SESAME (SEquence Sorter & AMplicon Explorer): genotyping based on high-throughput multiplex amplicon sequencing
Texte intégral
Figure
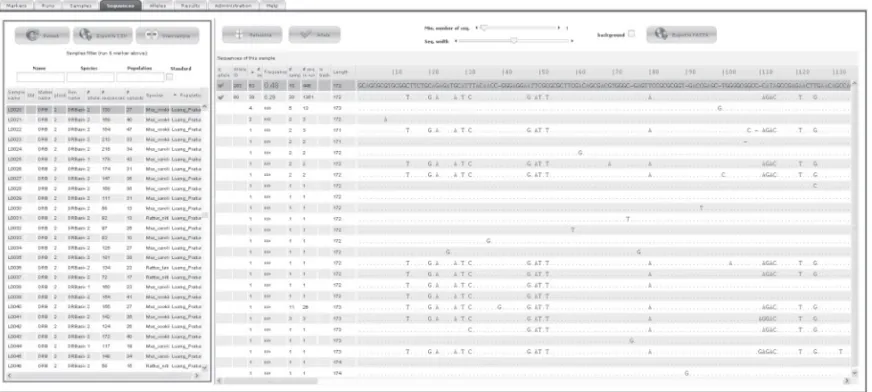
Documents relatifs
TABLE 2 Numbers of sequences for 12 pathogenic OTUs observed in wild rodents, negative controls, and positive controls, together with T CC and T FA threshold values
PCR amplification, library preparation and sequencing Each of our seven multiplexing experiment corresponds to an Illumina library composed of single-sequence or mock community
In the present study, we compared the performance, with respect to microbial community profiling, of two com- monly used broad range 16S rDNA primer pairs with 16S rDNA gene
SVJedi was applied on a real human ONT PromethION 44x dataset for the individual HG002 as well. Finally, we considered a real short read dataset for the HG002 individual, 2 X 250
B- Comparison of bacterial and fungal diversity detected in two sputa (2 patients, P1, P2) spiked with 10 5 conidia of Aspergillus section Fumigati and Aspergillus section Nigri
Local assembly [200] is more sensitive at the event (expressed SNP, alternative splicing forms) scale. A gene or a gene family corresponds to a bi-connected component in the graph,
SVJedi : Structural vari- ation genotyping using long reads.. HiTSeq 2019 - Conference on High Throughput Sequencing, Jul 2019,
Pearson correlation coefficients among variables measured during two seasons and computed across seasons (Comb) in the DH population inoculated with strain PSS4