Genomic selection of dairy cows
Texte intégral
Figure


Documents relatifs
Summary - A very simple breeding scheme for milk yield was generated by a Monte- Carlo method in order to evaluate the potential impact of bovine somatotropin (BST)
These genomic regions harbored several candidate genes (full gene names of all gene sym- bols are in Table S2 [See Additional file: 1 Table S2]) in- cluding SPATA6 and FAAH in
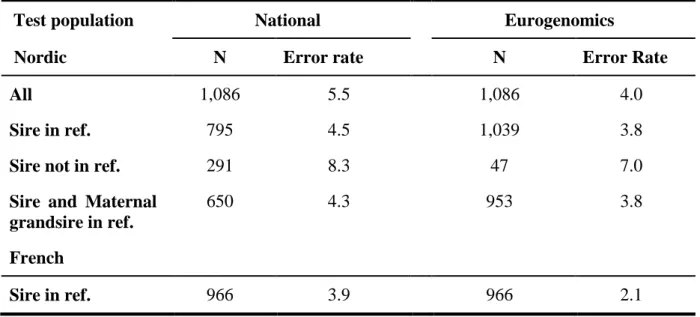
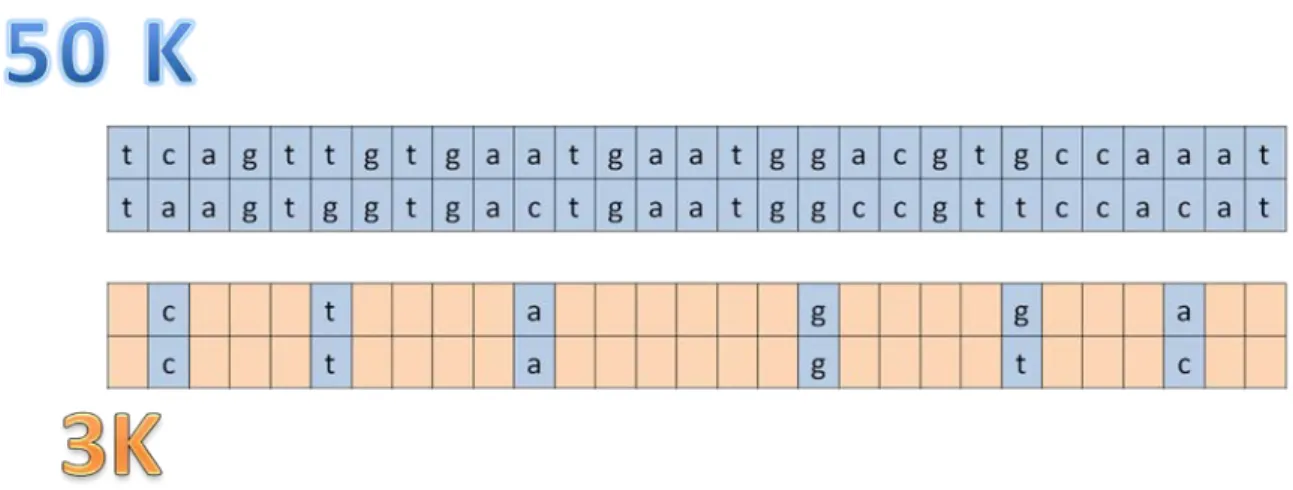
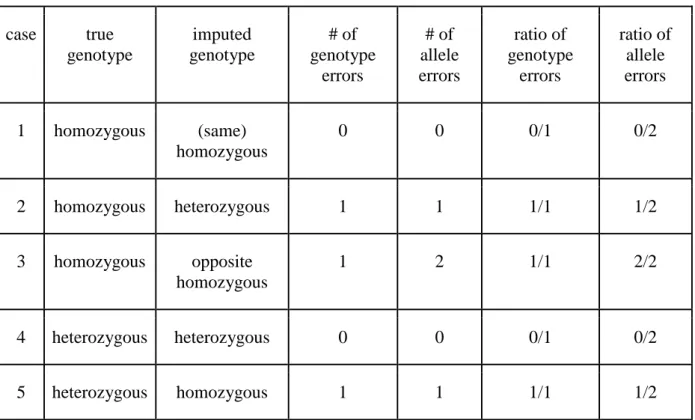
The genotypes of the bulls for these latter SNP being imputed using as reference population either the run4 population of the 1000 Bull Genomes project (scenario 2) or
Previous works showed that genomic selection led to an increase in annual genetic gain in major French dairy cattle breeds but, in some cases, the increase in genetic
Pascal Croiseau, Thierry Tribout, Didier Boichard, Marie Pierre Sanchez, Sebastien Fritz.. To cite
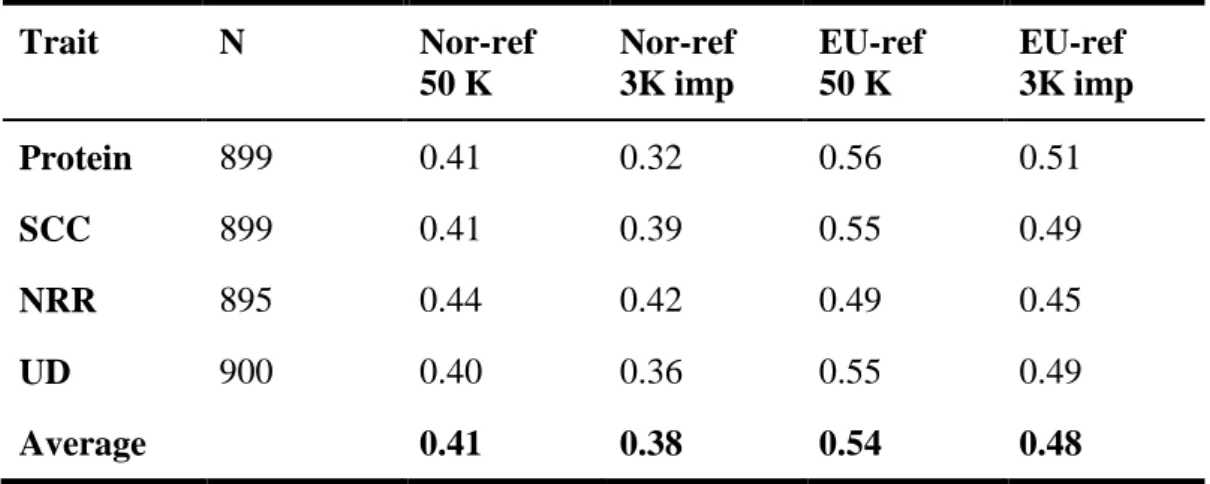
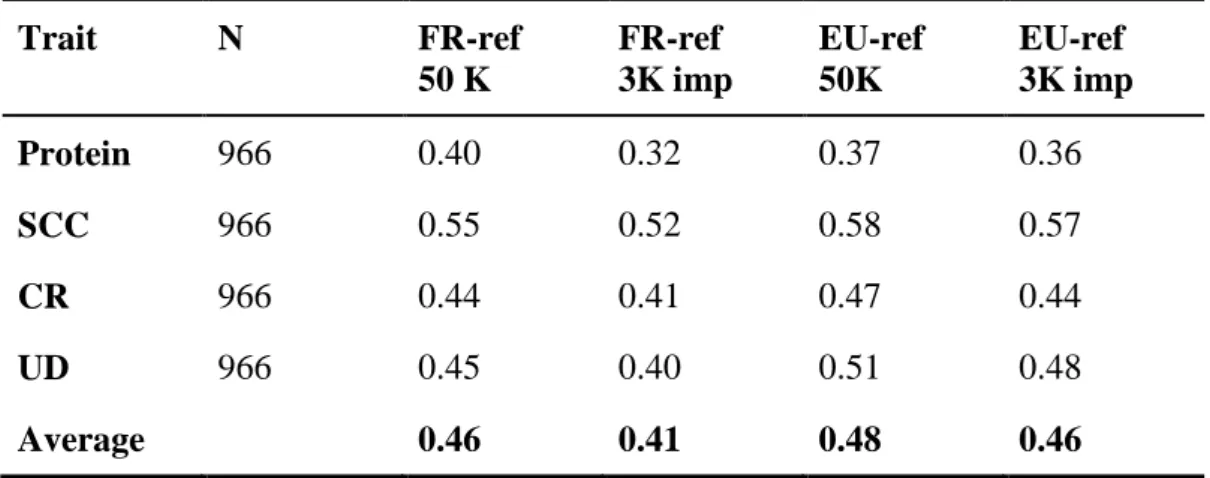
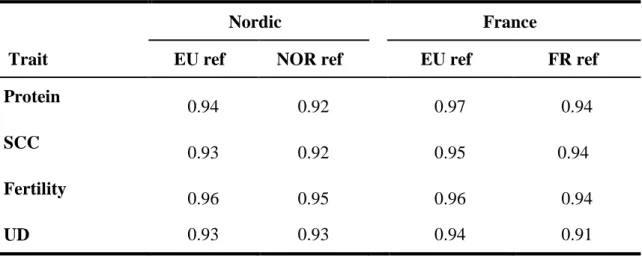
Extending the reference set by adding a large number of cows with genotypes and phenotypes increases the reli- ability of breeding values of young selection candidates and
Then, future industry selection strate- gies were evaluated by manipulating three features of the model: (1) change in the reliability of breeding values due to improved
Using the reference population that included all males and females, accuracies of genomic breeding values derived from prediction error variances (model accuracy) ob- tained