Development of thermostable affinity reagents for
low-cost, paper-based medical diagnostics
by
Eric Alexander Miller
B.S.H. Chemical Engineering, Stanford University (2012)
Submitted to the Department of Chemical Engineering
in partial fulfillment of the requirements for the degree of
Doctor of Philosophy in Chemical Engineering
at the
MASSACHUSETTS INSTITUTE OF TECHNOLOGY
June 2019
Massachusetts Institute of Technology 2019. All rights reserved.
Signature redacted
A u th o r ... . . . .. . . . . .. .Department of Chemical Engineering
May 15, 2019
Signature redacted
Certified by...
Accepted by...
MASSACHUSETTS INSTITUTE OF TECHNOLOGYMAY 2
3 2019
LIBRARIES
Hadley D. Sikes
Esther and Harold E. Edgerton Career Development
Associate Professor of Chemical Engineering
Thesis Supervisor
Signature redacted
...
Patrick S. Doyle
Robert T. Haslam Professor of Chemical Engineering
Chairman, Committee for Graduate Students
Development of thermostable affinity reagents for low-cost,
paper-based medical diagnostics
by
Eric Alexander Miller
Submitted to the Department of Chemical Engineering on May 15, 2019, in partial fulfillment of the
requirements for the degree of
Doctor of Philosophy in Chemical Engineering
Abstract
The timely diagnosis and treatment of disease in resource-constrained settings requires the development of robust point-of-care (POC) diagnostics, which provide accurate re-sults and can be employed by users with minimal medical training and limited access to basic infrastructure. One of the most common POC diagnostic formats is the im-munochromatographic rapid diagnostic test, which traditionally uses nitrocellulose-immobilized IgG antibodies as binding proteins for the capture of disease biomarkers from patient samples. However, these antibodies are expensive to produce and struc-turally complex, and are prone to thermal denaturation. In contexts where continuous cold chain storage may be infeasible, the resulting loss in binding activity can manifest as diminished assay sensitivity, leading to adverse clinical outcomes and eroding patient trust in the diagnostic format.
In the interest of replacing diagnostic antibodies with a more cost-effective, robust class of binding proteins, this thesis explores the development of thermostable affin-ity reagents based on the hyperthermophilic scaffold protein rcSso7d. Given its native microbial host, minimalist structure, and high wild-type melting temperature (98'C), rcSso7d represents a viable alternative to antibodies in in vitro POC assays.
To assess the applicability of the rcSso7d scaffold in this context, protein engineer-ing techniques were used to rapidly select analyte-specific bindengineer-ing variants from a yeast surface display library of >109 members. A high-affinity rcSso7d binder was identified, produced in high yield in a bacterial host, and readily purified in a single chromato-graphic step. The in vitro activity and thermal stability of this engineered binder were characterized in the context of a low-cost, paper-based assay, and significant improve-ments in stability and production economics were observed for rcSso7d-based assays, relative to assays featuring a representative antibody reagent.
Additionally, general strategies were developed to improve the diagnostic perfor-mance of paper-based assays employing rcSso7d-based reagents. In one instance, chimeric protein constructs in which rcSso7d variants are fused to a cellulose-binding domain were found to bind to unmodified cellulose in an oriented fashion and with high effi-ciency. This substrate anchoring approach permits the rapid, high-density
immobiliza-tion of the rcSso7d species in paper-based assays, and yields significant sensitivity en-hancement by enabling both the depletion of the soluble analyte from the sample, and the processing of large sample volumes within clinically relevant timescales. Detection reagents incorporating rcSso7d binders were also developed, using novel fusion con-structs which enabled in vivo labelling while preserving analyte binding activity. These techniques were applied in the context of a urine-based tuberculosis biomarker, and may one day permit the development of multiplexed assays targeting a suite of these analytes. Such a development would enable point-of-care diagnostic testing without requiring the production of sputa, facilitating disease detection in otherwise inaccessi-ble patient populations (e.g. children under five, the elderly, and immunocompromised patients).
Thesis Supervisor: Hadley D. Sikes
Title: Esther and Harold E. Edgerton Career Development Associate Professor of Chemical Engineering
Acknowledgments
History is replete with examples of virtuosic geniuses who toiled away at their craft in thankless solitude, mustering all required inspiration and motivation from within and rejecting the company of their fellow humans in order to render their visions manifest in a pure, unpolluted form.
The author of this thesis is not one of these virtuosos, and this thesis is far from the product of a singular effort (and is better for it). Rather, this research program would have been completely inconceivable without the input of a whole host of gracious con-tributors, funders, collaborators, and supporters; it would perhaps be more efficient to call out those few misanthropic individuals in the greater Boston metro area who didn't contribute substantively to this process.
However, there is a smaller subset of individuals whose contributions have been ab-solutely pivotal in this effort, and I would like to thank them for their support. This will be wordy and over-long (tag-line for this whole thesis), but as it is the only PhD thesis I ever intend to write, I want to take this opportunity to recognize the many people who have played a part in its making.
Firstly, I want to thank Professor Hadley Sikes (whose tenure decision came back in the well-deserved affirmative just today!). I sought to join Hadley's lab all those years ago because I could immediately tell that our values were aligned -Hadley is steadfast in her commitment to using her research program as a platform for good, pursuing research that 1) can be translated into the real world and 2) can make an impact in under-served communities. Combine that translational focus with her strong emphases on student welfare, technical mentorship, and scientific integrity, and Hadley is very much the ideal advisor. Though every thesis has its periods of full-court-press effort, I never once felt under-valued or over-burdened by Hadley. To the contrary, she took pains to ensure that my focus could remain on research and scholarship (even in the throes of compiling her own tenure package), making it so that I rarely had cause to wonder what logistical, fi-nancial, and political challenges she was navigating in the background. Over these past five years, Hadley has been many things for me - a mentor, a friend, a role model, a
source of encouragement and inspiration, a voice of pragmatism and focus, and an ex-cellent scientific collaborator with whom to ideate and invent. I am immensely grateful for her mentorship throughout this process, and her support as I've explored potential commercialization options for our work.
I would also like to thank the members of my thesis committee, who generously
con-tributed their time and scientific expertise over the years. Professor Dane Wittrup not only invented the brilliant process upon which all of this work is based, and provided us with the yeast library to screen, but also served as a valued sounding board for our ideas and a sterling example of scientific leadership and collegiality. The initial insight to in-troduce a sacrificial protein mass to reduce steric occlusion was also his suggestion, and
it set us off on the fruitful path of exploring alternative fusion constructs for the rcSso7d
species. Professor Matt Shoulders has been a constant source of inspiration -his course on chemical biology and his own innovative research program further piqued and ce-mented my interest in this area, and his input has always lent clarity to the molecular view of our system, as well as specificity to our description of it. Lastly, Professor Chris Love's incisive questions have always cut straight to what was important in our process. Professor Love has also consistently provided much-needed encouragement and feed-back on how to define a cohesive narrative arc for this thesis.
Additionally, much of our work has been supported by academic and clinical collab-orators, and I'd like to thank them for their partnership and trust. Balaji Rao and Carlos Teran-Cruz provided us with initial insights and materials for exploring the Sso7d scaf-fold. Madhukar Pai has been our guardian angel throughout this process, connecting us with members of his professional network, including the head of the TB control pro-gram in India and relevant stakeholders in a host of global health NGOs. Drs. Antonio Campos-Neto and Nira Pollock were two of these early connections, and as the initial discoverers of the TB biomarkers we are pursuing diagnostics for, they were instrumen-tal in providing the plasmids, purified protein, and guidance for how to produce those analytes. Michael Traxlmayr and Professor Dane Wittrup initially produced the rcSso7d scaffold for in vivo work, and generously shared that resource -over a year in the mak-ing -with us for our research. Thanks as well to the entire Wittrup lab, who put up with
us regularly moonlighting on their FPLC. Justin Paloni, Carrie Mills, and Professor Brad Olsen have been consistently excellent collaborators, proposing novel applications for our binding scaffold, and pursuing those stories with dogged determination. Likewise, Qifan Zhang, Lukas Zeininger, Kosuke Yoshinaga, and Professor Tim Swager were won-derful partners, and I appreciate them exploring the use of our binding species in their agglutination assays. Lastly, thanks to Professors Bryan Bryson and Andrew Kruse for providing materials that will be used for further validation of our platform.
The shared physical research resources at MIT are world-class, but the scientists who support those facilities are the truly valuable resource. I am hugely grateful for the help and insight of the staff scientists in the Flow Cytometry Core (Glenn Paradis, Merve Saturno-Condon, Michele Griffin, and Mike Jennings), high-throughput core (Jaime Cheah and Christian Soule), and biophysical instrumentation facility (Brad Turner and Debo-rah Pheasant). More than the manuals and research articles I read while trying to on-board processes in their respective cores, their expertise and patient training has been absolutely instrumental (pun intended).
Likewise, none of our research would be possible without the logistical support of our administrative staff -thank you to Andre Puca, Gwen Wilcox, Gail Monahan, Jazy Ma, Shivangi Misra, Cory Harris, Amy Davis, and Mariann Murray for putting up with my inane requests, slow response timelines, and general haplessness. Andre has been a par-ticularly critical part of our team since October of 2014 -without him, our group's work would have ground to a screeching halt. Likewise, a huge thanks to our departmental support staff throughout the years -you are the reason that our department has the co-hesive, friendly, and inclusive culture that it does. Thank you to Suzanne Maguire (our first departmental mom), Joel Dashnaw (our first sustainability champion), and Brian Smith (our eternally friendly, helpful EHS coordinator), and to everyone in the student office -to Eileen Demarkles, Sharece Corner, Suzanne Ronkin, and Barbara Balkwill.
I would also like to acknowledge the programs and people who have financially
sup-ported this thesis: the NIH Biotechnology Training Program, the Tata Center for Tech-nology and Design, the Deshpande Center, the Sandbox program, and the Singapore-MIT Alliance for Research and Technology. In particular, I'd like to single out
program-ming staff from the Tata Center (Rob Stoner, Diane Rigos, Jason Prapas, Chintan Vaish-nav), who have crafted an incredibly helpful fellowship program to prepare us for trans-lation of our technology into products designed for under-served communities. I've been incredibly grateful for this team's mentorship and friendship, and for their cease-less support. Thanks also to our collaborators at the Booth School of Finance (Amy Altchuler, Ariadne Souroutzidis, Connie Fan, and Atsushi Yamazaki), who conducted an
extremely informative business case study on our front-line TB diagnostics.
Likewise, our mentors at the Deshpande Center and the Sandbox program have pro-vided us with a platform for identifying the key value propositions of our approach, and for thinking about how our technology can be translated more broadly, into a range of different products and industries. I've greatly appreciated the perspectives and insights of Karen Golmer, Leon Sandler, Anna Voronova, and Lori Pressman, as well as those of my Sandbox mentors, Maxine Jonas, Patrick Rivelli, and Rajeev Surati. The team that we've been connected with at the Technology Licensing Office has also provided helpful guidance and support -my sincere thanks to Jon Gilbert, Tod Woolf, and Diana Borgas.
I would also like to thank the teaching team with whom I was a TA, for 10.01
(In-tro to ChemE) - Professors Kris Prather and Barry Johnston are inspirational pictures of engaged and dedicated teaching and mentorship, and were wonderful exemplars of how to actively seek student feedback to better tailor your teaching style to their needs. Sunny Kang was a great co-TA -I really appreciate her support and positive attitude as
we coordinated this course.
Much of my graduate experience has been crafted and improved by the people I've most closely shared it with, and I'd like to take a moment to acknowledge my labmates. Kaja Kaastrup, Brandon Heimerl, Joe Lim, Ji-Sam Wong, and Kara Huang all laid the groundwork for my thesis, teaching me most everything I know about biological engi-neering research. Thank you for establishing our lab's warm culture, for being so gen-erous with your time and expertise, and for doing the hard work of setting up our lab in its early days. Shefali Lathwal was a consistent source of humor and conversation, and I greatly enjoyed our lab hangouts -I'm so glad that we've been able to stay in touch following your graduation! Brooke Tam and Troy Langford were my cohort comrades in
the lab, and I'm hugely grateful to them for their friendship and commiseration when experiments were running awry (and for their patience when I inadvertently wrecked their samples). To Kassi Stein, a fellow sarcastic and great lover of Harry Potter -thank you for sharing your real talk, your wicked sense of humor, and your incisive technical feedback. To Ki-Joo Sung, thank you for sharing the Sso journey with me and for al-ways being willing and excited to entertain (and improve upon) my hare-brained tech-nical schemes and musings. To Emma Yee, for taking on the neglected mantle of lab social chair - you've provided/been the glue that's held us together these past couple of years, and I've really appreciated it (as well as your input on diagnostics-related is-sues). Thanks also to Peter Herzog and Alan Aguirre Soto -you were both only with us for a brief season, but you made a lasting impact on our lab, through both your techni-cal brilliance and your warm personas. Lastly, to Seunghyeon Kim, Lynn Hao, and Sun Jin Moon: you're all such hard-working and brilliant researchers, and I've been so im-pressed with what you've already achieved this early in your graduate degrees. It's been a great pleasure getting to know you, and I'm excited to see the great things you continue
to accomplish -you've got this!
There has also been one other critical set of people that I've had the good fortune of sharing the lab with over the years -the students who joined me as undergraduate researchers: Janice Ong wowed me with her fast grasp of complex cloning schemes; Sharon Wu brought characteristic warmth and persistence to her work, and I really en-joyed our lunchtime chats; Jackie Shen demonstrated a real willingness to dive into the molecular details of our work, and I was grateful to her for sharing her personal inter-ests with me; Daniel Osorio spent a fun summer working with me, and I appreciated his wide-ranging technical interests and his flexibility in the face of a discombobulated re-search agenda; Subha Baniya's enthusiasm, humor, dedication (with biweekly trips out from Wellesley), and fierce curiosity were infectious, and deeply appreciated; and Yara Jabbour al Maalouf, who has taken true ownership of her SuperUROP project, and has grown so much as a scientist -I'm continually in awe of your maturity in dealing with difficult circumstances, and the humor and high spirits you exhibit throughout. I've been so impressed with each of their accomplishments and arcs as scientists, and I am
immensely grateful for their help and interest.
Now to my family - Kelly, Rachel, Mom, and Dad, thank you for your unfaltering love, support, patience, and inspiration. It's an incredible gift to have grown up in a fam-ily of such high-achieving, considerate, fun-loving, and good people, and I credit most of what I achieve to your influence -every opportunity I've had has been because of the supportive environment I grew up in, and the incredible examples I had to learn from. I'm so grateful for each of you, and excited that we'll continue to grow in our relation-ships over the years to come. Likewise, Kevin and Rico, I'm grateful for your friendship, hospitality, and support, and I'm so glad that you've both become such integral parts of our family. Thank you also to Kevin, for sharing your insights and perspectives on academia with me, and for being yet another positive example of ideal mentorship. Kate and Dave, thank you for being the most supportive and engaged aunt-uncle duo I could have asked for. Your enthusiasm for my work always re-inspired my own, and I so appre-ciate you asking great questions, showing interest, and making the effort to understand our work. Likewise, to Uncle Chris -thanks for letting me regurgitate my thesis at you over one single-minded meal, and for being uniformly interested throughout. Before I'd voiced it to you, I don't think I'd yet conceived of the full narrative arc of my thesis, and it was hugely helpful to forcibly bounce it off you. To the rest of my extended family scat-tered across the US, thank you for your interest, love, and support, and for not asking me about my graduation time-line too pointedly. Lastly, Mina and Maya, when you find this footnote in some dusty attic decades from now, know that you lit up my years in grad school, and that your pictures hung above my desk as a reminder of all the happy times I've spent being your goofy fun uncle. I love you both very much (as I'm sure I will your soon-to-be-born little sister)!
Thank you as well to my housemates and friends, both in Cambridge and around the country. To my former and current housemates (Rob and Allie Hicklin-Strom, Annah Cat, Tom Luly, Dan and Jen Pisegna, Georgia Lagoudas and Scott Oleson, Jess Newfield, Meghan Brooks, and Sammy Malavarca), thank you for making our house a home, and for sharing your insights, humor, and many personal peculiarities with me. You have been the spice of my life for these past few years, and not just because I've often pilfered
your spices from the cupboard. Special shout-out goes to Marianna Sofman, who has been my friend since close to the very beginning of grad school when she stabbed me with a santoku -though I've become more of a curmudgeon in the past years, she has been ceaselessly positive, energetic, and punny, and I'm so grateful for her friendship. To my broader friend group, thanks to each of you for your good humor and compan-ionship -Nick and Eder (that inseparable unit), Kat and Felix (also a unit, only slightly less strongly bound than Nick and Eder), Heidi and Nancy (perpetual goofs and some of my first non-MIT pals), and Boris (my estranged roommate, entrepreneurial inspiration, and gatekeeper to the Boston Russian community/Kaya the husky) and Erica (intrepid hiker, brewer, nurse, social organizer, and possessor of a wonderfully sarcastic sense of humor, to boot). To my friends in the ChemE and BioE departments, thank you for your camaraderie and technical insight throughout this process. To Natasha, Orpheus, Yamini, Leia, Matt, and Kameron, thank you for your support during year one and be-yond -I definitively would not have made it through without you. And to my dear friends
from before grad school -Kryssi, Ian, Akwasi, Leslie, and Autumn, I've so appreciated your continuing friendship and the initiative you often take, and that you have allowed me to travel the world and experience my roaring twenties vicariously through you (and sometimes along with you). Each of you is such an inspiration in your unique pursuits and commitment to self-realization, and I'm grateful for your example and continuing friendship.
Lastly, and almost certainly not leastly, to Sarah Rosenkrantz and her kinfolk. First to her family -thank you to Eric, Renee, Jaclyn, for welcoming me into your home and being my family-away-from-family. You've shown me such support and love, and I'm so grateful that you've made such an effort to include me in your family's goings-on. Thank you as well to the Newfields, who have shown me such consistent warmth and hospital-ity (their particular brand of it), and to all of Sarah's extended family -the Rausch's and Rosenkrantz' and aunts-by-adoption who have hosted me, fed me, and generally smoth-ered me in affection. Secondly, to her wonderful friends, who have likewise accepted me into the fold -thank you for extending your friendship to me, and for ceaselessly sup-porting and advocating for both of us.
Finally, thank you to Sarah. You've been the best part of these past five years, and if at the end of this process I had nothing to show for it but a loving relationship with you, I'd consider it time well-spent. I'm so grateful to you for all of your support -for celebrating my little wins when I thought they were too small to celebrate, for letting me vent to you when my cells wouldn't pellet or my gel wouldn't run, and for letting me rest assured that no matter what happened during any given day, I'd return home to love, kindness, empathy, and humor. You have kept me grounded, well-balanced, and healthy during this often-difficult and non-ideal chapter of my life, and have done so with such patience, grace, and selflessness. I am so proud of and inspired by the amazing things that you've accomplished during this time, as well as the driven, genuine, just, and loving person that you are. I am beside myself with gratitude that you've chosen to be in my life - I love you so much, and I am so excited to share the many chapters to
Contents
Abstract . . . . Acknowledgements ... List of figures ... List of tables. ... 1 Introduction 1.1 Introduction ...1.2 The need for point-of-care diagnostic assays
1.3 Thermal stability of point-of-care diagnostics
1.4 Development of thermostable affinity reagents
1.5 rcSso7d library construction details . . . . 1.6 Yeast surface display . . . . 1.7 Diagnostic case study: tuberculosis . . . . 1.7.1 Disease scope and impact . . . .
1.7.2 Diagnostic delays . . . .
1.7.3 Status quo diagnostic techniques . . . .
1.7.4 Emerging diagnostic techniques . . . . .
1.7.5 The impact of novel TB diagnostics . . . 1.7.6 Tuberculosis biomarkers . . . .
1.8 Thesis overview . . . . 1.9 References . . . .
2 Barriers to the adoption of rapid diagnostic tests in global health
2.1 A bstract . . . . . . . . 1 . . . . 3 . . . . 16 . . . . 20 23 . . . . 23 . . . . 24 . . . . 25 . . . . 26 . . . . 2 8 . . . . 30 . . . . 33 . . . . 33 . . . . 36 . . . . 36 . . . . 38 . . . . 40 . . . . 42 . . . . 45 . . . 46 57 58
2.2 Introduction . . . .
2.3 Technical barriers . . . . 2.3.1 Susceptibility to heat and humidity . . . .
2.3.2 Inadequate characterization of affinity agents and antigens .
2.3.3 The prozone effect . . . . 2.3.4 Subjective interpretation of test lines . . . . .
2.3.5 Time-dependence of signal development . . .
2.3.6 Insensitivity to low disease loads . . . .
2.3.7 Unsuitability for pathogen quantitation . . . .
2.4 Biological barriers . . . .
2.4.1 Non-ideal biomarkers . . . .
2.4.2 Lack of biomarkers/drug resistance . . . .
2.4.3 Cross-reactivity in complex fluids . . . .
2.5 Social barriers . . . . 2.5.1 Non-compliance with negative RDT results
2.5.2 Non-adoption due to cultural determinants
2.6 Infrastructural barriers . . . . 2.6.1 Inadequate training and instructions . . . . .
2.6.2 Supply chain disruption . . . .
2.7 Regulatory barriers . . . . 2.7.1 Product evaluation . . . .
2.7.2 Establishing testing standards . . . . 2.7.3 Quality assurance . . . . 2.7.4 Positive control wells . . . .
2.8 Economic barriers . . . . 2.8.1 Cost-effectiveness and pricing . . . .
2.9 Conclusions and future directions . . . .
2.10 References . 59 . 61 . 64 . 66 . . . . 70 . . . . 71 . . . . 72 . . . . 73 . . . . 76 . . . . 77 . . . . 77 . . . . 83 . . . . 83 . . . . 84 . . . . 85 . . . . 85 . . . . 86 . . . . 86 . . . . 87 . . . . 88 . . . . 89 . . . . 90 . . . . 91 . . . . 92 . . . . 92 . . . . 93 . . . . 94 95
3 Activity-based assessment of an engineered hyperthermophilic protein as a
cap-ture agent in paper-based diagnostic tests 111
3.1 A bstract . . . 112
3.2 Introduction . . . 113
3.3 Materials for the development and characterization of a streptavidin-binding rcSso7d variant . . . 117
3.3.1 Reagents . . . 117
3.3.2 Biological materials . . . 118
3.3.3 Consumables . . . 118
3.3.4 Cell culture media and buffers . . . 119
3.4 Binder selection and characterization . . . 120
3.4.1 Combinatorial yeast library revival . . . 122
3.4.2 Combinatorial library screening via magnetic bead sorting . . . 123
3.4.3 Combinatorial library screening via fluorescence-activated cell sort-in g . . . 12 5 3.4.4 Library sequencing and stable clone generation . . . 126
3.4.5 Affinity characterization via yeast surface display and flow cytometry 129 3.5 rcSso7d-SA production and characterization . . . 132
3.5.1 Plasmid cloning of streptavidin-binding rcSso7d variant . . . 134
3.5.2 rcSso7d-SA plasmid transformation and heterologous expression . 136 3.5.3 rcSso7d-SA purification . . . 137
3.5.4 SDS-PAGE characterization . . . 138
3.5.5 Production cost analysis . . . 139
3.6 Paper sample production and processing . . . . 139
3.6.1 Eosin 5'-isothiocyanate coupling reaction . . . 139
3.6.2 Aldehyde-functionalized paper preparation . . . . 141
3.6.3 Paper-based assay development . . . 141
3.6.4 Fluorescence microscopy and sample processing . . . . 144
3.7 Activity-based characterization . . . . 144
3.7.2 Thermal challenge antigen-binding assay . . . . 147
3.7.3 Curve fitting and parameter determination . . . . 149
3.8 Conclusion . . . . 151
3.9 References . . . . 152
4 Paper-based diagnostics in the antigen-depletion regime: high-density immobilization of rcSso7d-cellulose-binding domain fusion proteins for efficient target capture 159 4.1 Abstract. . . . .. . . . . 160
4.2 Introduction ... 162
4.3 M aterials and Methods . . . . 165
4.3.1 Yeast surface display selection and characterization of rcSso7d-based binding variants . . . . 165
4.3.2 Combinatorial library screening . . . . 166
4.3.3 Production of gene constructs . . . . 167
4.3.4 Recombinant protein expression, purification, and characterization 169 4.3.5 Fabrication and testing of biofunctional cellulose test zones . . . . 170
4.3.6 Fluorescence microscopy . . . . 171
4.3.7 Quantification of surface-immobilized CBD fusion proteins . . . . 171
4.4 Results and Discussion . . . . 173
4.4.1 Derivation of the exact analytical solution for a monovalent binding system ... 173
4.4.2 Derivation of the pseudo first-order rate constant model . . . . 178
4.4.3 Selection and characterization of rcSso7d binding variants . . . . . 185
4.4.4 Characterization of rcSso7d.SA-CBD cellulose-binding activity . . . . 191
4.4.5 Characterization of assay sensitivity using cellulose-immobilized rcSso7d.SA-CBD ... ... 193
4.4.6 Identification of the antigen-binding regime . . . . 196
4.5 Conclusions . . . . 206
4.6 References . . . . 206
5 Design principles for enhancing sensitivity in paper-based diagnostics via large-volume processing 5.1 Abstract . . . . 5.2 Introduction ... 5.3 Materials and Methods ... 5.3.1 Estimation of physical parameters . . 5.3.2 Modeling and assay simulation . . . . 5.3.3 Cellulose test zone preparation . . . . 5.3.4 Large-volume sample processing . . 5.3.5 Fluorescence microscopy . . . . 5.3.6 Curve fitting . . . . 5.4 Results and Discussion . . . . 5.4.1 Dimensionless number analysis . . . Finite-element model: Instantaneous transport . . . . Finite-element model: Instantaneous capture . . . . . Pseudo first-order rate constant (PFORC) analysis . . Sensitivity enhancement via large-volume processing Concentration dependence of analyte capture . . . . . 211 . . . . 212 . . . 213 . . . 216 . . . 216 . . . . 216 . . . 221 . . . 222 . . . . 223 . . . . 223 . . . 224 . . . . 224 . . . . 227 . . . . 230 . . . . 232 . . . . 236 . . . . 238
5.4.7 Estimation of immobilized binder abundance on oxidized paper . . 5.4.8 Integrated guidance for large-volume processing . . . .
5.5 Conclusions . . . . 5.6 References . . . .
6 Conclusions and Future Directions
6.1 Sum m ary . . . . 6.1.1 Validation of the rcSso7d scaffold for in vitro assays . . . . 6.1.2 High-abundance immobilization of the rcSso7d species on paper
via fusion to a cellulose-binding domain . . . . 5.4.2 5.4.3 5.4.4 5.4.5 5.4.6 240 242 245 247 251 251 252 253
6.2 Future research directions . . . . 254
6.2.1 Development of specific detection reagents for multiplexed diagnostic assays . . . . 254
6.2.2 Applicability of rcSso7d fusion constructs in standard diagnostic formats . . . . 255
6.2.3 Scaffold oligomerization for enhanced activity . . . . 256
6.3 rcSso7d: A viable replacement for antibodies in in vitro diagnostic tests . . . . 256
List of Publications 259 A Engineering hyperthermostable rcSso7d as reporter molecule for in vitro diag-nostic tests 261 A .1 Abstract . . . . 262
A.2 Introduction . . . . 263
A.3 Results and discussion . . . . 265
A.4 Conclusions . . . . 274
List of Figures
1-1 Representative alternative scaffolds and their drawbacks . . . . 27
1-2 The structure and sequence of rcSso7d . . . . 29
1-3 Yeast surface display schematic . . . . 32
1-4 India's diagnostic algorithm for pulmonary tuberculosis . . . . 37
2-1 Standard configuration and mode of action for an immunochromatographic RDT... 62
2-2 Malaria RDT anomalies encountered in production lots. . . . . 71
2-3 Characteristics of the ideal biomarker of infectious disease . . . . 78
2-4 Multiplexed detection using silver nanoparticles (AgNPs). . . . . 81
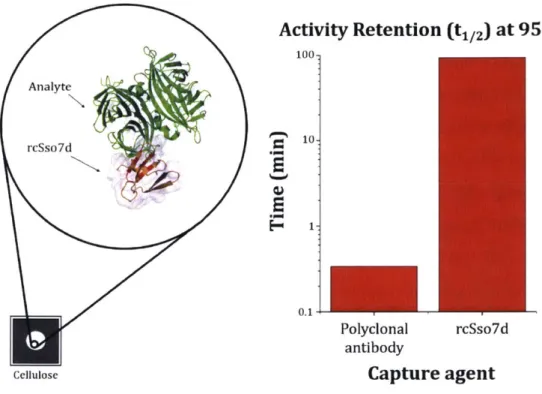
3-1 Relative activity retention of polyclonal antibodies (pAbs) and rcSso7d . . 112
3-2 Characteristics of the ideal alternative binding protein for point-of-care diagnostic biosensors . . . 115
3-3 Schematic representation of experimental approach used for Sso7d-based binder developm ent. . . . 116
3-4 Equilibrium binding titration curve for determining the affinity of the yeast surface-displayed streptavidin binding protein, rcSso7d.SA . . . 121
3-5 Cytometry plots from Rounds 1-5 of FACS library screening . . . 127
3-6 Representative cytometry plots from the single-color and secondary bind-ing controls . . . 131
3-7 Representative chromatogram from the IMAC purification of rcSso7d-SA . 133 3-8 Binding activity of BSA, pAbs-SA, and rcSso7d-SA under equimolar condi-tion s . . . 143
3-9 Representative fluorescence images of paper-based assays . . . . 145
3-10 Titration of paper-immobilized rcSso7d-SA with streptavidin-eosin
conju-gate ... ... 146
3-11 Activity-based accelerated degradation study for streptavidin-binding
poly-clonal antibodies (pAb-SA) and rcSso7d-SA . . . . 150
4-1 Graphical representation of the benefits of the rcSso7d-CBD fusion
con-struct ... ... 161
4-2 Schematic representation of the rcSso7d.SA-CBD genetic construct . . . . 164
4-3 FACS plots for yeast-surface display selection of Rv1656-binding variants
ofrcSso7d ... ... 167
4-4 FACS plots for yeast-surface display selection of Rv1656-binding variants
ofrcSso7d ... ... 172
4-5 Comparison of ligand capture efficiency at equilibrium for the analytical
solution and PFORC model . . . . 181
4-6 Comparison of t9 9 values for analytical solution and PFORC model . . . . 182
4-7 Deviation between the exact analytical solution and PFORC model in the
predicted proportional ligand capture at equilibrium . . . . 183
4-8 Deviation between the exact analytical solution and PFORC model in the predicted value of t9 9 . . . . 184 4-9 Affinity determination for rcSso7d.Rv1656 via yeast-surface display titration 186
4-10 15% SDS-PAGE gel of all purified recombinant products . . . . 188
4-11 Activity time course for different species under dry incubation at 40'C . . 189
4-12 Activity end points for different species following 4 months' dry incubation
at40 C ... ... 190
4-13 Time course of primary incubation . . . . 192
4-14 Comparison of SA-AF647 titration curves in rcSso7d/rcSso7d-CBD formats 194
4-15 Comparison of Rv1656b titration curves in rcSso7d/rcSso7d-CBD formats 195
4-17 SA-E titration curves for various applied soluble concentrations of
rcSso7d.SA-CBD ... ... 198
4-18 Limits of detection for various applied concentrations of rcSso7d.SA-CBD 200 4-19 Micro BCA assay data for the absolute quantification of surface-immobilized rcSso7d.SA-CBD . . . 201
4-20 Micro BCA assay data indicating the adsorption efficiency of rcSso7d.SA-CBD on non-functionalized cellulose . . . . 202
4-21 Theoretical binding profiles generated using the exact analytical solution 204 4-22 Depletion of antigen from 10-pL samples of SA-AF647 at 256 nM . . . . 205
5-1 Graphical representation of sensitivity enhancement through large-volume processing . . . . 213
5-2 Syringe-based assay format . . . . 219
5-3 Set-up of COMSOL proportional analyte capture model . . . 220
5-4 Set-up of COMSOL diffusion model . . . . 221
5-5 Confirmation of fluid flow across the entire assay cross-sectional area . . . 225
5-6 Finite-element modeling data . . . . 228
5-7 Proportional binder occupancy at varying concentrations and volumetric flow rates . . . . 229
5-8 Proportional binding curves predicted by the finite-element model in the diffusive lim it . . . 231
5-9 Correlation of flow rate, binder concentration, analyte capture, and Da, . 234 5-10 Deviation between finite-element analysis and PFORC model . . . 234
5-11 Damkbhler master curve . . . 235
5-12 Binding isotherms under PFORC treatment . . . . 236
5-13 Sensitivity enhancement through large-volume processing . . . . 237
5-14 Titration curves near the point of signal onset . . . . 237
5-15 Comparison between analyte titration curves for rcSso7d-CBD at varying local concentrations . . . . 239
5-16 Comparison between small-volume titration curves for rcSso7d-CBD at
lo-cal concentrations of 400 pM and 40 pM . . . . 240
5-17 Calculation of immobilized protein abundance on functionalized paper . 241 5-18 Assay performance for varying flow rates and total processing times. . . . 243 5-19 Linear regression slopes correlating the number of recirculations and the
degree of signal development . . . . 244
5-20 Representative manual titration curve using streptavidin-eosin as the
sol-uble analyte . . . . 245
A-I Schematic of assay format and signal enhancement . . . . 263 A-2 Genetic constructs and representative protein illustrations for all Sso.TB
variants . . . . 267 A-3 SDS-PAGE of all purified recombinant proteins . . . . 268
A-4 Biotin quantitation results, approximate yields from avidin column
purifi-cation, and approximate product yields from chemical conjugation . . . . 269
A-5 Exploration of signal intensity associated with various Sso.TB species . . . 271 A-6 Demonstration of analyte-specific signal via colorimetric methods . . . . 273
List of Tables
3.1 Sort conditions for FACS rounds 1-5 . . . . 125
3.2 Primary protein sequences of observed streptavidin-binding clones. . . . 128
3.3 Primary protein structure of wild-type Sso7d and the selected
streptavidin-binder, rcSso7d-SA . . . . 129
3.4 Experimental conditions for the determination of the KD of yeast-surface
displayed rcSso7d-SA . . . . 130
3.5 Parameter values for the calculated line of best fit for the combined
yeast-surface display titration data . . . . 132
3.6 Reagents used in the protein expression and purification process . . . . . 134
3.7 Cost analysis of the production and purification of rcSso7d-SA . . . . 134
3.8 Oligonucleotide sequences of primers used in sequencing reactions and
plasmid cloning of selected binder rcSso7d-SA. . . . . 135
3.9 Experimental conditions for the determination of the apparent KD of
paper-imm obilized rcSso7d-SA . . . . 147
3.10 Parameter values for the calculated line of best fit for the combined
surface-imm obilized activity data. . . . . 151 3.11 Parameter values for the calculated line of best fit for the combined t1/2
activity data following incubation at 95 . . . . 152
4.1 Oligonucleotide sequences of primers used in plasmid cloning of selected
binders rcSso7d.SA and rcSso7d.Rv1656 . . . . 168
4.2 Primary protein structure of selected rcSso7d binders . . . . 185
5.2 Fitting parameters for broad-range titration curves . . . . 223 5.3 Fitting parameters for low-concentration (LOD) titration curves . . . . 224
Chapter
1
Introduction
1.1 Introduction
The high incidence rates of infectious disease in the developing world represent one of the most pressing global health challenges currently facing humanity. It is estimated that in 2010, just under 10 million deaths were attributable to bacterial, viral, or parasitic infections, the vast majority of which occurred in low- and middle-income countries. Of these deaths, approximately 40% -3.83 million -were due to just three diseases: tuber-culosis, malaria, and HIV/AIDS.[1]
Robust, simple diagnostic tests are needed at the point-of-care (POC) in order to ad-dress the growing burden of disease in resource-constrained, rural communities. The decentralizaiton of diagnostic resources has been heralded as an effective means of overcoming geographical and financial barriers to diagnostic consultation, by provid-ing patients and local health care practitioners with timely, accurate, and actionable diagnostic data which can inform clinical decisions and ultimately improve patient out-comes. These broadly-distributed diagnostic resources are also critical for creating the accurate epidemiological models needed to efficiently direct resource distribution and elimination campaigns, and play a crucial role in preventing presumptive treatment and the further spread of drug-resistance in pathogenic populations. POC diagnostics may also be compatible with emerging telemedicine applications, which leverage India's broadly-distributed 4G cellular network to enable patients to remotely seek the input
and interpretation of medical professionals. [2]
The deployment of diagnostic tests to resource-constrained settings ultimately re-quires robust assay formats which can be employed by users with minimal medical training and limited access to basic infrastructure. One of the most common diagnostic formats that potentially meets these criteria is the immunochromatographic rapid diag-nostic test (RDT), which traditionally uses nitrocellulose-immobilized IgG antibodies as binding proteins for the capture of disease-relevant biomarkers from patient samples. However, these antibodies are prone to thermal denaturation, which can render test re-sults unreliable. In infrastructure-poor contexts where continuous cold chain storage may be infeasible, this silent test invalidation may result in adverse patient outcomes, eroding patient trust in the diagnostic format.
This thesis focuses upon the development of engineered non-antibody binding pro-teins, and their integration into robust, rapid, and affordable diagnostic assays. It also explores modular approaches for the enhancement of device performance, enabling the sensitive detection of disease biomarkers without the need for centralized, complex lab-oratory equipment or trained clinicians.
1.2 The need for point-of-care diagnostic assays
In order to effectively address the profound diagnostic challenges posed by the global spread of infectious disease, novel technologies must be designed for the contexts and the end use scenarios in which they will be employed. The World Health Organization has established a list of practical design specifications which candidate diagnostic tests should ideally satisfy in order to see widespread adoption in the difficult environments where they are most needed. The criteria for this ideal diagnostic assay, termed an
"ASSURED test," include 1) affordability, 2) sensitivity, 3) specificity, 4) a user-friendly
configuration requiring minimal training, 5) rapid delivery of results, 6) a robust design compatible with local environmental constraints, 7) minimal equipment and infrastruc-ture requirements, and 8) distribution networks which allow consistent delivery of the
These design criteria are not currently being satisfactorily met by available diag-nostics. In March of 2009 a meeting was convened by M6decins Sans Frontieres, the Treatment Action Group, and Partners in Health to characterize the ideal diagnostic test that would meet these ASSURED criteria. It was determined that the minimum specifications for an impactful point-of-care (POC) diagnostic are as follows: the test should cost less than $10 in production (and should be available under concessional pricing schemes); it should exhibit sensitivity and specificity of at least 95%; it should be amenable to facile, infrastructure-free operation and interpretation with a throughput of twenty tests a day; and it must be stable in high humidity at temperatures of 300C for
up to two years, and at higher temperatures for shorter periods of time. [41 None of these benchmarks are trivial to meet, but one that is particularly challenging, yet has received relatively scant attention, is this final design criterion: the need for tests to be robust and thermally stable in challenging environments.
1.3 Thermal stability of point-of-care diagnostics
Point-of-care diagnostics are frequently exposed to extreme temperatures when deployed to rural communities. Studies tracking shipments of malarial rapid diagnostic tests
(RDTs) to sites in Cambodia found that devices were exposed to temperatures in ex-cess of 30'C for 28% of the time spent in transit and storage - roughly 3,400 hours. [5] Likewise, in a study conducted in Senegal, devices left at one site experienced temper-atures in excess of 40'C for 18.2% of the time, and were stored at over 300C for 89.3% of
the time. Similar results were found in Ethiopia, where devices in one site were stored at temperatures over 30'C for 80.3% of the time.[6] Extended exposure to these condi-tions are thought to drastically reduce the shelf life of diagnostic tests, leading to warp-ing of device membranes, dehydration and degradation of fluid transport channels, and perhaps most importantly, denaturation of the capture protein. This in turn results in reduced sensitivities - a site in Brazil found that after fifteen months' storage at room temperature, P falciparum-specific and P vivax-specific test lines on one device exhib-ited sensitivities of 79.7% and 85.7%, respectively. This diminished performance was
attributed to extended storage in conditions outside of the recommended temperature range. [7]
These concerns were further validated by the results of the World Health Organiza-tion (WHO) RDT survey, which assessed the heat stability of 48 malaria RDTs available on the market. The devices were subjected to 60 days' incubation at 45'C and 75% hu-midity, and were then exposed to a low-parasitaemia sample, at 200 parasites/pL. Out of 45 P falciparum-specific test lines, 57.8% maintained viability under these conditions, and out of 20 P vivax/pan-malarial test lines, only 10% remained operative. This re-duced efficacy suggests that for many rapid diagnostic tests, thermal stress may indeed result in the diminished reliability of test results. [8] Furthermore, the variability between test lines suggests that the incorporated IgG affinity reagents may be responsible for this diminished functionality. The WHO has since identified ideal protocols by which to as-sess device stability and inform test procurement, many of which implicate the reduced activity of IgG affinity reagents under thermal stress. [9, 10]
1.4 Development of thermostable affinity reagents
Relatively few published studies have documented the development of robust capture proteins for diagnostic applications,[11, 12] and historically the adopted approach was to develop an antibody with established binding activity and to attempt to enhance its thermostability. This, however, is a daunting prospect, as protein stability arises from a complex interplay between intramolecular salt bridges, atomic packing density, hy-drophobicity, and proline content,[ 131 and the impact of remote mutations upon these factors is difficult to predict and characterize. In light of this fact, the focus of this thesis has been approaching the problem from the opposite direction -using small, innately-stable protein species as a scaffold which could be endowed with binding activity. In order to do so, we sought to move away from using antibodies and antibody fragments,
and undertook instead to assess the broad range of alternative scaffold proteins.
A number of alternatives to antibodies have been investigated since the advent of
relatively small, robust, and amenable both to use with cell-surface display technolo-gies for protein engineering, and to high-yield production in bacterial culture. For the purposes of producing point-of-care diagnostics, the ideal scaffold would also be tremely thermally stable, and would be devoid of disulfide bonds that would require ex-pression under oxidizing conditions. The list of potential non-IgG binding scaffolds in-cludes anticalins (small-molecule transport proteins), designed ankyrin repeat proteins (wholly-synthetic, modular binding proteins), fibronectins (integrin-binding proteins), affibodies (variants of the Protein A binding domain), knottins (a peptide motif related to scorpion toxins), a whole host of antibody fragments,[15], aptamers (RNA/DNA oligos with distinct folded structures),[16] and many more.[17, 181
PDB: 1R18 PDB: 1ULL
Single-domain camelid antibodies DNA aptamers
- Low expression yields - Black box development process - Poor solubility - Frequent false positives - Native disulfide bonds - Low affinities
Figure 1-1: Representative alternative scaffolds and their associated drawbacks
Although the thermal stability and small molecular footprint of these species were promising, many of these scaffolds featured native cysteine residues, low expression yields in bacterial culture, or a tendency for non-specific binding. In light of these short-comings, the search for an ideal binding scaffold led to one species in particular: Sso7d. This 7-kDa protein is native to the hyperthermophilic archaeon Sulfolobus solfataricus, and in its natural host, it acts as a chromatin analogue, participating in DNA compaction and stabilizing the DNA duplex at elevated temperatures. [191 The binding face of this species has been well-characterized, and its wild-type melting temperature has been
measured at 98'C, making it an ideal candidate for point-of-care affinity applications. Rao et al also showed that the species was amenable to binder development projects, having successfully selected for binding molecules for six different antigen targets, and showing that the species' high melting temperature was not adversely affected by the development of this binding activity. [20]
Thus, in order to enable the development of thermally stable, point-of-care immunoas-says, we have opted to use Sso7d as our non-IgG binding scaffold of choice. Thus, it was determined that Sso7d would be the non-IgG binding scaffold of choice, and that a combinatorial library of Sso7d variants would be used to select for binders to diagnostic biomarkers of interest.
1.5 rcSso7d library construction details
Our work has employed a combinatorial library based on the Sso7d scaffold, which Traxlmayr et al developed and integrated into the yeast surface display plasmid, pCTCON-2.[21] A number of modifications were made to this species in order to render it more suitable for affinity applications. Initially, 22% of the amino acids found in the 63-residue protein were positively-charged lysine residues, giving the scaffold a net formal charge of +7. In order to reduce non-specific adsorption to negatively charged surfaces, Traxl-mayr et al created a "charge-neutralized" variant of the scaffold, removing two lysine residues found at the C-terminus, and mutating a further four lysine residues to un-charged amino acids, to yield a net formal charge of + 1. [22] Truncating the scaffold at
the C-terminus also served to flatten the profile of the DNA-binding surface, eliminat-ing potential steric interactions which might occlude the bindeliminat-ing interface.
A combinatorial library of this charge-neutralized species was generated, in order to
produce functional diversity from which to select novel binding reagents. Nine amino acid positions within the binding face were chosen as targets for saturation mutagen-esis, based on the residues' participation in the native DNA-binding interaction, their relative proximity to one another, and the favorable, outward orientation of their side chains. Selection based upon these criteria serves to increase the likelihood that the
library members' side chain moieties will interact with target antigens, as well as to en-sure that the introduced mutations do not disrupt the herringbone packing of aromatic residues which serves to stabilize the core of the scaffold. [23]
1 10 20 30 K T Q K K KK W V QMS T Truncated to remove K62 & K63 40 AT R A K K27Q K6T K8Q K39A
Figure 1-2: The structure and sequence of rcSso7d, as adapted by Traximayr et al.[22] Mutated lysine residues are in light green, conserved lysine residues are in light blue, and the nine amino acids in the DNA-binding face are in red. K62 and K63, which were removed in the charge-neutralized library, are shown at left. PDB ID: 1SSO
It was also determined that the suite of amino acids which could be introduced into these nine positions should be limited to eleven, to allow for a greater proportion of the maximum theoretical diversity to be sampled. Randomization of nine residues with eleven potential amino acids yields a maximum diversity of 2.36 x 109 different variants, which is of the same order of magnitude as the library diversity accessible via yeast sur-face display. Furthermore, Traximayr et al found that by carefully selecting which amino acids to include within the suite, a suitable range of functional diversity could be main-tained, including polar, nonpolar, and charged residues. This choice was also informed
by the analysis of Bogan and Thorn, who assessed the frequency with which each amino
acid was involved in known binding interactions; [24] those species which were more
50
K
60
K KK
highly implicated in facilitating binding were chosen for inclusion in the amino acid
suite. Cysteine and proline were specifically ruled out, as cysteine can cause dimeriza-tion via disulfide bond formadimeriza-tion, and the unique structure of proline can introduce hairpin turns which can destabilize the protein core.
Having defined the specific complement of amino acids to be included, trimer phos-phoramidites were used to generate DNA sequences which were randomized at the se-lected positions. The use of trimer phosphoramidites made it possible to ensure that the eleven amino acids would be represented in equimolar ratios, eliminating the codon bias that occurs when using NNN- or NNS-randomization. This also made it possible to prevent the incorporation of premature stop codons, creating a high-quality library of full-length mutants. The random library created by Traxlmayr et al was cloned into the broader Sso7d scaffold with the pCTCON-2 yeast surface display plasmid, and was transformed into the EBY100 strain of S. cerevisiae, through twenty separate transfor-mation reactions. These reactions were pooled, and serial plate dilutions showed this library to have a diversity of 1.40 x 109, or approximately 59.3% of the total diversity
ac-cessible via this mutagenesis scheme. This large library is on par with typical library sizes achieved in the phage display format.
1.6 Yeast surface display
Yeast surface display is a robust platform for the selection of protein variants with en-hanced binding function. This platform permits the physical linkage of protein pheno-type (i.e. the binding function of a given protein variant) with the associated genopheno-type (i.e. the physical DNA encoding for that functional variant). In order to do so, the ran-domized library of rcSso7d genes is integrated into a plasmid construct, in which the rcSso7d framework is fused to the 3' end of the a-agglutinin Aga2p protein subunit, a cell surface receptor which mediates cell-cell contacts during yeast mating. Following galactose-based induction, Aga2p is exported to the exterior of the yeast cell, where it covalently binds via a pair of disulfide bonds with a partner embedded in the cell wall, (Agalp). [25] By virtue of being genetically fused to this Aga2p species, the rcSso7d
vari-ant is also anchored to the exterior of the yeast cell, and thus each yeast cell acts as a vehicle for the rcSso7d variant encoded on its particular copy of the pCTCON-2 plas-mid. On average, roughly 50,000 copies of a given rcSso7d clone are expressed on the exterior of a strongly displaying yeast cell. [26]
Induced yeast cells can be sorted using either magnetic bead sorting (MBS) or fluorescence-activated cell sorting (FACS). Magnetic bead sorting is commonly used to quickly deplete a naive library of non-binders, enriching the population for protein variants which in-teract -even marginally -with the analyte of interest. This process involves the anchor-ing of the analyte to the surface of a magnetic bead (typically via a biotin-streptavidin linkage), and the subsequent incubation of these analyte-coated beads with the yeast surface display library. Yeast clones featuring functional rcSso7d variants will avidly as-sociate with the analyte immobilized on the surface of the beads, and can be withdrawn from the broader population using a strong magnet. Likewise, negative selection can be conducted, by incubating a sub-library of clones with the bare beads and retaining yeast cells which are not withdrawn by the magnet. [27]
Following the MBS process, which typically reduces the library size from over a bil-lion members to a more manageable number of distinct clones (-1 milbil-lion), yeast li-braries are further refined via flow cytometric cell sorting. The full-length display of each rcSso7d variant can be queried by way of primary antibodies specific to the hemagglu-tinin (HA) and c-Myc epitope tags which flank the rcSso7d species, followed by fluo-rescently labeled, species-specific secondary antibodies. Likewise, binding function of each rcSso7d variant can be assessed using a labelled form of the analyte of interest, fea-turing a hexa-histidine tag or biotin moiety. The association of the analyte can then be detected using a fluorescent secondary antibody or streptavidin species, respectively. [21]
By screening this population via FACS, cut-off thresholds can be established on the
ba-sis of the intensity of the protein display and antigen-binding signals. Orthogonal sets of fluorescent reagents can be used, switching between subsequent rounds of sorting in order to avoid applying a coherent selective pressure for binders against the sec-ondary reagents. Over the course of multiple rounds of FACS, the selection stringency is typically increased, in order to selectively enrich the population for the binders with
the strongest analyte-specific affinity. This process permits precise control to be ex-ercised over the binder development process, via constant tracking of both analyte-specific and off-target binding signal, across multiple rounds of cell sorting. Addition-ally, this process is amenable to counter-selection steps, enabling the development of binding reagents which differentiate between closely related target variants.
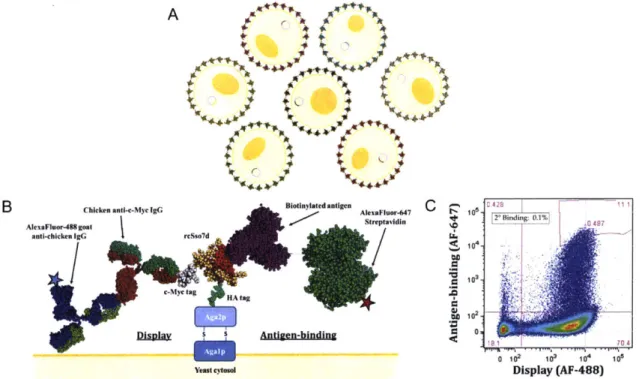
A AA A LA P~LA
B Chicke...nti..Myc IgG Biodinylated antigen A4x~nr-4 0
AtexaFloor-488 goal Streptavidin 2* Blnding; 0.1%, 0487 anti-chicken RGC rcSso7d
IV
103-C-Myc tag OR"a
HA tag
Disp 5 S Antdgen-bindin r 0
470
4 j0 410 6
Yeast cylow Display (AF-488)
Figure 1-3: Yeast surface display schematic. A) In a naive yeast library, each yeast cell contains the gene for a different variant of the rcSso7d scaffold, each encoded on a
pCTCON-2 yeast surface display plasmid. B) The expressed rcSso7d variant is fused
to the Aga2p yeast surface receptor, which is exported to the exterior of the yeast cell
and anchored via disulfide bonds to the Agalp protein. The rcSso7d variant is fused be-tween two peptide tags (an N-terminal HA, and a C-terminal c-Myc tag), which can be targeted with epitope -specific antibodies to assess the efficiency of full-length rcSso7d display. Clonal binding to a biotinylated form of the antigen is queried using fluores-cently labeled streptavidin. C) A representative fluorescence- activated cell sorting dot
plot. Each dot represents a single yeast cell, and its position on the x- and y-axes
rep-resents the display efficiency and antigen binding activity, respectively, of the rcSso7d variant displayed on the surface of the yeast cell.
This thesis strongly relies on the robust and flexible nature of this protein
engineer-ing platform, and demonstrates its use for the development of rcSso7d variants which specifically bind to a) a model analyte (streptavidin), and b) soluble biomarkers of
tu-berculosis. It should be noted, however, that the findings and methods documented herein constitute generalizable guidance, and that the core conclusions of this research can be applied to affinity reagent development for protein-based biomarker of disease. Likewise, the modular scaffolds developed and principles explored over the course of this thesis are broadly applicable for the development of any affinity-based diagnostic assay.
1.7 Diagnostic case study: tuberculosis
In order to anchor our discussion of point-of-care diagnostic assays, we must first typify the experience of patients in rural communities, and the limitations of existing diagnos-tic networks. To do so, we will focus primarily upon the burden imposed by tuberculo-sis (TB), and the extant need for point-of-care diagnostic assays enabling the detection of active TB cases. We will first introduce tuberculosis in the global context, and we will then consider the specific healthcare system, socioeconomic context, and current TB case-finding apparatus of India (the country with the greatest TB burden), in order to further narrow our focus and to provide an anchor with which to contextualize this work.
1.7.1 Disease scope and impact
According to the 2018 WHO Global Tuberculosis Report, there were an estimated 10 mil-lion incident cases of tuberculosis (TB) in India in 2017, amounting to 27.4% of all cases globally. These cases resulted in 1.3 million deaths, rendering tuberculosis the second leading cause of death due to a single infectious agent, after HIV. It is estimated that a further two billion people harbor a latent TB infection, which, while not contagious, will ultimately transition into an active infection in 5-10% of cases. International attempts to curb the spread of this disease are complicated by a number of factors, including the rise of multi-drug resistant (MDR) TB, which was implicated in 475,000 new cases in 2017, and the high rates of TB co-infection in HIV-positive individuals, observed in 920,000 of the ten million incident cases. [281
The staggering global toll of this disease becomes even more apparent when one considers the associated count of disability-adjusted life years (DALYs). The WHO de-fines a count of DALYs as a population-wide index measuring the years of healthy life that are lost due to premature death and non-fatal illness or disability. Applying this measure within patient populations for tuberculosis reveals a grim situation -within the context of individually-addressable conditions, tuberculosis represents the ninth leading source of DALYs, and among infectious disease, tuberculosis is the third largest source of DALYs. The WHO estimates that in 2010, TB (or TB/HIV co-infection) caused 64.3 million DALYs - 2.58% of the global total. [29]
The magnitude of these values illustrates not only the high rates of incidence and the lingering health effects of these diseases, but also the disproportionate impact they have on young children. In 2017, there were an estimated 1.01 million incident cases of tu-berculosis in children under the age of 14, which ultimately caused 234,000 deaths. [30] In addition to representing an urgent humanitarian crisis, this disproportionate impact on children serves to perpetuate the cycle of poverty and insufficient education in de-veloping countries. It has been proposed that these high death rates may indirectly con-tribute to higher fertility rates within households, as parents seek to compensate for the potential loss of a child. This compensation often results in larger families and reduced per-capita investments in education, which disproportionately affects young girls. The demands associated with raising a large family may also result in children being taken out of school to provide additional labor and familial support. [31]
On the household level, the costs of contracting tuberculosis can be economically catastrophic. Studies of the financial burden imposed upon Indian patients by the TB diagnostic process indicate costs which vary from 10-35% of an individual patient's an-nual income. These costs can reach as high as 50% of their anan-nual income for patients in the lowest wage quintile. More than 70% of Indian patients took out loans in order to pay for these services, given that 57% of all health-care expenditures are paid for out-of-pocket.[2, 32] Additionally, health-care access is significantly stratified by socioeco-nomic status and proximity to urban centers - only 3% of patients with major illnesses in urban settings remain untreated, whereas 12% of patients with major illnesses in
![Figure 1-2: The structure and sequence of rcSso7d, as adapted by Traximayr et al.[22]](https://thumb-eu.123doks.com/thumbv2/123doknet/14744557.577734/30.917.144.684.323.615/figure-structure-sequence-rcsso-adapted-traximayr-et-al.webp)