Advances
in
the
detection
of
plant
pathogens
by
polymerase
chain
reaction
edited by
Charles Manceau
ktboratory
of plant pathology, INRA, Beaucouzé, Franceand
Josef Spak
Instirute of
plant
molecular biology, Ceske Budejovice, Czech RepublicCOST
823
New technologies to improve phytodiagnosis
Workshop
of
the
nucleic
acid-based
technology
working
group
Ceske
Budejovice,
Czech
Republic
Part one - PCR DETECTIoN oF VIRUSES
USE OF DEGENERATE PRIMERS FOR RT.PCR
DETECTION
OF APPLEAND
PEARTREE VIRUSES
J.
KI'MMERT*,
G. RTIFFLARD, V. DEALMEiDA
MARINHOCentre National de Phytovirologie IRSIA-Unité de Phytopathologie,
Faculté
Universitaire desSciences agronomiques, B-5030 Gembloux, BELGIUM
Kev words : Apple chlorotic leafspot virus (ACLSV), Apple stem grooving virus (ASGV),
Apple stem pitting virus (ASPV), degenerate primers, RT-PCR detection.
ABSTRACT
Apple chlorotic leafspot virus (ACLSV), Apple stem grooving virus (ASGV), Apple stem
pitting virus (ASPV) isolates and complexes containing these viruses have been transferred and
multiplied
on
herbaceous host plantsin
the
greenhousefor
the
developmentof
RT-PCRdetection methods using degenerate primers. Comparative analysis
of
nucleotide and deducedamino acid sequences published
for 2
isolates of ACLSV, one isolate of ASGV and one isolateof
ASPV showed the presenceof
several short stretchesof
homologous amino acids, althoughthe three viruses belong to different phytovirus genera. Only amino acid sequence homologies
corresponding
to
the
translation product
of
the
3'-terminal
part
of
the
putative
RNApolymerase gene are sufficiently conserved and have a sufficient length to allow the design
of
different sets
of
degenerate primers. These primers have been usedfor
amplificationof
viralsequences
from
reverse transcribedtotal RNA
preparationsof
virus-infected
leavesof
Chenopodium quinoa or
Nicotiana
occidentalis.As
the sizesof
the amplified products arequite similar
for
the 3 viruses, the precise identificationof
the responsible virus' would requirethe combination
of
PCR and hybridizationwith
specific probesfrom
clonedcDNA.
ClonedPCR products have been sequenced
to
identify the causal agents and to design more efficientor more specific primer pairs
in
order to allow the delectionof
the various isolatesof
viruses(specially ASGV and ASPV). The RT-PCR protocols thus obtained
will
be adaptedfor
use onwoody material and specially mother trees with the aim to define certification scheme
for
virusfree propagating material.
INTRODUCTION
Apple chlorotic leafspot virus
(ACLSV),
Apple stem grooving virus (ASGV) and Applestem pirting virus (ASPV) are three latent viruses with elongated particles infecting pome
fruit
trees (apple and pear),
ACLSV
isolates also infects stonefruit
sometimes inducing, severesymptoms (pseudo-pox ; bark split).
The detection
of
thesefruit
tree viruses relies on biological indexing by grafting on woodyindicators
in
thefield, or
eventuallyin
the greenhouse. These techniques which consdfute the"baseline" tests for the certification of
fruit
tree planting material are working quite well, but arecumbersome, lengthy and expensive to perform (time, space and manpower required), they are
also not always very specific and may not differentiate mixed infections
with
several relatedviruses.
Part one - PCR pBrecrloN oF vrRUsEs
There seems now to exist a good evidence that some
fruit
tree viruses could be assayed,with
equalreliability
to biological methodsby
laboratory tests that are becoming increasinglysensitive and refined with the accumulation of experience
in
serology and molecular biology.Serological detection by ELISA tests is possible for ACLSV and ASGV with commercially
available antisera,
but
accurateonly
duringa
shorttime
periodin
the
spring;no
availableantiserum exists for ASPV, to day.
On the other hand, the transfer of isolates of these viruses on herbaceous host plants, in the
greenhouse, is possible, rendering them available for multiplication, characterization and use all
year long, although purification
of
virus particles, even from these herbaceous hosts remainsdifficult.
As the genetic organization and thus the classification
of
these viruses was confused andquite ambiguous
till
recently, and the precise identificationof
the reference isolates collectednot
sure,we
aimedat
developing RT-PCR techniquesusing
degenerate primersto
obtainamplification products which would be specific
for
virus-infected plants.The protocols developed can be used
for
direct detectionof
virus infectionby
analysisof
amplified products
in
ethidium bromide stained agarose gels.Specific amplification
products have been clonedfor
further
use,as
sequencingfor
taxonomic identification
of
the isolates and designof
more specific primersor
primers moreadapted
to
copewith
thevariability
of
the concernedviral
agents. The cloned products alsoallow the development
of
non radioactive probes to be usedin
dot blot hybridization testsfor
virus detection.
The present communication is dealing with the development of RT-PCR amplification tests
for
detection of ACLSV, ASGV and ASPVin
herbaceous hosts by using polyvalent degenerateprimers
and
analysingthe
amplification productsby
electrophoresisin
ethidium
bromidestained agarose gels, and of
it
further appraisal for use with woody infected samples from appletrees.
MATERIAL
AND METHODSTransfer
ofthe
virus isolates on herbaceous hosts plantsYoung
leavestaken
on
treesin
orchards,or
developedon
shoots takenduring
thevegetative rest, stored at 4oC and put into growth
by
dippingin
water or nutritive solutions at20-24"C under light, are grinded in 4 parts (w/vol) potassium-sodium phosphate buffer 0.04
M
pH
7.2 containing 0.01M
sodium diethyl carbamate and2
7o nicotine sulphate. The solutionand
the
inoculum are
maintainedon melting ice.
Inoculation
is
madewith
gloves
onCarborundum dusted leaves, and plants are immediately rinsed
with
distilled water. The testsplants used
for
virus multiplication
are Chenopodiumquinoa
andNicotiana
occidentalis,grown
in
the greenhouse with supplementedlight
(16 h).Part one - PCR nnrecrroN oF vrRUsEs
Infected reference trees
Next
to
the collection
of
virus
isolates transferredon
herbaceous hosts,we
disposeof
10 apple-trees prepared by the research stationof
Gorsem plantedin
soilin
Gembioux andinfected
by
1 or more generally by a complex from 2 to 3of
the viruses studied (Table 1). Anextensive collection
of
infected treesis
also available at the research station onfruit
treesof
Gorsem.
Table
I
- Apple trees plantedin
soil in Gembloux36
Identification Viruses determined by biological indexing
9L-325 9r-327
9t-328
73058 10291t0392
1t043
J.C.D. 1390 4.TH.1385 LP68O ASPV ACLSV + SED* ACLSV ASPVACLSV+ASGV+SED
ACLSV + ASGVACLSV+ASGV+SED
ASPV ASPV ASPV**
"Spy epinasty and decline" diseaseRNA
extraction and cDNA synthesisTotal RNA was extracted from 100 mg samples
of
leaf materialof
herbaceous host plantsor
apple trees using the Tripure reagentfrom
Boehringer, accordingto
the
manufacturer'sprotocol.
Single stranded
cDNA
was synthesizedfrom
1to
5
pg of
purified
totalRNA
using theSuperscript preamplification system for
first
strandcDNA
synthesis(Life
Technologies) or theExpandrM reverse transcriptase (Boehringer), with oligo-dT primers. The resulting cDNAs were
then diluted 5-fold
with
sterile water. Five microliters from these preparations were then usedfor
the PCR amplifications.In
some cases, speciallyfor
total RNA
preparationsfrom
applematerial,
or
for
sequencingof
PCR amplified products, moredefined
degenerateor
specificPart one - PCR osrscrloN oF vlRUsEs
PCR experiments
Amplification from the oDNA were carried out with 5
pl of
cDNA preparationsin
a totalvolume
of
50or
100pl of
PCR buffer (10mM
Tris-HClpH
9.0, 50mM Kcl, 0.1
VoTnton
X-i00),
containingMgCl2
(1.5mM),
each dATP, dCTP, dGTP, dTTP (200mM),
0.05 or0.1 nmol
of
each upstream and downstream primers and1 unit
of
Taq
DNA
polymerase(Boehringer). Thermal cycling was realized
in
a triothermoblock cycler (Biometra, Gôttingen)or
a PTC 200 cycler from M.J. Research. Taq polymerase was added during a5
min stage at72oC after a denaturation step of 5 min at 94"C.
For
short degenerate primerswith
added adapter sequences,cycling
was thefollowing
:template denaturation at94"C
for
30 sec, primer annealing at 40 or 42oC (cycles 1-5) or 50oC(cycles 6-35)
for
1 min andDNA
synthesis at 72oC for2
min. For degenerate primers (23 to27 nucleotides) without adapter sequences, primer annealing temperature
of
50'C
was used for30 cycles.
A final
15 min elongation step was performed at the endof
the 30 or 35 cycles and10
pl
of
the reaction mixfures analyzedby
electrophoresisin
I
Vo agarosegel
stained withethidium bromide (Sambrook et
al.,
1989). For some more specific primers, particular cyclingconditions may be used which are specified in the text.
Cloning and sequencing of PCR amplified fragments
After
electrophoresis, the amplifiedDNA
fragmentsof
expected sizes were excised andelured
with
theQIAEX
gel extractionkit
(QIAGEN). The amplified fragments obtained withthe
first
degenerate primers were cloned in pBluescript vector (Stratagene) using the EcoRI andBamHI
restriction sites
locatedin
the
primers
(Sambrooket
al.,
1989).
The
amplifiedfragments obtained
with
further selected degenerateor
specific primers were clonedin
theplasmid pCRtr using the
TA
cloningkit
(Invitrogen) according to manufacfurer's instructions.Double-stranded
DNA
sequencingby
the
dideoxy
chain
termination method
wasperformed
with
fi
DNA
polymerase (Pharmacia) accordingto
manufacturer's instructions.Long
template amplification products are sequencedwith
the help
of
the
double-strandednestàd deletion
kit
from Pharmacia, using exonucleaseIII
and S1 nuclease.RESULTS AND DISCUSSION
Multiplication
of virus isolates on herbaceous hostsAll
the isolates usedin
this study have been transfened and multipliedin
C. quinoa and/orN. occidentalis in
the greenhouse. Table 2 presents the origin and composition of the differentisolates and the symptoms induced on the tests plants used.
The constitution of this collection
of
virus isolates has shown that the identification basedon the
resultsof
serological tests andpartial
sequencingof
PCR amplification
producfsconducted on these herbaceous hosts may be quite different from that given
for
the trees or theshoots when received, and most generally based on reactions observed on woody indicators.
Part one - PCR DETECTIoN oF vrRUsEs
.38
Table 2
-
Isolates maintained on herbaceous host plants in the greenhouseOrigin P863 91-300
9t-297
Gorsem 77 10311 10604 TO77L VDM879 PVO199 PA66 ASPV-J 3536 PSA-H isolateACLSV
ACT^SVACLSV
+ASGV
ASGV
ASGV ASGVASGV
ASGV
ASGV ASPV ASPV ASPVP\
a/
-rnosymptoms;LL:locallesions;Mo:mosaic;S:spotsorblots;c:chlorotic;n:
necrotic;d:deforming;r:reddening;t:translucid;e:epinasty;0:labileoroccasional
symptoms. N.T.: not tested.
-;-:
inoculated leaves; systemic symptoms.Reference isolates received
in
herbaceous hosts from:
(1) : CandresseT.
(INRA, Bordeaux,Fr.) ; (2) : Jelkmann
W.
(BBA, Dosenheim, Ge) ; (3) : Leone G. IPO/DLO, Wageningen,NL)
;(4) DSM, (Braunschweig, Ge)
Design of degenerate primers
for
PCR amplifïcationOur
approachof
RT-PCRis
basedon
the useof
degenerate primers consistingin
themixture
of
mostor all
the potential nucleotide sequences codingfor
conserved amino acids inthe products
of
corresponding genes from related viruses.This approach has been shown
of
practical use in our laboratory asit
allowed the detectionand
further
identification
of
4
different
potyvirusesinfecting
sweet Potato(Colinet
andKummert, 1993
;
Colinet etal.,
L994).It
can be used to detect most orall
the isolatesof
oneparticular virus, different related viruses within a genus, as shown for potyviruses (Langeveld et
aI.,
l99I)
luteoviruses (Robertson etaI.,
1991) or geminivimses (Rybicky and Hughes, 1990),and maybe also viruses from related genus in a family like Potyviridae or Potexviridae.
Although the
3
viruses belongto
different virus
genus, several stretchesof
sequencehomologies were found
in
the amino acid sequencesof
the translation productsof
the putativeRNA
polymerase geneof
ACLSV
(plum isolate:
German etal.,
1990; apple isolate : Sato etal.,1993),
ASGV (Yoshikawa et al.,1992) and ASPV (Jelkmann, 1994), allowing the designof
degenerate primers
for
PCR experiments(fig.
1).j
Symptoms observed on C.quinoa
N. occidentalisLLt;
(Mo)LLc-n;Mo
LLC;
Mod
Sc,r;
(Mo)
LLC
;Mod
Sc,r
; Sc,r
(LLc);Mod,e
-(LLC);Mo
Sc; ScLLC
; Mod, eLLC;
Mod,e
N.T.
LLC;Mod,
e
s.,
fa,)ruro-
Sc,r;Mo
Sc,r;Mod
-
Mod;
Mod apple apple apple apple apple(+) pear(2) pear(2) Pear(3) pflrne(l) apple apple apple Peartz;Part one - PCR DETEcrroN oF vrRUsES
39-AcI sv-a RÉFREFXONN GWSIQFRAEÀ GPNWKHPYRV NQA.I{SYEAIY:.PRHRMDDDIJII
a c I s v -p RS{RE FKGKN G!,ISNQFREEÀ C PII{KFP YKV NQÀI{SYI'ÀVY . PRmo'DDDr,T.
AsPV REDREFRIGD ITÎEQFÎODH SI(\IRGQEL.T V.C.ÀERYEÀIY Pzu{l(CTEiÀT
asgv KSKAEFQSVL GLSÀQF,LD!.G KNGCKIDT.. .r,pr.nnôuw ignoesobrc:
1 900
195 0
2000
2r00
aclsv-a :FlÀj\IKNRLR FDNrÂ1'lNYÀ.K'r:FKÀÀrSRcKy IÀtWFri&{VEi
IKCCRDQRLL
a cl sv-p ir,eerri<nrn. r,ouveivwex'rxeeesnoKy irxrr,i,crw t*"àroô*ii
aspv ruinvxxnr,! rsspeeenex,ËneexeecKF LTJDTFÉKRVP UrssmËna,t
ASgv NWÀGVQKRIR::KSNI.IRRSKSK::EEIFESQ6KE DLQEEISIGPJ PEFKVNiKEI
190 1 19 51
a c I sc -a DaCRO EÉiEsJ KÉSKIÀATIG MSORSDSD9{ : pinki etnir,::SOÉêr.ffi elGj
à c I sv -p . DecRe EEÈR! KLSKSÀÀTTG À}{SOASDJD!{., plDKrF_tFl{,:;.gO!4T4qEKR}
ASPg QEÀVHÀFEEI( KLSKSI{ÀTIE NHSGRSCEDW.PVDI(ÀLITIN SQrcISIDNN;
asgv EDGEKSFLEK RKLrGEtC.lr{À MESERSDIDW,KI/DgÀFLF1,I( SQYCTI€GKI;i'
+ 2001 aclsv-a F{EÀKÂêQÎÈ::ÀCFPHKILVE acl sv-p !TLx\I(!icQTLi|:ÀCJPHi(ILVE aspv :ERSAKÀGQTL::ACF(}{SVLCR a sgv rIEAKÀcQTrJ :ÀCFQHIVLTR .'" :::i:::-i:r+; 2 051 â cI sv-a acl,sv-p a sgv a s9v 20 50 FSPTCRYTEK VLfÀNLPDNY : ÎIÉORJCIFSE
FsF,{cRyrEX vr,tlllr,eoNf ..irionrd'rese
FIPY!{RYrES KW9VIreKls.. rr-sscKNrDD
FgqI!BÀIE.S ÀFLRsccDSf,t:X-T}tSc{ryRFC
a cl sv-a Ë.EDFÀKR. . F SNGS{CVESD :iYÎÀFDVSQDHI TIIjAFEVETjIJ KH!!1{DOISL
â c I sv-p,LEDFÀRR. . F SNCSICVESP,::YqÀ!4ySODHi ÎI.$rF-VELl* nHeCVTOOeW
aspv,!:AÀlll\/m..9 KFNGVCTESDi:tEÀFDÀSQDHaFIIjÀIELÉW KTIGLPSDLI
Àsgv LDSFVTKNÀS VFDGFSTESD.yTÀf.DsSOoH'vrr.esEi{ÀLE: qvldiærerç
2101 215 0
a c I sV -a OSYI KMKCTL GC'RI.G6FAN,T. RFIEEFSIFL. ENT!ÀNUI..F! FCRYEI/. PPG à cI sv-p QSYIKMKCII. GCRIGGFÀDI'RF]EEFSTFL. FNIT{1NMVFi .FcRii'1I. PDG a spv ÀDYTFrK,rH! icsKtcglln( :RF.!qe4STFL: FNTUÀNr{LFti,et&lpL. NCR
asgv TJDyI,RLKLTL::Cq_RLGSr,À.n{ .EIÎG:EFSTS!, FNTFNT}(I.FT:QLr,Sr.IDqBA
2151 2200
TPICFÀGDOI.{' CAITRNL1 . RE IDTHECII,SK: LSIJKÀK\ lRî:l KVPIFCGI.{Rû:
r p rceeeoi:u,, ceiruL: . ne r DûenrLirl lsrKÀr{NFf,
l: Ki/piâeq{B!.i
EÀICFÀC DDÀI : CÀNSR!' . KV INR.F SNFLDKI I KLKAJWQ FÎ À1 glFCGHqi
HR rLpÀcoDH. csLss LKRIR ceFÀrRL:'!(s Fs LTÀvErvR KF p{'(Fccwit
.{-220t 2250
a cl sv-a CFDGTJTKEPC LiYERLOVAT EivG'!.MDVrD' SYFLEPSFI',:]KLGERrÈSHLI àc I sv-p ccDGLTKEpC LrVUn QVlr ErrCRl.rfDVrD SrrleÈSrey f,I,oERLysH.L.
aspv ceacyrireo LIGERLQIÀR ETRNLa\clDlNyerwsèÀyr,xirceNLi{rrL:
a sgv SPYGMSPK LtwÀ.RIn{uS ERQLLKECVD, NYLFSÀIgÀY :RLGER!EII!r,
a cI sv-a ac1 sv-p aspv as9v acl sv-a aclsv-p aspv asgv 2251 2300
EIEQLIIYHQV LTRFFIRNIGI. LLâCDSRHNI SÛ,E. . .. ,. WLSDEDSDDO
EIæLNYHoV LTRF€IRNla{l t!RcDsR!û.{I, Selgr. .. .. HLSDEDCDND
IPQEVDÀHYN CVR.trIVQm{H; LLKSN;RDLF KG.E;. .. .. $,PiSS. ...
KEEDF SY!{TL VIRFFVRNSK' ILÎCLSK3L !i FEICEC IGSK 9iÉSsTSfÀ.sS
2301 215 0
KGSQIEDRR CYS!E1{CEru QNLF. . . .
KGæIEDRRR GYSI'lCl'rGelG QNLf . .. .
RRSNLQT SKL UI,SRPQS FTR I{Q PîSNQîC L IÀSKGLIQTS RF PLDLWÀS
Figure
I
- Alignment of amino acid sequences of translation products from the 3' end of theputative RNA polymerase gene of ACLSVp (German et
al.,
1990), ACLSVa(Sato er al., 1993), ASGV (Yoshikawa et al, 1992) and ASPV (Jelkmann, 1994). Arrows indicate the selected degenerate primers used for this work.
Adapter sequences containing
a
restrictionsite
were addedto
the
shortfirst
selectedprimers
(fig.
2a),for
subsequentcloning
of
the amplifiçation products thus obtained. Theyalso
allow
a higher temperaturefor
annealing from cycle 6in
the polymerase chain reaction.The
availability
of
theTA
cloningkit
(Invitrogen)for
PCR amplified products allowed thefurther selection
of
longer primers, v/ithout adapter sequences (fig. 2b).Sequencing
of
amplification productsfrom
reverse transcribedtotal RNA
preparationsfrom plants infected by different isolates from the same vims allow the design of more specific
and
mainly
moreefficient primer
pairs basedon
the comparisonof
nucleotide sequences.Fig. 2c
shows the sequencea
sucha
primer pair definedfrom
the sequencesof five
ASGVisolates. The same approach
will
be used for ASPV.!
t I
Part one - PCR ostncrloN oF vIRUsEs
-40
ÀCGP1a
FLFMKS
5' 3'
gEcgaÂIlc qrN rrY ÀrG AÀR Tc
MFCGW 3, 5, TÀC ÀÀÀ ACÀ CC$f ACCÈaddaEc r(L) F M K S A 5, 3, aÈgclgçagMrN rrc ATG ÀÀR Tq/ CÀÀ FÀGDDMC 3' 5'
ÀÀR CGN CCI{ CfR CrR TAC ÀCcagçlgatc
Y E ÀV(I) Y P R H
5' 3'
TÀT GÀR GCT ÀTÀ ÎÀY Ccs.I AG, CÀ
FÀGDDMCÀ
3' 5'
AAR CGÀT CûV CTR CTR TAC ÀCF, CG
AKÀGQTLÀC
5' 3,
CY ÀÀR GCN GGH CÀR ACI YT,l GCY TG
ÀIMRFTGE 3, 5, Cç NTÀ DTA CT( T,IAÀ RTG NCC NC! 5' 3, GÀR GCW ÀjlÀ GCT GGY CÀÀ (nc 3925 to 3942) l' 5,
TAG CGç.r CAY Cr! CTC C4y (nE 4485 Eo 44711 ÀccP6
ÀsG1/lF
ÀsculR
Figure 2 - Primers designed for RT-PCR reactions from cDNA lranscripts from total RNA
preparations from infected plants.
(a-b) : Selected conserved amino acid motifs found in the C-terminal part of the
putative RNA-polymerase of ACLSV, ASGV and/or ASPV, and nucleotide
sequences of deduced degenerate primers conesponding to either the coding or the complementary strand. Lower case letters represent nucleotides not derived from
the viral sequences (adapter sequences)
(c) : degenerate primers corresponding to the nucleotide sequences of cloned
amplification products obtained with primers ACGla, ACG2a for plants infected
by 5 ASGV isolates,
R=
AorG; W=AorT; Y=CorT; M=AorC;
K=
G orT;H=4, TorCi
Part one - PCR onrucrloN oF vrRUsES
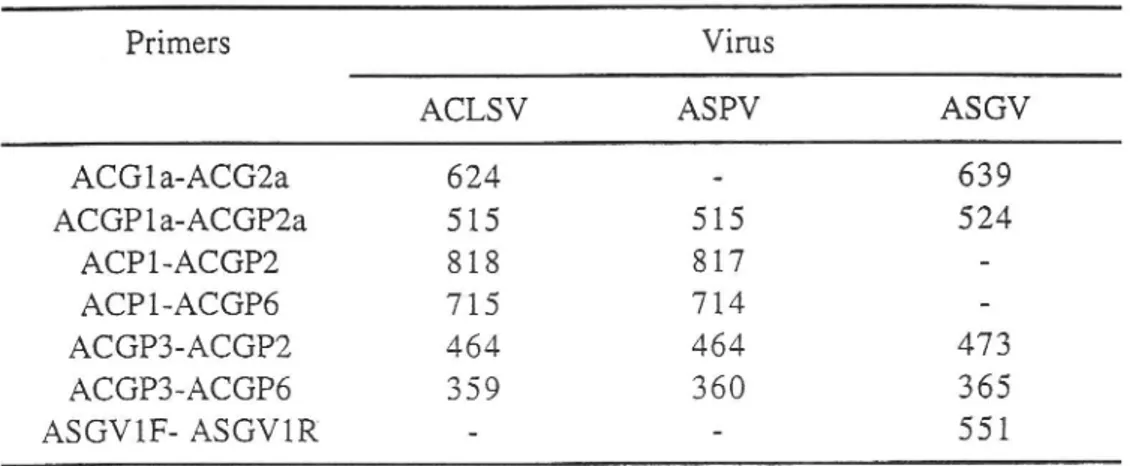
Table
3
presents the expected sizesof
ampiification products obtainedfor
the differentselected primer pairs.
Table 3
-
Expected sizesof
the amplified products (bp)Virus ACLSV ASGV
-41
ACGIa-ACG2a ACGPIa-ACGP2a ACPl-ACGP2 ACPl.ACGP6 ACGP3-ACGP2 ACGP3-ACGP6ASGVlF- ASGVlR
624 515 818 715 464 359 5158t7
714 464 360 639'?o
473 365 551Size expressed as
the
numberof
base pairs.For
sequencesof
the
primersand
theirlocalization
in
the
3'
terminalpart
of
the gene codingfor
theviral RNA
polymerase, seerespectively figures
2
and LRT-PCR experiments
Combined assays
of
reverse transcription and polymerase chain reaction (RT-PCR) havebeen performed
on total RNA
preparationsfrom
symptomatic leavesof
C.
quinoa
andN. occidentalls infected with isolates of ACLSV, ASGV, a mixrure of both viruses, or ASPV. As
the genomic RNA of the targeted viruses possess a polyadenylated 3' end, reverse transcription
has been primed with oligo-dT, and the PCR reaction conducted with the different primer pairs
defined
in
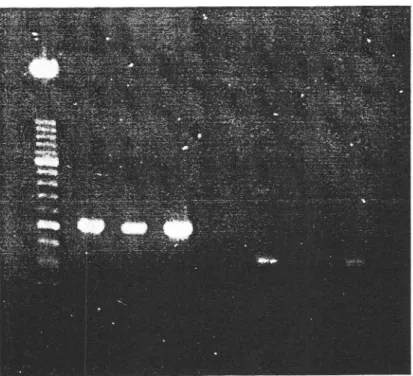
figure 2.Figure 3 shows the results of agarose gel electrophoretic analysis of amplification products
obtained
from
transcriptsof
totalRNA
preparationsof
leavesof
C. quinoa plants inoculatedwith
anACLSV
isolate (91-300), anASGV
isolate (10311) and a complex containing bothviruses (91-297)
with four
different pairs
of
degenerate primers.The
respective sizesof
amplification
products obtained(-
520,630, 360or
470 bp) correspondto
those expectedfrom
the positionof
the different setsof
degenerate primers on the sequences publishedfor
ACLSV and ASGV. The absence
in
this assay of a specific amplification productfor
the ASGVinfected material
with
primers ACGPIa-ACGP2a illustrates the less consistent amplificationPart one - PCR D[rltcrroN ot- rlRLrsES 42
Figure 3
-
A-uarose gel electrophoretic anal!sis ol PCR amplification products fromtranscripts ol total
R\A
preparations from leaves of C. quinoa infected with isolates 9l-300 (ACLSV: lines1.5,9,
l3),9l-291
(ACLSV + ASGV; lines2.6,
10, l-+). 10311 (ASG\'; Iines-i,7,
ll,
l5). Lines4,8,
12, 16 correspond to the negative control (no cDNA added). Primer pairs wereACGPIa-ACGP2a (lines l-1t. ACGIa-ACG2a (lines 5-8), ACGP3-ACGP6
(lines 9-
ll)
and ACGPi-ACGPI (lines l3- 16).trl
= molecular weightmarker ( 100 bp DNA ladder, Lit'e Technolcrgies ).
Figure.l - A_sarose -eel electrophoretic anal!sis
oi
PCR amplilication products fromtranscripts ol total RNA preparations irtrm leaves ol C. quinoa inoculated
rvith ASGV isolates l03l
I
(linesl.
5. 9,l3).
10391 (lines 2,6,10,14),<>r lO11l (lines -3. 7.
li.
l5). Negative ;ontrol without added cDNA (lines4.8,
12,l6).
Prirner pairs wcre ACCIa-ACG2a (lines 1-4),ACGPIa-ACCP2a (lines ,5-$;. ACGP,1-ACGPI tlines 9-12) and ACGP3-ACGP6
(lincs l-l-16). \1 = rnolecular rvcight marker
tl00
bp DNA ladder, LifePart one - PCR orrrcrtoN oF vtRUsES
43-Figure
4
shows the resultsof
agarose gel electrophoretic analysisof
amplified productsfrom
transcriptsof
total RNA
preparationsof
leavesof
C.
quinoa plants inoculated with3 different ASGV isolates. Here also, the sizes of the amplified products obtained with primers
ACGIa-ACG2a, ACGP3-ACGP2 and ACGP3-ACGP6, correspond to those expected
from
theposition
of
the respective primers on the sequences publishedfor
an ASGV isolate. Asfor
theexperiment
reported
in
figure
3,
the primer pair
ACGPIa-ACGP2adid
not allow
theamplification
of
one virus specificDNA
fragment from transcriptsof
totalRNA from
plantsinfected by ASGV.
Figure 5 shows the results
of
agarose gel electrophcretic analysis of amplification productsobtained from transcripts
of
total RNA preparationsof
leaves of N. occidentalis plants infectedby ASPV isolates 3536 and ASPV-J using primers ACGPIa-ACGP2a and ACGP3-ACGP6. The
sizes
of
the
amplification
products, respectively-
520
and 360bp,
correspondto
thoseexpected
from
the positionof
the degenerate primers on the sequence publishedfor
ASPV.The absence
of
visible amplification productswith
primers ACGP3-ACGP6for
transcriptsof
total RNA preparation from the leaves stored at
J"C (initial
inoculum for ASPV-J) may be dueto degradation of the virus particles.
Agarosc gel elcctrophtrrctic anllvsis
ol
PCR amplilication products from transcripts of total RN,\ prcparllions lrom icrve s ol N'. occidentalis infectedby isolate 35-i6 (lincs 1.5). ASPV-J:
initill
inoculum received as freshlcavcs nrateriai and conscrved .rt -1'C tlrncs 2. 6t, ASPV-J after several successivc pi-rssâ-scs ()n lobaccL) in thc grr'r'nhouse (lincs 3,7). Iines 4,8 =
ncsativc control rvithout lcldcd ;DN"\. \1 = ntolccular weight marker XIV
l'rorn Boc'ltringcr ( I(X) bp
D\.\
laddcr, Printcrs pair ACGPIa-ACGP2atlincs
I
-1 r. ACCPI-'\CCP6 {lrnc' -j-li iPart one - PCR oerpcrroN oF vIRUsEs
Figures
4
and
5
show that the
first
selectedshorter sets
of
degenerate primersACGIa-ACG2a and ACGPIa-ACGP2a may lead to inconsistent amplification
of
specific viralsequences. Furthermore, due
to
the relativelylow
temperature (42oC) usedfor
annealingof
these short degenerate primers during the first cycles
of
amplification, non specific small sizedamplification products are
often
observed which emphasizes the needto
include molecularweight markers
in
the gel to control the correct sizeof
amplification products in order to avoidmisinterpretation
of
RT-PCR tests dueto
incomplete or unspecific amplification when usingdegenerate primers.
As a
matterof
fact,
the productionof DNA
complementaryto
plantnucleic acid sequences when using primers designed
for
the amplificationof
viral nucleic acid .sequences has been reported
by
several authors and,for
example, when using degenerate.primers
for
the detectionof
potyviruses. Pappu etal.,
(1993) and Pearson etal.
(L994) haveshown that
with
some plant species the amplificationof
discrete fragmentsof cDNA
may notbe indicative of virus infection.
This observation
let
us consider the researchof
more specific primers, although able tocope with
the
variability of the different virus isolates of the same virus.For ASGV, amplification
products obtainedwith
the primerspair
ACGIa-ACG2afor
5
different isolates have been cloned and sequenced. Comparative analysisof
the sequencesthus obtained allowed the design
of
a primers pairASGVIF-ASGVlR
(figure 2c) based on thepresence of strechtes
of
nucleotide sequences with a minimum of variations.Figure
6
shows that the useof
these primers allows the amplificationof
one specificproduct
with
the expected size,from
transcriptsof
totalRNA
preparationsfrom C.
quinoaleaves infected with the different ASGV isolates tested.
Figure
7
shows that the same primers also give amplification productsfor
transcriptsof
total RIIIA preparations from ASGV infected apple trees. This test was performed during a dry
hot period (samples collected, and total RNAs extracted the
3i.05
and 10,6.96). The bandof
expected size was
faintly
visible
in
the ethidium bromide stained agarosegel,
but
clearlypresent
by
Southern blotting and detectionwith
a digoxigenine labelled probe synthesized byPCR
from
cloned amplification productof
isolate 10311with
primers ACGIa-ACG2a. Thecollected leaf material gave positive resuits in ELISA tests with ASGV antiserum,
Concerning cDNA trancripts from total R-,\A preparations from apple leaves, virus-specific
amplifications have been occasionally but inconsistently observed
for
material taken on budsdevelopped
on
dormant shoots grownin
nutritive
solutions andon
material takenon
appletrees
in
may andjune
1995 but not injuly
and august of the same year; the primers pairs usedwere ACGPIa-ACGP2a and A52-A53 (developped by T. CANDRESSE, INRA-Bordeaux, for
ACLSV) (results not shown).
Part one - PCR oprgcrroN oF vTRUSES
Figure 6 - Agarose ge) electrophoretic analysis oi PCR amplilication products obtained with primers ASGVIF and
ASGVlR from transcripts ol total RNA preparations from leaves of C. quinoa inlected with ASGV isolates 10391 (lines l, 4), 101 11 (line 2).
l0i7l
(lrne -1), or from R\A exrracted from purified virus preparation(isolare 103 1l; line 5). Negative control \r'rthour added cDNA (line 61. tut = molecular weight marker XIV
lrom Boehringer (100 bp DNA laddcr).
t3 kç cT
Figure 7 - Agarose gel electrophoretic (A) and Southern blot (B) anal."sis of PCR amplification products obtained with primers ASCVIF ancl ASGVIR lrom tran:cripts of totei RNA preparations from leaves of C. quinoa infècted with ASGV isolatc "Corscm 77" (line ,3) or from leaves of apple trees 10291 (lines 1,4) or 10j92 (llnes 1.5); the 2 srmplcs corrcsponded to tqo dillerent total RNA preparations made at l5 days inrervnl. Line 7 = ncgittivc conrrol w'irhout atidcd
cD\.\ \l
= molecular weight marker XIV from Bttehringcr ri00 bpD\,\
i.idticrl. DIC llbcllcti pnrl-re uscd tor:outhern blot was obtained from cloned PCRlmpliiicetion product obtlincd for .{SGV r:oirtc
l()ll1
urth primers ACGIa-ACG2a; labelling and;olLtrirn.'trie dctcctior, \\L.rù pcrt()riIlcrl .rceiir,lrn: l0 mlnuiiei,:er's intructtons (Boehringer).
45
B
Part one - PCR DETECTToN oF vrRUSEs
-46
As
the useof
degenerate primers targeting theRNA
polymerase genedid not
allow
areproducible and sensitive detection
of
latent viruses(ACLSV, ASGV,
ASPV)in
fruit
treematerial, experiments were conducted to amplify a large fragment corresponding to the 3' end
of the RNA genome (from 3' end
of
the polymerase gene to the polyA
tail), including the coarprotein gene
of
different ASGV and ASPV isolates,for
sequencing. Comparative analysisof
these sequences
will
allow us to define specific primers to be used for the sensitive and specificRT-PCR detection
of
eachof
these virusesfrom
woody plants (apple and pear).For
thatpurpose,
cDNA
was synthesizedfrom
total RNA preparations from ASGV-infected C. quinoaand
ASPV-infected
N.
occidentalisusing oligo-dT
+
adapter andthe Expandru
reverse transcriptase (Boehringer).For
ASPV, PCR was conductedwith
thecDNA
thus obtained,with
ACGP1or
ACGP3degenerate primers and
a
primerwhich
was complementaryto
the adapter sequenceof
theoligo-dT
primer usedfor
thecDNA
synthesis, using Expandrulong
template PCR system(Boehringer). Figure
8
shows the analysisof
the productsof
Expand long template RT-PCRamplification
from a total RNA
preparationfrom
leavesof
N.
occidentalis infected withASPV-J. The discrete
DNA
bandof
approximatively 3600nt
has been purifiedfrom
the geland cloned
in
pCRII plasmidfor
sequencing. Similar products have been obtainedwith
ASPVisolate 3536.
For ASGV, the use
of
degenerate primers ACGPIa or ACGP3, togetherwith
the reverseprimer complementary to oligo-dT
+
adapter, did not allow the identification of a discrete bandcorresponding
to
the expected long template amplification product(-
2600 nt). Although, theuse
of
2
specific forward primers
designedfrom
analysisof
ACGP3-ACGP6 amplifiedproducts
for
different ASGV isolates allowed the obtentionof
long template PCR amplificationproducts
for
5
different
ASGV
isolates
multiplied
in
C.
quinoa,
and also
from
theASGV-infected tree 10291,.
Sequencing
of
all
these long template amplification products, clonedin
pCRIIis
acnrallyPart one
.
PCR onrscrroN oF vtRt;sE.s 47Figure
8 -
Electrophoretic analysisol
Expandlong
template PCRamplilication producr from a rotal RNA preparation from leaves
of ltl. occidenralis inlecred with ASPV-J using primer ACGPIa and M2 (consisting in oligo-dT and an adapter sequence for
cloning and sequencing ol the PCR product). M = molecular
weight marker (mixture oi
Hindlli
fragments of I,DNA andHaelll fragments olôX171. Lile technologies).
CONCLUSIONS AND PERSPECTIVES
Different
setsof
degenerate primers have been designed allowing the specific RT-PCRamplification
of
fragmentsof
the 3'-terminal partof
theRNA
polymerase geneof
ACLSV, ASGV and/or ASPV from transcripts of total RNA preparations from herbaceous host plants.Although
the
same primersdid
not allow
the consistent and sensitiveamplification of
specific viral sequences from infected apple and pear trees, this approach seems
of
valuable usefor
the prime detection and identificationof
poorly characterized viruses. The further cloningand sequencing
of
the amplification products thus obtained allow the study of the variabilityof
these viruses and the desi-en
of
more specific primers for the developmentof
sensitive RT-PCRdetection protocols applicable to naturally infected (woody) host plants.
This study has also demonstrated that degenerate primers may be used
for
long templateRT-PCR amplification allowing the sequencin-e
ol
lon-e fragmentsof
the genomeof
differentPart one - PCR oerscrloN oF vtRUsEs
ACKNOWLEDGENIENTS
We
thankP.
Simon (Research Stationof
Gorsem, Belgium)for giving
the
shootsof
virus-infected apple trees and preparing
the
treesplanted
in
Gembloux.T.
Candresse(INRA,
Bordeaux, France),W.
Jelkmann(BBA,
Dossenheim, Germany)and
G.
Leone(IPOIDLO-W'ageningen, The Netherlands) kindly provide us with characterized virus isolates on
herbaceous test plants. This work was financially supported by the UIRSIA", Brussels, Belgium. REFERENCES
Colinet D., Kummert J., 1993. Identification
of
a sweetpotato feathery mottle virus isolate fromChina (SPFMV-CH)
by
the polymerase chain reactionwith
degenerate primers. Journalof
Virological Methods45,
159-169.Colinet
D.,
Kummert J., Lepoivre P., SemalJ.,
L994.Identifciationof
distinct potyviruses inmixedly-infected sweetpotato
by
the polymerase chain reactionwith
degenerate primers.Phytopathology 84, 65-69.
German S., Candresse
T,,
LanneauM.,
Huet J.C., Pernollet J.C., DunezJ.,
1990. Nucleotidesequence and genomic organisation
of
apple chlorotic leaf spot closterovirus. VirologyL79,
r04-r12.
Jelkmann
W.,
1994. Nucleotide sequenceof
apple stempitting
virus andof
the coat proteingene
of
a
similar virus
from
pear
associatedwith vein
yellows
diseaseand
theirrelationship with potex- and carlavirus. Journal
of
General Virology 75, t535-1542.Langeveld S.A., Dore J.M., Memelink J., Derks A.F.L.IYI., Vand Der Vlugt C.LM., Asjes C.J.,
Bol
J.F.,
1991. Identificationof
potyviruses using the polymerase chain reaction lvithdegenerate primers. Journal
of
general Virology 72,l53l-I541.
Pappu S.S., Brand R., Pappu H.R., Rybicki 8.P., Gough K.H., Frenkel M.J., Niblett C.L., 1993.
A
polymerase chain reaction method adaptedfor
selective amplification and cloningof
the
3'
sequencesof
potyviral
genomes: applicationto
dasheen mosaic virus. Journalof
Virological Methods 41, 9-20.Pearson
M.N.,
Thomas J.E., Randles J.W., 1994. Detectionof
an unidentified potyvirus fromRoystonea
regia
palm using the polymerase chain reaction and degenerate, potyvirusspecific primers and potential problems arising from the amplification of host plant
DNA
sequences. Journal
of
Virologicai Methods 50, 211-218.Rybicki E.P., Hughes F.L., 1990. Detection and typing of maize streak virus and other distantly
related
geminivirusesof
grassesby
polymerasechain reaction amplification
of
aconserved
viral
sequence. Journalof
General Virology 7I,2519-2526.Robertson
N.L.,
French
R.,
Gray S.M.,
1991. Use
of
group-specificprimers
and
thepolymerase chain reaction
for
the detection and identificationof
luteoviruses. Journalof
General
Virology 72,
1473-1477.Sambrook
J.,
FritschE,F., Maniatis
T.,
1989. Molecularcloning
:
A
laboratory manual.New-York, Cold Spring Harbor Laboratory.
Sato K., Yoshikawa N., Takahashi
T.,
1993. Complete nucleotide sequence of the genome of anapple isolate
of
apple
chlorotic leaf
spot
virus.
Journalof
GeneralYirology
74,1927
-r931.
Yoshikawa N., Sasaki
8.,
Kato M., TakahashiT.,
1992., The nucleotide sequence of apple stemgrooving Capillovirus genome.