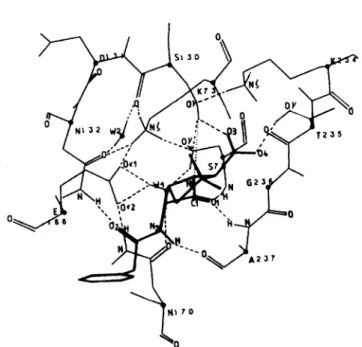
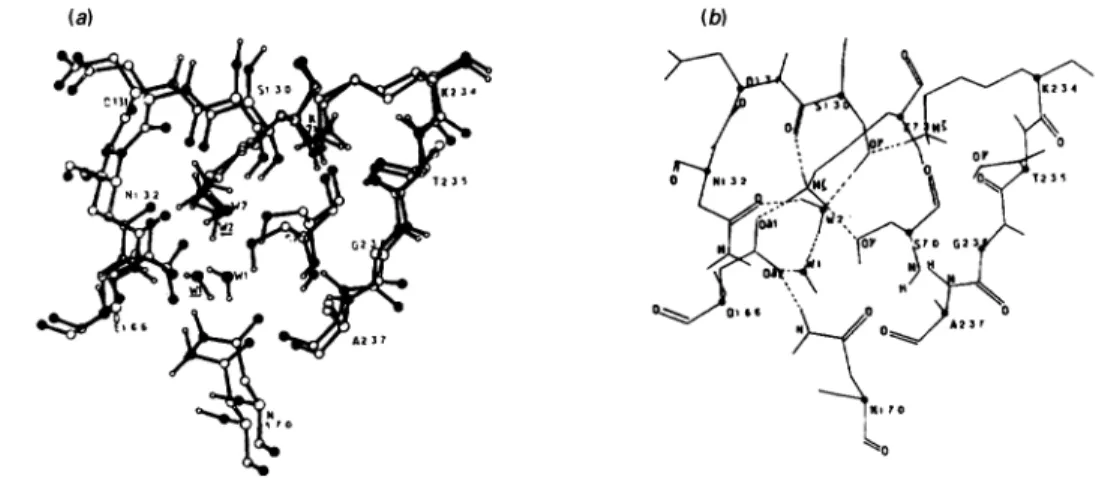
Benzylpenicillin binds to the active site of the wild-type /?-lactamase (Fig. 2) via formation ofhydrogen bonds between:
0-3 of penicillin and they-OHofSI30; 0-4ofpenicillin and the y-OHof T235;0- Iofpenicillin and the backbone NHgroupsof S70 and A237; N2-H ofpenicillin and the backbone carbonyl
group of A237; and 0-2 of penicillin and the side-chain NH2
Fig. 2. Diagramof thehydrogen-bondingnetworkof theMichaeliscomplex
formed between benzylpenicillinand theactive siteof the wild-type
S. albus G 8-lactamase
group of N132. Formation of the Michaelis complex has no or little effect on the pre-existing hydrogen-bonding network of the ligand-free active site, except that the hydrogen atom of
SI
3OyOHno longer interacts with W2, but is oriented towards N-1 of penicillin [as shown in Fig. 2; 0.311 nm (3.11 A);
168.5°1.
TheMichaelis
complex has other remarkable features. The oxygen atom ofS700yOH
is 0.28 nm (2.8 A) from C-I of penicillin; the angleC/i-Oy (of S70)... C-I (of penicillin) is 1080; the hydrogen atom of S7OyOH is in interaction with OeiE166 via WI (the triadS700yH-.WIH-..0eIE166)
and, more loosely, withOe2E1
66. Hence the conditions are fulfilled to allow: proton abstraction of theS7OyOH by OelE166 via Wl; attack ofC-I of penicillin byO-y
of S70; polarization of C-1=0-1 of penicillin located in the oxyanion hole formed by the backbone NHgroups ofS70
and A237; and proton back-delivery toN-I of penicillin by the W2,NCK73,
NMK234
and Sl3OyOH hydrogen-bonding subnetworks, thus achieving formation of theserine-ester-linkedpenicilloyl-enzyme.
From the foregoing it follows that the El 66D mutationaffects the presumed proton abstractor, whereas the
S130G,
S130A, N132S and K73R mutations affect, one way or another, the presumed machinery of proton transfer from E166 to S130. The hydrogen-bonding rearrangements induced by these mutations (in the ligand-free active sites and Michaelis complexes) are expressed in terms ofX... H bond distances and YH...X bond-angle values (X and Y are heteroatoms) in Tables 1 and 2. These rearrangements, especially the fate ofW1 and W2, are briefly described in the ensuing sections. They are illustrated in Figs. 3-7. A 0.25 nm (2.5 A) distance is regarded as the upper limit for a hydrogen bond.The effects caused by the selected mutations on the enzyme
activity are expressed in terms of
kcat
/Km values (Table 3). It is assumed that the K73R andE166D
mutations have effects onthe Streptomyces,/-lactamase comparable with those for the highlyhomologous
,-lactamase
I of Bacillus cereus 569/H (Gibson et al., 1990), whose assumed catalytic residues areconserved,except for T235, which is replaced by S235. Thekcat./Km
values (equivalent to the second-order rate constant of enzyme acyl-ation) show that the S130A and K73R mutations and, to a still greater extent, the El 66D mutation, are profoundlydetrimental,irrespective of the /J-lactam ligand used (benzylpenicillin or 6-APA). The Si
30G
and Ni 32S mutations decrease selectively the enzyme activity towards 6-APA (to various extents, the S130Gmutation being more deleterious than the Ni 32Smutation), but do not affect significantly the activity towards benzylpenicillin.
Wild-type f-lactamase
Unlike benzylpenicillin, 6-APA and mecillinam have no exo-cyclic amide bond (Fig. 1). Thereby binding of 6-APA and
mecillinam to the wild-type /-lactamase active site generates Michaelis complexes that lack the bond NI32N8H. 0-2(in the case of 6-APA) and bonds N132N8H ...O0-2 and N2-H-
--O=CA237
(in the case of mecillinam). Apart from the fact that these bonds are absent, the hydrogen-bonding networks have features comparable with those found in theMichaelis complexformed with benzylpenicillin (Fig. 2; Table 1, rows 1-4). Con-sistently, benzylpenicillin, 6-APA and mecillinam have almost identical substrate activity
(kcat./Km
3000mM-.S130G
andS130A enzyme mutantsIn the wild-type enzyme, the S130C=O interacts viaW2 with the side-chain carbonyl groupofN 132(Fig. 2), and the S30OyOH
is thought to be involved in proton back-donation. Hence the
S130G
and S130A mutations are expected to eliminate the-'ITN rl CIA CIA N 00 cn N- "C tr
NC C N- N- 00 N NC N- N N 'IC N- 00 NC NIC ON O
NC N N N~~~~'C t) ~C 'IC NIC NIC NC C N- NC N- N'IC 'IC
ON IC C C C V)Cl Cl N
-r
=-CCl-C N- m N- N- ON - N N NIC NIC 'IC
N-M 00) 0 00 - ON 00 NIC 00 N oNl 00
r, N1)NC X c)"Cc)-N 'I.- ~'I .>'I N N N
'IC 'IC 'IC N0 NCIC NIC NIC 'IC NIC 'INC
Cl ) 'IC " t 00 'IC n NIC N- 00 t ON n
00 ON, ON ON ON 00 ON 00 ON ON 0 -0NON - 00 - N~-- ~ 0 00NCl- C O N ON C r C N '
*o
NC rcNOrl~0
00 -0Cl0N0~~~~~~t~--
O00e 0000 00O .-- 00 OO Cl 4 - Cl Cl Cl~~~~'I 00 - 00 -I - 0 -*.*Cl*C> l 0 0 ,I -CC " r * *,, lc0~~~~~~~~~~~~~~~~~~~~o
C~~~00C~~~~NC~~~~0 ~ ~~Cl~~~t~~~Nr~~~ClC rON NC 'ItN,CWI~ W) W ClO 0 00 0r~~~9C. ONI I I Cl00Cl~~ClCl -~~ ClON c~~~~~~Cl- Cl O~~~~~~~~~~~~~~~~~N Cl -00 ,I ON 00 0Cl Cl nON O ON 0= NC - ) "lC 00 Cl Cl -"C 0 -1 - 'I Cl ON - C 0 NCC0 enC 0 N O Cl N cn C ) NC -:)Cl-ON0l0~~N NerN I 0O- -00 0rf NC 0 - 0 ON~~~~~~~t- 0IW 00 - - - N - - 00en Cl cn ON 00 - -C -- o-
*c
r O )r ON "C 00 Cl N N ON 00 W' N- ON N l C Cle~~~~ - ~~~ ON ~~N NC C 00 en rn Cl N- "OC - N 00 ON~~*0ON00~~~r00NC 00. 000000k0 00 00 00N-00 tN.-0000O 0 ONON 0 0~~~O Cl ON f ON NC 00 - C C N 00 ON 00 C, NllNC C> N NC, 0- "C N 0- N NCC NC C 00 NC el c rON N 0 N - -ClCO~ 00 "CNC 00 r- 00 N"C 0 0 N Cl O enC N - ON,Cl 00C NC>NC ON 0 0IT- NCM ND NC N0 e 0 00 0 -" 00> 0 O r-00 00)~N -0 e WN)N 0 N NCN 00 ~~~~~ClON ON N N- MC ON Cl~ 0 0N NC) NC N C.N C.NC 00~~~~~~~c -C. n 00 r "C NC 0 0- C4 0Cl C. NC "C NC " H+ + + + + + + M + ± + ~Z+ NC+ -Mlr .Z tn NC N 00 ON 0 -M "T NC 00Molecular modelling of class A f-lactamase mutants 191
eo en NuNC 0 0 0 0 . QV * 00
.
0
X l ** N 7 C) z 'It 0 cn 1-) ~)0 C .0 ~c C C 1-6 III C) Ce
c) -O C m 0 0 2 N = 2 5 'U 0 0R
0 'U 'U 01 'U C 0 01 0 'U ": .5 I- -0*. o "o 'U'U 0 'U A C = .-. 'UN . :5 'U0 01 01 'U Ca z CI .l - -00 )0_q _ z °) NC)0
NC N -"lo NC NC xo C eo C 0 0 N 00CAO
= r-NLL=0 = Z 3 0J. Lamotte-Brasseurand others
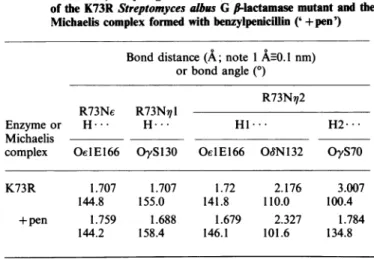
Table 2. X...H distances and YH.... X bond angles (X and Y are
heterotoms)ofhydrogenbonds around R73withintheactivesite of the K73RStreptomyces albus G Ii-lactamasemutantand the Michaeliscomplex formed withbenzylpenicillin ('+pen')
Bond distance(A; note 1AnO.Inm) orbondangle(0)
R73N,12
R73Ne R73N 1
Enzyme or H... H... H1 H2...
Michaelis
complex Oe1E166 OyS130 Oe1E166 OWN132 OyS70
K73R 1.707 1.707 1.72 2.176 3.007
144.8 155.0 141.8 110.0 100.4
+ pen 1.759 1.688 1.679 2.327 1.784
144.2 158.4 146.1 101.6 134.8
Table 3.Catalytic efficiency(kcatl/Km)of theS.albusG and B.cereus
wild-type/I-lactamasesandfi-lactamasemutants
The catalytic-centre activities of the Streptomyces and Bacillus wild-typefl-lactamasesareidenticalwith respecttobenzylpenicillin
(kcat 2800s-1 as against 2200 s-1) and of the same order of magnitudewithrespectto6-APA(720 s-'asagainst260s-'). The
differences in thekcat/Km valuesaremainlyduetodifferences in the
Kmvalues. Results for S.albusG arefromJacobetal. (1990a,b);
those forB.cereus arefromGibsonetal.(1990).
kcat./Km
(mM1Enzyme Substrate... Benzylpenicillin 6-APA Mecillinam
S. albusG Wild-type 2800 3700 2600 S13OG 480 4 S130A 70 3 N132S 1500 100 2500 B. cereus Wild-type 34000 170 K73R 550 0.4 E166D 77 2.2
proton donor and to perturb the W2 hydrogen-bonding sub-network.
Molecularmodelling ofthe S130G mutant enzyme (Figs. 3a and3b;Table 1,row5)shows that W2stillcloselyinteracts with
ON1 32 and forms a W2H...O=CG1 30 bond (similar to the
W2H...O=CS130 bond in the wild-type enzyme), but W2 is
displaced 0.29nm(2.9 A) towards theG130DN132 motif. Re-placement of S130 by G also affects the WI subnetwork.
Thoughstillclosely interactingwithO01 E166,WI isdisconnected from Oe2E 166 andO0N170aswellasfromS700y,thusdamaging
the S700yH-.WlH-*OelE166 proton-abstractor triad.
Molecular modelling of the S130A mutant enzyme (Figs.
4a and 4b; Table 1, row 8) shows that the triad S700yH-+
WlH-.c0E166 remains as strongly hydrogen-bonded
as it is in the wild-type enzyme but, as a result of the steric effect of the Al30 methyl group, W2 is displaced 0.14nm (1.4 A)sothat itforms,with N132 andWI,astrongly
hydrogen-bonded N13280. HW2H WI triad.
Docking6-APAinthecavityof bothmutantenzymesleadsto
optimized structures in which theconfiguration of the
proton-abstractor machinery (Table 1, rows 7 and 10) remains
in-compatible with any significant catalytic activity.
By contrast, dockingbenzylpenicillin in the active site of the S13OG mutant enzyme shows that, in the Michaelis complex, a strongly hydrogen-bonded S70OyH-+WlH-.Oe1E166 triad is reformed (Fig. 3c; Table 1, row 6). Moreover, in the tetrahedral intermediate, W2 undergoes such a displacement that it is located
atapproximately the same position as that occupied bySI3OyOH
in the wild-type enzyme and has one of its hydrogen atoms oriented towards N-I of penicillin (Fig. 3d). These induced configuration changes are probably able to restore a functional proton-abstraction-donation network, though its configuration
differs, atleast in part, from that of the wild-type
fl-lactamase.
With reference to the wild-type enzyme, the S13OG mutant enzyme hydrolyses benzylpenicillin effectively, with a 6-fold-decreasedkcat./Km
value.Conversely, the presence of the additional methyl group in the S130A mutant results in a less efficient positioning of the W2 molecule, explaining an additional decrease of the rate of acylation by benzylpenicillin.
N132S mutant enzyme
Changing N132 is another way to perturb the S130C=O... HW2H...
ONN132
triad and, asa consequence ofthis,the W2 andWI hydrogen-bonding subnetworks. Modelling
of the N132Smutantenzyme(Figs.5aand 5b; Table 1, row11) shows that W2 is ininteraction with O=CS130
(as
in thewild-typeenzyme),butnotwith
OyS132,
andthat WI is ininteractionwith Oe1E166 (asinthewild-typeenzyme),butnotwith
S700y.
Dockingbenzylpenicillin (Fig. Sc; Table 1, row
12)
andmecil-linam(Fig. 5d;Table 1,row14)in themutantenzyme generates
Michaelis complexes whose WI and W2
hydrogen-bonding
subnetworksre-adoptaconfiguration similar tothat formedin the benzylpenicillin-, or mecillinam-,wild-type-enzyme Michaelis
complex. 6-APA (Table 1, row 13), however, fails to restore a
functionalconfiguration. These observationsareconsistent with the data of Table 3.
K73R mutantenzyme
Inthe wild type
fl-lactamase,
K73NC iscentraltotheW2-W1hydrogen-bonding subnetworks throughthebonds that it forms withO=CS130,O0N132, OeE166and
OyS70
(Fig. 2; Table 1,row 1). Replacement of K73 by R73 perturbs the original configuration (Fig.6aand6b;Table 1,row 15,and Table2). In
thenewly formedconfiguration,W2still interacts with O=CS130,
but it is connected to
S13OyOH,
not to ON132. WI still interacts withS7OyOH,
but it is linked much more closely toOe2E166
thantoOeE 166.Uponbenzylpenicillin binding
(Table
1, row 16, and Table 2), the N13280. HW2H - O=CS 130triad is regenerated (asit occurs in the wild-type
,1-lactamase),
but WI remains in close interaction with 0e2E166, thus
pre-ventingrestoration of the 'normal'
S700yH-.WIH-OelEl66
proton-abstractor triad.Consistently the K73Rmutantenzyme
has avery weak activity towards benzylpenicillin.
E166D mutantenzyme
Shortening the side chain of the carboxylic acid at position 166, i.e. the proton abstractor, has drastic effects (Figs. 7a and 7b; Table 1, row 17). WI isdisconnected from
S7OyOH
and is hydrogen-bondedto082DI66, notOdID166;
W2 stillinteracts withON132,
but it is disconnected fromOySl30
and forms withWIahydrogen-bondedW2-H...WIdyad. Benzylpenicillinbinding
fails to regenerate aconfiguration
compatible
with proton abstraction-donation (Table 1, row 18). The E166D(a) (b)
(c)
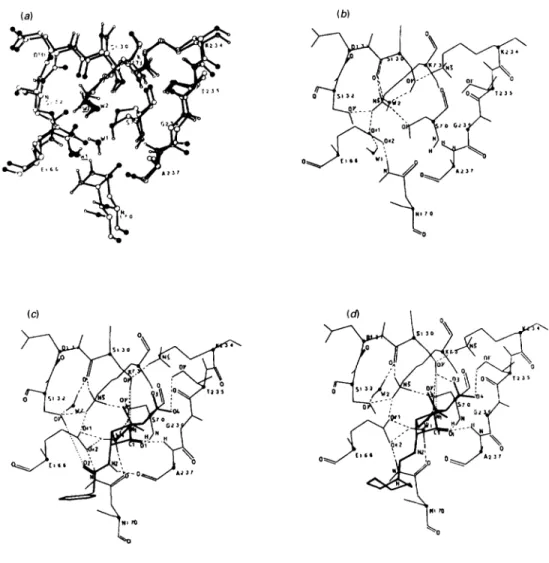
Fig.3.S. albusG/I-lactamaseS13OGmutant
(a)Superimposition ofthe cavity of the mutant(black polypeptide backbone) and the active site of the wild-type enzyme (white polypeptide
backbone). Key toatoms: 0, oxygen;
(,
nitrogen atoms. Theunderlined WI and W2 belong to the mutant. (b) Diagram of thehydrogen-bonding network of the ligand-freecavity of the mutant. (c) Michaelis complex formed with benzylpenicillin. (d) Tetrahedral intermediate leading topenicilloylation ofS70.
(a) (b)
Fig. 4. S. albus Gfl-lactamaseS130Amutant
(a)Superimposition ofthecavityofthemutant(blackpolypeptide backbone) and the active site of thewild-typeenzyme(whitepolypeptide backbone).Keyto atoms: 0,oxygen; O,nitrogen. The underlinedWI and W2belongto the mutant.(b) Diagram of the hydrogen-bonding
J. Lamotte-Brasseurand others
(a) (b)
(c)
Fig. 5. S. albus Gf,-lactamase N132Smutant
(a)Superimposition of the cavity of the mutant (black polypeptide backbone) and the active site of the wild-type enzyme (white polypeptide
backbone)Key to atoms: 0, oxygen; @, nitrogen. The underlined W1 and W2 belong to the mutant. (b)Diagram of the hydrogen-bonding network of the ligand-free cavity of the mutant. (c) Michaelis complex formed with benzylpenicillin. (d) Michaelis complex formed with
mecillinam.
(a) (b)
Fig.6.S.albus-Gfi-lactamaseK73Rmutant
(a)Superimposition of thecavityof the mutant (blackpolypeptide backbone) and the active site of thewild-type enzyme(whitepolypeptide backbone). Keytoatoms: 0,oxygen; O,nitrogen. The underlined WI and W2belongtothemutant. (b)Diagramof thehydrogen-bonding
networkof theligand-freecavityof themutant.
(a) (b)
Fig. 7. S. albus GII-lactamaseE166D mutant
(a) Superimposition of the cavity of the mutant (black polypeptide backbone) and the active site of the wild-type enzyme (white polypeptide backbone). Key to atoms: 0,oxygen; 0, nitrogen. The underlinedWI andW2 belong to the mutant. (b) Diagram of the hydrogen-bonding
network oftheligand-free cavity of the mutant.
mutant enzyme has a 500-fold-decreased kcat /Km value with respect to benzylpenicillin.
DISCUSSION
Ithas beenproposed (Lamotte-Brasseuretal., 1991)thatthree
amino acids, namely S70, E166 and
S130,
and two watermolecules, W1 andW2,arecentral totheStreptomycesalbus G
,/-lactamase-catalysed
hydrolysis ofpenicillin. WI transfers the protonfromS7OyOHtothe abstractorOIE166,allowing
attack of thecarbonylcarbonatomof the/3-lactamringby
S700y.
W2 is an integral component ofthe relay systemthrough
which aproton is delivered back to the nitrogen atom ofthe
fl-lactam
ring, viaS130y0H
(thus achieving rupture of the amide bondand formation of the serine-ester-linkedacyl-enzyme).Totestthe mechanism,theeffectsof mutationsaffectingE166 andS1 30(the
proton abstractor and a residue involved in proton
donation)
andN132 and K73 (involvedin theconfigurationoftheWl and W2 hydrogen bondingsubnetworks)
have been examinedby
optimizingthecomplexesformed
by docking
benzylpenicillin,
6-APAand sometimesmecillinam in themutated-enzyme
cavities. The observedchangesinduced in thehydrogen-bonding
networkby each of themutations studied well
explain,
atleast to afirstapproximation, the effects that these mutations have on the
catalyticactivity of the,J-lactamase.
The present studiesalso showthat,.owingtothehigh density
of the hydrogen-bonding
network,
the enzymecavity
is astructureofhigh plasticityboth
structurally
andmechanistically.
Local modifications thatcausethedisappearance
orweakening
of any hydrogen bond may propagate its effects far from themutated amino acid and
modify
theentireconfiguration
ofthe cavity.Acavity, damaged
by
mutation,
mayregain functionality
upon binding of aproperly
structuredfl-lactam compound,
either byreadopting
ahydrogen-bonding configuration
com-parable with that of thewild-type
enzyme orby
utilizing
analternaterouteof proton shuttle from El66tothe
nitrogen
atomof the
fl-lactam
ring. To all appearances,however,
an intact, stronglyhydrogen-bonded
S70y0H-+WlH-.O01E166
triad isessential tothe
catalytic
mechanism.Adaptation of the
,1-lactamases
to the environmentalcondi-tions is well documented.,?-Lactamasesshow hysteretic kinetics.
A unique conformation is induced in the enzymes by each of severalclosely related ,-lactam substrates, and the enzymes can adjusttounfavourable modifications in the substrate (Samuni & Citri, 1979).
This work was supported, in part, by the Belgian programme on
Interuniversity Poles ofAttraction initiated by the Belgian State, Prime
Minister's Office, Science Policy Programming (PAI n°19), an Action concerteewiththe Belgiangovernment (convention 86/91-90), the Fonds de la Recherche Scientifique Medicale (contract n°3.4537.88), and a Convention tripartite between the Region wallonne, SmithKline
Beecham, U.K., and the University of Liege. G. D. and F. J. are
fellows ofthe Fonds National de la Recherche Scientifique (FNRS,
Brussels).
REFERENCES
Ambler,R. P., Coulson,A. F. N., Frere, J. M., Ghuysen, J. M., Joris, B., Forsman, M., Levesque, R. C., Tiraby, G. & Waley, S. G. (1991) Biochem.J. 276,269-272
Dewar,M. J.S., Zoebisch, E. G., Healy, E. F. & Stewart, J. J. P. (1985) J.Am. Chem. Soc. 107, 3902-3909
Dideberg, O., Charlier, P., Wery, J. P., Dehottay, Ph., Dusart, J.,
Erpicum,Th., Frere, J. M. & Ghuysen, J. M. (1987) Biochem. J. 245,
911-913
Ghuysen,J. M. (1991)Annu. Rev. Microbiol. 45, 37-67
Gibson,R. M.,Christensen, H. & Waley, S. G. (1990) Biochem. J. 272, 613-619
Herzberg, 0.& Moult, J. (1987) Science 236,694-701
Herzberg,0.(1991) J. Mol. Biol. 217, 701-719
Jacob, F., Joris,B., Dideberg, O., Dusart, J., Ghuysen, J. M. &Frere,
J. M.(1990a) ProteinEng. 223, 114-120
Jacob, F., Joris, B., Lepage, S., Dusart, J. & Frere, J. M. (1990b) Biochem. J. 271,399-406
Lamotte-Brasseur,J.,Dive, G.,Dideberg,O.,Charlier, P., Frere,J. M. &Ghuysen,J. M.(1991) Biochem. J.279,213-221
Moews, P.C., Knox, J.R., Dideberg, O.,Charlier, P. & Frere,J. M.
(1990)Proteins7, 156-171
Samuni,A. &Citri,N.(1979)Mol.Pharmacol. 16,250-255
Weiner, S.J., Kollman, P.A., Case, D.A., Singh, U.C., Ghio, C.,
Alagona, G.,Profeta, S.,Jr. &Weiner, P.(1984)J. Am.Chem. Soc.
106,765-784
Vol. 282