Direct evidence for cancer-cell-autonomous
extracellular protein catabolism in pancreatic tumors
The MIT Faculty has made this article openly available.
Please share
how this access benefits you. Your story matters.
Citation
Davidson, Shawn M et al. “Direct Evidence for
Cancer-Cell-Autonomous Extracellular Protein Catabolism in Pancreatic
Tumors.” Nature Medicine 23, 2 (December 2016): 235–241 © 2017
Nature America, Inc., part of Springer Nature
As Published
http://dx.doi.org/10.1038/NM.4256
Publisher
Nature Publishing Group
Version
Author's final manuscript
Citable link
http://hdl.handle.net/1721.1/116946
Terms of Use
Creative Commons Attribution-NonCommercial-NoDerivs License
Direct evidence for cancer cell-autonomous extracellular protein
catabolism in pancreatic tumors
Shawn M. Davidson1,2,3,+, Oliver Jonas1,4,+, Mark A. Keibler5, Han Wei Hou6, Alba Luengo1, Jared R. Mayers1, Jeffrey Wyckoff1, Amanda Del Rosario1, Matthew Whitman1, Christopher
R. Chin1, Kendall J. Condon2, Alex Lammers1, Katherine A. Kellersberger7, Brian J. Stall7, Gregory Stephanopoulos5, Dafna Bar-Sagi8, Jongyoon Han5, Joshua D. Rabinowitz9, Michael Cima1, Robert Langer1,4, and Matthew G. Vander Heiden1,2,3,10,*
1Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology,
Cambridge, Massachusetts, USA
2Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA 3Broad Institute of MIT and Harvard University, Cambridge, Massachusetts, USA
4Department of Radiology, Brigham and Women’s Hospital and Harvard Medical School,
Cambridge, Massachusetts, USA
5Department of Chemical Engineering, Massachusetts Institute of Technology, Cambridge,
Massachusetts, USA
6Department of Electrical Engineering and Computer Science, Massachusetts Institute of
Technology, Cambridge, Massachusetts, USA
7Bruker Daltronics, Inc. 40 Manning Rd, Billerica, MA 01821
8Department of Biochemistry and Molecular Pharmacology, New York University, New York, New
York, USA
9Department of Chemistry and Integrative Genomics, Princeton University, Princeton, New
Jersey, USA
10Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School,
Boston, Massachusetts, USA
Abstract
Protein scavenging by macropinocytosis can serve as a source of nutrients for pancreatic cancer cells. We provide direct evidence that extracellular protein is a fuel source for pancreatic cancer cells in vivo. We demonstrate that albumin-derived peptides and amino acids accumulate in tumors in a Kras-driven mouse model of pancreatic ductal adenocarcinoma. In addition, we implement a device to deliver large molecules directly into the tumor and observe protein catabolism and macropinocytosis by cancer cells within pancreatic tumors. Local release of a macropinocytosis
*Correspondence: Matthew G. Vander Heiden, Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA 02139, USA,
HHS Public Access
Author manuscript
Nat Med
. Author manuscript; available in PMC 2018 February 01.Published in final edited form as:
Nat Med. 2017 February ; 23(2): 235–241. doi:10.1038/nm.4256.
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
inhibitor leads to a drastic reduction in amino acids levels in tumor tissue arguing that the direct uptake and catabolism of extracellular protein is necessary to provide amino acids to pancreatic cancer cells in tumors. These data provide evidence for albumin catabolism by tumors and also suggest a method for testing therapies that take advantage of the propensity of pancreatic cancer cells to scavenge extracellular protein.
INTRODUCTION
Mammalian tissues rely on a variety of nutrients to support physiological function1. Altered metabolic processes are involved in the pathogenesis of cancer, but which nutrients support inappropriate cancer cell growth within intact tumors is incompletely understood2–4. Amino acids, particularly glutamine and serine, have been shown to be essential nutrients for many cancer cell lines5,6. Some cancer cells can also obtain key amino acids through catabolism of extracellular protein via macropinocytosis to fuel anabolic pathways required for
growth3,7,8. However, the source of amino acids contributing to cancer cell proliferation within intact tumors is less well studied.
Albumin is the most abundant extracellular protein in blood and in tissues, and has been previously shown to be internalized into lysosomes of cells in tumors9,10. Hypoalbuminemia is observed in many patients with cancer and epidemiological studies suggest that levels of serum albumin prior to therapy can be a predictor of survival11,12. Decreased albumin levels observed in cancer patients and tumor-bearing animals has been posited to be a result of decreased hepatic synthesis or increased degradation by the liver or other organs such as skeletal muscle13–16. Direct catabolism of albumin by the tumor itself might also contribute to hypoalbuminemia, but the extent to which this occurs in vivo remains controversial due to the lack of reliable methods to trace albumin fate.
Macropinocytosis is a conserved, actin-dependent, endocytic process that results in non-specific bulk internalization of extracellular material, including protein and other macromolecules, into the cell7,8,17. Pharmacological and genetic inhibition of
macropinocytosis can inhibit cell line proliferation and xenograft tumor growth, presenting a potential therapeutic target for a subset of tumors7,8. Protein scavenging has been posited based on levels of amino acids observed in human pancreatic tumors7, but direct evidence that protein is catabolized in a tumor-autonomous manner in vivo is lacking.
We utilize miniaturized plasma exchange to deliver labeled albumin to tissues in live mice and demonstrate albumin breakdown contributes to free amino acids in pancreatic tumors in vivo. In addition, we deliver albumin directly into the tumor using an implantable
microdevice, adapted and modified from [18], and directly observe protein catabolism and macropinocytosis in situ by cancer cells in pancreatic tumors but not in adjacent non-cancerous pancreatic tissue. Intratumoral inhibition of macropinocytosis results in a decrease in amino acid levels. Taken together, these data suggest pancreatic cancer cells consume extracellular protein, including albumin, as a source of amino acids in spontaneously arising pancreatic tumors in vivo.
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
RESULTS
Isotope-labeled mouse serum albumin [15N]MSA was produced using the yeast Pichia pastoris17,19,20 (Supplementary Fig. 1a–c) in order to determine whether albumin could be used as a nutrient source by tumors in vivo. Exchanging endogenous albumin in the plasma for labeled albumin is necessary to obtain sufficient enrichment of labeled protein over reasonable infusion times to trace the fate of labeled albumin in tissues. Because albumin has a large volume of distribution in the body and slow turnover (approximately 1–2% total per day)11,21, we designed a plasmapheresis approach to exchange endogenous albumin with [15N]MSA using a modified perfusion based microfluidic strategy22,23 in mice with and without spontaneously arising pancreatic ductal adenocarcinoma (PDAC) (Supplementary Fig. 1d, e). PDAC was initiated by pancreas restricted Cre-recombinase (Pdx1-Cre)
expression in KrasLSL-G12D/+; p53loxP/loxP (KP) mice to induce mutant KrasG12D expression and loss of p53.
Following plasmapheresis of control (WT) and KP mice, we achieved an initial plasma enrichment of approximately 45% +/− 9.2% labeled albumin (Fig. 1a) with minimal total protein loss from the plasma (Supplementary Fig. 2a). Despite slightly lower basal levels of protein in KP plasma as compared to WT mice, albumin levels were unchanged even in end-stage disease in mice with high tumor burden (Supplementary Fig. 2b). We reasoned that if albumin is being catabolized, [15N]-labeled amino acids derived from albumin may be present in the plasma of these animals after plasma exchange (Fig. 1b). Increased levels of labeled alanine, leucine, and phenylalanine were observed in the plasma of tumor-bearing KP mice as compared to WT control mice approximately 12 hours after plasma exchange indicating the presence of albumin-derived amino acids in the blood and supporting previous work suggesting that whole body protein turnover is increased in KP animals with
pancreatic tumors24 (Fig. 1b). To determine the site(s) of albumin catabolism, we first examined labeled albumin peptides and free amino acids in normal pancreas from WT mice and PDAC tissue from KP mice. Labeled albumin peptides were increased in tumor relative to normal pancreas tissue, suggesting that labeled albumin uptake is increased in tumors (Fig. 1c). Next, we examined free amino acids in tumor and normal pancreas following plasma-exchange. We observed a significant increase in many labeled free amino acids in the tumor relative to the normal pancreas demonstrating that amino acids from the breakdown of labeled albumin are enriched in the tumor relative to normal pancreas (Fig. 1d).
To determine whether albumin-derived amino acids might be generated outside the pancreas through catabolism at distant sites, we examined labeled peptides and free amino acids in the liver, lung, and muscle of normal (WT) and tumor-bearing (KP) animals (Fig. 1e, f and Supplementary Fig. 2c–f). In contrast to the findings in pancreatic tumors compared to normal pancreas, we observed no significant increase in albumin peptides in the livers of mice with pancreatic cancer relative to mice without pancreatic cancer (Fig. 1e). Some 15 N-labeled free amino acids were increased in the livers of animals with pancreatic cancer (Fig. 1f) and we also observed elevation in some labeled free amino acids in the lungs of tumor-bearing animals (Supplementary Fig. 2d). However labeled amino acids were not increased in skeletal muscle of mice with PDAC, and neither the lungs nor the skeletal muscle showed
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
an increase in albumin peptides (Supplementary Fig. 2c, e). Together, the findings suggest that pancreatic tumors uptake more albumin than normal pancreatic tissues, and amino acids from the breakdown of albumin contribute more to the tumor than normal pancreas. The presence of labeled amino acids in the liver and other organs (Supplementary Fig. 2c–f) leaves open the possibility that the labeling of amino acids in pancreatic tumor tissue results from a combination of local albumin catabolism and/or the uptake of free labeled amino acids derived from albumin breakdown in non-tumor tissues.
To determine whether tumor-autonomous albumin catabolism is a feature of pancreatic tumors, we used an implantable device for delivery of small and large molecules into solid tissues (Fig. 2a). The device was adapted from [18] by modifying its diameter and reservoir sizes to allow for more effective release of large molecules into the pancreas. This device allows direct, controlled delivery of materials into the tumor microenvironment in a defined location that can be tracked via microscopy. Macropinocytosis can be visualized by tracking the fate of fluorescently labeled high molecular weight dextran (70kDa) a previously demonstrated marker of macropinocytosis25. Protein degradation can be assessed by employing self-quenched, BODIPY-labeled albumin (DQ-BSA), which becomes fluorescent upon enzymatic cleavage in the lysosomes of cells26.
Devices loaded with 1μg of Rh-Dextran and DQ-BSA were implanted into subcutaneous xenografts derived from the MIA PaCa-2 (mutant RASV12) and BxPC-3 (WT RAS) human PDAC cell lines. These two cells lines have been reported previously to differ in their use of macropinocytosis for proliferation in vivo8. The implanted devices allowed local release of Rh-Dextran and DQ-BSA adjacent to reservoirs after one day (Fig. 2b). We tracked the fate of DQ-BSA and 70kDa Rh-dextran delivered by the micro-devices to investigate whether internalization of proteins by macropinocytosis in cells can be observed in these xenograft tumors (Fig. 2b, c, d). 24h after device implantation, high levels of dextran uptake and DQ-BSA fluorescence are observed to ~650μm from the device in MIA PaCa-2 tumor tissue (Fig. 2b). Dextran was also released into BxPC-3 tumors, exhibiting a similar gradient of intratumoral release as in MIA PaCa-2 tumors (Fig. 2c). Combined local release with EIPA, an agent known to inhibit the formation of macropinosomes27, eliminated dextran uptake in MIA PaCa-2 tumors (Fig. 2c). We monitored DQ-BSA fluorescence over two days to determine whether the observed Dextran diffusion is accompanied by cellular uptake by tumor cells and to further examine the kinetics of this degradation. Bright narrow regions of fluorescence observed in MIA PaCa-2 tumors indicate local protein degradation that appeared approximately 150μm from the device by 24h after device implantation (Fig. 2d). At 48h, fluorescent regions appeared at approximately 400μm from the device (Fig. 2d). The observed diffusion distances are congruent with previously observed intratumoral release studies of several large and small molecules18. In contrast, intratumoral fluorescence from DQ-BSA was not observed when identical devices were placed into BxPC-3 xenograft tumors (Fig. 2d). These findings argue that local delivery of DQ-BSA allows determination of intratumoral albumin catabolism by macropinocytosis.
To determine if we can observe uptake and catabolism of protein in real time, we utilized live multiphoton imaging of fluorescence from both DQ-BSA and 70kDa Rh-Dextran released from micro-devices in MIA PaCa-2 and BxPC-3 tumors. We observed uptake of
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
dextran and an increase in fluorescent signal from DQ-BSA in cells within xenografted MIA PaCa-2 tumors in mice (Fig. 2e). Dextran uptake is observed in the majority of tumor cells present within the exposed region at one-hour post-device implantation. Uptake of albumin by tumor cells occurs at steady rates, with approximately 2/3 of the cells in an exposed region showing internalized Rh-Dextran and DQ-BSA fluorescence within 3 hours of device implantation (Fig. 2e), a time consistent with the presence of albumin-derived amino acids observed in tumors in mice (Fig. 1). In contrast, the BxPC-3 cell line exhibited Rh-Dextran labeling only of cell outlines, and few punctate spots from DQ-BSA were observed
indicating minimal albumin degradation in these tumors (Fig. 2e). Images from the DQ-BSA studies were also used to determine “macropinocytic index”, a metric previously described to quantitate macropinocytosis in fixed samples28. By this metric, MIA PaCa-2 cells exhibit 4-fold higher levels of macropinocytosis relative to BxPC3 cells (Fig. 2f).
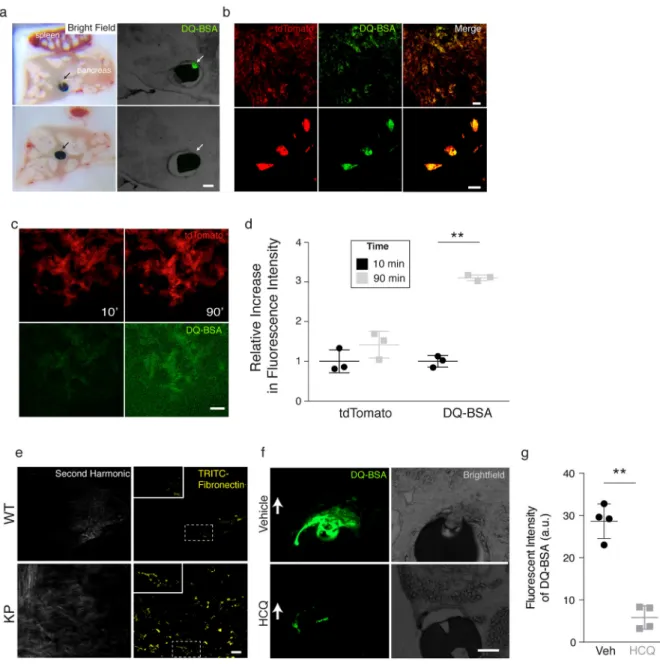
To assess macropinocytosis and local protein degradation in KP tumors, microdevices containing DQ-BSA were surgically implanted directly into the pancreas of 8-week old KP mice harboring autochthonous pancreatic tumors using a biopsy needle. In 8-week old KP mice the pancreas appears as a heterogeneous array of tumor and normal tissue24,29 (Fig. 3a and Supplementary Fig. 3a). 24h after device delivery, tissue adjacent to device reservoirs containing DQ-BSA exhibits fluorescence only when reservoirs were adjacent to tumor tissue (Fig. 3a). No fluorescence of DQ-BSA was detected in non-tumor pancreas regions, suggesting an absence of albumin degradation at these sites. We next sought to determine the kinetics of dextran uptake and DQ-BSA degradation and whether these processes occurred in cells with K-rasG12D activation within the pancreas. To distinguish Cre-expressing cells, multi-photon live microscopy was utilized in a KP mouse that also harbored a
tdTomatoLSL/+ allele as a fluorescent marker of Cre-recombinase activity and, therefore, K-rasG12D activation. Using multi-photon intravital microscopy, we observed a high degree of co-localization of fluorescent tdTomato signal and fluorescent DQ-BSA signal (Fig. 3b) and an increase in fluorescence due to DQ-BSA degradation was observed over a 90-minute time period in tumor cells with tomato fluorescence (Fig. 3c, d). When a microdevice is
introduced into a KP mouse lacking the tdTomato allele comparable fluorescence from DQ-BSA is observed in tumor tissue, arguing that the observed DQ-DQ-BSA is not the result of overlap in the tdTomato and DQ-BSA emission/excitation spectra (Supplementary Fig. 3b). Based on DQ-BSA fluorescence, we further observed that the spontaneous KP tumors exhibit a level of macropinocytosis comparable to that observed in MIA PaCa-2 xenografts (Supplementary Fig. 3c).
One hallmark of pancreatic tumors, including tumors found in KP mice, is a dense stroma that is rich in extracellular matrix (ECM)30,31. To determine whether tumor cells can internalize ECM-derived proteins, we utilized the microdevice to deliver TRITC-labeled fibronectin (440kDa), one of the largest protein components of the extracellular matrix. Using intravital microscopy in live mice, we observed clear uptake of fibronectin in tumor tissue in KP mice and not in the WT pancreas (Fig. 3e). Taken together, these observations suggest that cells in spontaneously arising pancreatic tumors can consume protein in their environment.
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
Hydroxychloroquine is a lysosomal inhibitor being studied as a potential therapy for PDAC32. To determine whether we could inhibit catabolism of DQ-BSA, we delivered devices with reservoirs containing DQ-BSA and vehicle or hydroxychloroquine. We observed decreased DQ-BSA signal around reservoirs with hydroxychloroquine as compared to vehicle reservoirs (Fig. 3f, g), suggesting that the lysosome is involved in the catabolism of DQ-BSA in PDAC cancer cells in tumors.
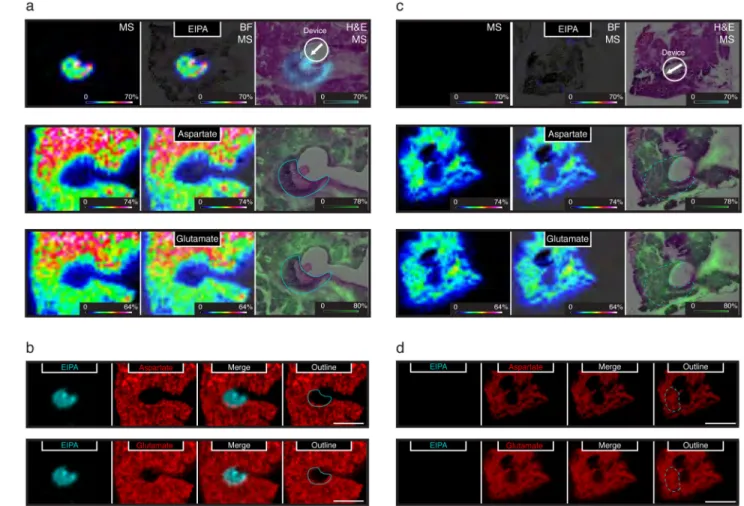
To determine whether inhibiting macropinocytosis with EIPA had an effect on amino acid levels in pancreatic tumors, we implanted devices containing EIPA or a vehicle control into the pancreas of 7–8 week old KP mice for 24 hours. After tumor harvesting, tissue slices adjacent to EIPA or vehicle containing reservoirs were analyzed using matrix-assisted laser desorption ionization coupled to imaging mass spectrometry (MALDI-IMS). This enabled the simultaneous measurement of EIPA distribution from the device into tumor tissue and how relative levels of amino acids might differ spatially within the tissue. We observed no change in the relative concentrations of choline, a metabolite that is not expected to be affected by EIPA treatment, in tumor tissue regions that were exposed to EIPA
(Supplementary Fig. 4a). A robust signal for the amino acids glutamate, aspartate, glutamine and histidine were also detected in the tissue slices. The relative concentrations of all four amino acids were decreased in tissue exposed to EIPA as compared to the adjacent tissue that was not exposed to EIPA, and amino acid levels were not changed in tumor tissue exposed to vehicle alone (Fig. 4 and Supplementary Fig. 4b). Importantly, the regions of tissue analyzed were confirmed to be tumor tissue by H&E stain, with relative
concentrations of the amino acids detected found to be anti-correlated with regions of tumor exposed to EIPA suggesting that exposure to this compound resulted in local amino acid depletion (Fig. 4a, b and Supplementary Fig. 4b). In tumor regions adjacent to vehicle control reservoirs, the relative concentration of aspartate and glutamate were uniform as compared to the rest of the tissue section (Fig. 4c). Using the orientation of device placement, we examined the approximate region of vehicle delivery to tumor tissue (400– 500mM), and no differences in aspartate or glutamate levels were observed around the predicted region of vehicle delivery (Fig. 4d). Thus, the decreased levels of amino acids in the EIPA-exposed regions of the tumor are consistent with the hypothesis that tumors derive a meaningful fraction of their amino acids from catabolism of extracellular protein obtained by macropinocytosis.
EIPA inhibits macropinocytosis at the concentrations used for these studies7, 8; however, EIPA is also an inhibitor of Na+/H+ exchange and may affect amino acid levels by other mechanisms. To test this possibility, we determined whether EIPA affected branched-chain amino acid (BCAAs) levels under conditions where past work had shown acquisition of BCAAs is or is not dependent on macropinocytosis33,34. We found that BCAA levels were decreased when pancreatic cancer cells were cultured in media lacking BCAAs, that BCAA levels could be rescued by providing cells with excess albumin, and the ability to rescue BCAA levels with albumin was inhibited by EIPA while EIPA had no effect on BCAA levels otherwise (Supplementary Fig. 4c). We also assessed whether EIPA affected intracellular amino acid levels in pancreatic cancer cells in culture where amino acids are freely available, and whether EIPA affected uptake of labeled amino acids from the media. EIPA had no effect on levels of most amino acids under these conditions (Supplementary
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
Fig. 4d, e), including glutamine, glutamate and aspartate, three of the amino acids found to be depleted by EIPA in tumors. These data argue that at the doses use, EIPA has minimal effect on the uptake or exchange of free amino acids and strengthens the conclusion that macropinocytosis is important to maintain amino acid levels within tumors.
In contrast to previous studies examining albumin synthesis and degradation by different organs in tumor bearing animals13,15,35, we provide direct evidence that pancreatic tumors catabolize albumin and derive a fraction of amino acids from the breakdown of extracellular protein. While these findings remain consistent with breakdown of host tissue proteins as a nutrient contributor to pancreatic tumors24, they further suggest that cancer cells in these tumors catabolize host-derived proteins in their environment. Hypoalbuminemia was not observed in KP mice, suggesting that the quantity of albumin catabolized by tumors is not sufficient to directly result in hypoalbuminemia over the time scales this process was studied in mice. However, increased turnover of whole body protein stores and albumin
consumption by tumors may eventually affect circulating albumin levels in patients with pancreatic cancers12.
We report two methods to interrogate albumin catabolism that could be extended to other solid tumors. Labeling of albumin on specific amino acids could also enable finer mapping of the fate of protein-derived amino acids in central carbon metabolism. Using albumin as a carrier for chemotherapeutics has been a successful therapeutic strategy in the clinic9,36, possibly owing to the propensity of some tumors to use macropinocytosis. Albumin conjugation and preferential albumin-uptake can allow for both better solubility and improved tumor delivery37. The approaches used here to study local protein catabolism may also allow for direct testing of therapeutic hypotheses in an individual tumor, and could be used at the time of biopsy to identify patients that might benefit from treatments that take advantage of increased protein catabolism by tumors.
METHODS
Animal Statement
All animal studies were approved by the MIT Committee on Animal Care. Mice were housed under standard 12-hr light/dark cycles and fed RMH 3000 (Prolab) ad-libitum until experiments were conducted. All experimental groups were assigned based on genotype and experiments conducted blinded. Plasma exchange experiments were conducted at the same time of day. Cohort sizes were based on anticipated results from previous studies in vitro8.
Production of [15N]MSA
The MSA cDNA was cloned from the liver of a wild-type mouse using the following PCR primers: (F-GAAGCACACAAGAGTGAGATCGC;
R-TTAGGCTAAGGCGTCTTTGCATCTA). The cDNA was cloned into an expression vector containing α-factor and a GAPDH promoter (pGAPZα) for subsequent transformation and expression in Pichia pastoris (Invitrogen). P. pastoris expressing MSA was cultured in a glucose fed-batch bioreactor (Sartorius) at 30°C for 7 days with [15N]labeled nutrients using conditions described previously38. Media containing MSA was separated from the cellular
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
fraction after centrifugation at 15,000xg for 3 hours at 4° C and supernatant containing MSA was concentrated using a rotary evaporator. Concentrated supernatant was dialyzed using a 45,000 Dalton molecular weight cutoff dialysis cassette (Thermo-Fisher) and further purified using a Q-Sepharose High Performance anion exchange column (GE). Purified MSA were subsequently lyophilized and resuspended in PBS and filtered through 0.45μM filters for use in animal experiments. Additional [15N]MSA was acquired from Albumin Biosciences, INC.
Microfluidic Plasmapheresis Device
The plasma exchange microfluidic device is based on the concept of cell margination and was previously developed for bacterial clearance in a mouse model of sepsis22,23. The device is made of polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning, Midland, MI) and consists of 3 channels; each 6 mm long, 20μm × 40μm (W×H) with 3 stages of bifurcation at 2 mm channel interval. A filter region with an array of square pillars (100μm gap) is added upstream of the margination channels to trap large thrombi or cell clusters to ensure smooth flow within the device. As blood flows through the margination channel, deformable RBCs migrate axially towards the channel center, leaving a layer of plasma next to the channel wall which is removed via the smaller side channels. To minimize RBC loss, the side channel skimming volume is progressively lower (15%, 10%, 5%) at each bifurcation stage with a total volume of ~30%. Therefore, the microdevice is capable of exchanging up to ~30% of plasma from the incoming blood with labeled albumin, which is infused to the filtered blood with a separate syringe pump. During the extracorporeal plasma exchange in mice, blood is drawn from an arterial (carotid) catheter using a peristaltic pump (P720, Instech Laboratories, Inc., Plymouth Meeting, PA) for ~25 minutes (30μLmin−1) into the device which would partially replace the filtered blood with labelled albumin (10μLmin−1) prior returning to the animal through a venous (jugular) catheter.
Mouse Pancreatic Cancer Models
For the autochthonous genetically engineered mouse model of pancreatic ductal
adenocarcinoma (PDAC), tumors were initiated using a pancreas-restricted Pdx1 promoter to express Cre-recombinase (Pdx1-Cre) in KrasLSL-G12D/+; p53loxP/loxP animals from a mixed 129/Sv and C57Bl6/J background. For imaging studies, mice harboring a tdTomatoLSL/+ allele were used to allow visualization of individual cells where Cre was expressed. Chronic catheters were implanted in the jugular vein and carotid artery when male mice were between 6–8 weeks of age with detectable tumors. For xenograft studies: two to four-month old male C57Bl6/J or nu/nu mice were used. Briefly, 2.5×105 established cell lines derived from C57Bl6/J KP mice, or 5×105 MIA PaCa2 or BxPC-3 human
pancreatic cancer cell lines, were injected into the flanks of recipient mice and experiments performed when total tumor volume was approximately 1cm3. All human cell lines were acquired from ATCC and cultured in 21% oxygen in DMEM containing 10% FBS and penicillin/streptomycin.
Metabolite Extraction
Tissues were weighed (wet tissue weight) and homogenized cryogenically (Retsch Cryomill). Metabolites were extracted from tissues weighing between 10–40mg in a
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
chloroforom:methanol:water extraction solution at a volume ratio of 400:600:300. Samples were vortexed and centrifuged at 10,000×g for 10min to separate aqueous and organic layers. Polar metabolites were dried under nitrogen gas and frozen at −80 °C for subsequent and analysis by gas-chromatography mass-spectrometry.
Measurement of Labeled Albumin Peptides
Mouse tissue was extracted and homogenized in ice-cold 8 M urea. Proteins were quantified using BCA assay (Pierce). Proteins were reduced (10 mM dithiothreitol, 56 °C for 45 min) and alkylated (50 mM iodoacetamide, room temperature in the dark for 1 h), then were subsequently digested with trypsin (sequencing grade, Promega, Madison, WI) at an enzyme/substrate ratio of 1:50 at room temperature overnight in 100 mM ammonium acetate pH 8.9. Trypsin activity was quenched by adding formic acid to a final concentration of 5%. Peptides were desalted using C18 SpinTips (Protea, Morgantown, WV) then lyophilized and stored at −80 °C. After being resuspended in 0.1% formic acid, each sample (1ug of tissue or 100 ng of serum) was separated by reverse phase HPLC using an EASY- nLC1000 (Thermo) over a 75-minute gradient before nanoelectrospray using a QExactive mass spectrometer (Thermo). The mass spectrometer was operated in a data-dependent mode. The parameters for the full scan MS were: resolution of 70,000 across 350–2000 m/z, AGC 3e6, and maximum IT 50 ms. The full MS scan was followed by MS/MS for the top 10 precursor ions in each cycle with a NCE of 28 and dynamic exclusion of 30 s. Raw mass spectral data files (.raw) were searched using Proteome Discoverer (Thermo) and Mascot version 2.4.1 (Matrix Science). Mascot search parameters were: 10 ppm mass tolerance for precursor ions; 0.8 Da for fragment ion mass tolerance; 2 missed cleavages of trypsin; fixed modification was carbamidomethylation; variable modification was oxidized methionine. We analyzed the following peptides and all possible isotopomers to determine plasma enrichment and quantity of labeled albumin present in each tissue: AADKDTcFSTEGPNLVTR (+2), AADKDTcFSTEGPNLVTR (+3), APQVSTPTLVEAAR (+2), ENYGELADccTK (+2), LVQEVTDFAK (+2), SLHTLFGDK (+2), TcVADESAANcDK (+2) (c =
carbamidomethylation). Fractional enrichment was determined for each peptide and each measurement represents the mean of the 7 peptides. The average fractional enrichment in plasma and tissues were normalized to the average fractional plasma enrichments of [15N]MSA post plasma-exchange.
Gas-Chromatography Mass-Spectrometry Measurement of Free Amino Acids
Dried tissue metabolites were dissolved in 10uL/10mg of 2% methoxyamine hydrochloride in pyridine (Sigma) and held at 37C for 1.5h. Tert-butyldimethylsilyl derivatization was initiated by adding 15uL/10mg of N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide + 1% tert-butyldimethylchlorosilane (Sigma) and incubated at 37C for 1hr. GC-MS analysis was performed using an Agilent 7890 GC equipped with 30m DB-35MS capillary column connected to an Agilent 5975B MS operating under electron impact ionization at 70eV. One microliter of sample was injected in splitless mode at 270C, using helium as a carrier gas at a flow rate of 1mL min−1. For measurement of polar metabolites, the GC oven temperature was held at 100C for 3mins and increased to 300C at 3.5C min−1. The MS source and quadrupole were held at 230C and 150C, respectively, and the detector was run in scanning mode, recording ion abundance in the range of 100–605m/z. MIDs were determined by
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
integrating the appropriate ion fragments and correct for natural isotope abundance using in house algorithms adapted as previously reported (METRAN, Matlab 2013b)8. Labeled amino acids are defined as the M+1 (one nitrogen labeled) and M+2 (two nitrogens labeled) isotopomers of examined amino acids for 15N labeled studies or M+n (where n= total carbon atoms composing each amino acid) for 13C labeled studies.
Albumin Measurements in Plasma
Plasma was collected in EDTA-coated tubes, aliquoted, and frozen at −80°C for further analysis. Plasma mouse albumin levels were determined using a mouse albumin ELISA assay according to manufacturers’ specification (Abcam #ab108791).
Device to Deliver FITC DQ-BSA, Rh-Dextran, FITC-Dextran, TRITC-Fibronectin, EIPA, and Hydroxychloroquine
Microdose drug delivery devices were manufactured as described previously18. In short, cylindrical, micro-scale devices with 820μm (diameter) × 4mm (length) were manufactured from medical-grade Delrin acetyl resin blocks (DuPont) by micro-machining (CNC Micromachining Center, Cameron). Circular reservoirs (2–6 per device) were shaped on the outer surface of devices in dimensions of 300μm (diameter) × 250μm (depth). DQ-BSA, Dextran, Fibronectin and/or hydroxychloroquine or EIPA (1μg per reservoir) were packed into device reservoirs using a tapered, metal needle (Electron Microscopy Sciences). Devices were implanted directly into the mouse tumor using a 19-gauge spinal biopsy needle (Angiotech) using the retractable needle obturator to push the device into tissue. Devices remained in situ for 6–48h. The tumor was then excised and the tissue containing the device was snap-frozen. Tissue was sectioned using a standard cryotome and tissue slices of 20μm thickness were collected from each reservoir and imaged on an EVOS microscope.
Multi-photon Imaging of DQ-BSA, FITC-Dextran, Rh-Dextran, TRITC-Fibronectin
Intravital multiphoton imaging was performed as described previously39,40 using a 25x 1.05NA water immersion objective with correction lens. Ninety-minute long time-lapse movies were analyzed for measuring and quantifying fluorescence intensity and cell characteristics in 3D and through time using NIH ImageJ41. Data were pooled from at least 3 mice per tumor group, with 4–8 fields imaged per mouse. Macropinocytic index was determined as previously described for xenograft tumors 28.
MALDI Imaging experiments
Samples were cryosectioned at 20 μm, thaw-mounted on glass slides, and serial sections analyzed. The orientation of specimens was shifted for serial sections analyzed in positive versus negative ion mode, with device location used to orient the sample in each serial section. Slides were then coated with HCCA (7 mg/mL, 50% methanol, 0.1% TFA) or 9-aminoacridine (10 mg/mL, 70% ethanol, 0.1% TFA) using a TM sprayer (HTX
Technologies, Carrboro, NC, USA). Serial sections were prepared for imaging analysis in both positive and negative ion modes on a 7T solariX-XR FTMS (Bruker, Billerica, MA, USA) equipped with dual ESI/MALDI source, SmartBeam II 2 kHz Nd:YAG (355 nm) laser, and paracell. Samples were analyzed with a raster width of 125 μm in positive ion
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
mode and 100 um in negative ion mode in the mass range of 80 – 2000 m/z at 2MW acquisition size. Images contained between 7200 and 10500 pixels. Data was visualized and co-registration of H&E images performed using FlexImaging 4.1. Compound identifications were made on the basis of accurate mass (<1ppm) and isotopic peak matching.
Cell Culture
Cell lines from murine KP pancreatic tumors were established by standard protocols24. Cells were cultured in complete DMEM with 10% fetal bovine serum and 5% penicillin/
streptomycin. For BCAA deprivation, DMEM with 5% dialyzed serum with or without BCAAs (leucine, valine, and isoleucine) was used in the presence or absence of 3% albumin and in the presence or absence of 20μM EIPA. Where indicated, 2mg/mL of a fully
labeled 15N or 13C labeled amino acid mixture (Cambridge Isotopes Laboratories) was included for 6 hours prior to metabolite extraction.
Statistical Analysis
Two-tailed unpaired Student’s T-test was performed for all experiments and data were normally distributed for all analyses conducted. Variances were not statistically different in any of the data. Results for independent experiments are presented as mean ± SD.
Supplementary Material
Refer to Web version on PubMed Central for supplementary material.
Acknowledgments
The authors wish to dedicate this paper to the memory of Katherine Kellersberger. We thank the MIT Department of Chemical Engineering and Jean Francois-Hamel for access to bioreactor equipment, Roderick Bronson for pathological grading of tumors, and Allison Lau for providing tumor-bearing animals. S.M.D. and A.L. received support from a National Science Foundation Graduate Research Award Fellowship, and support from T32 GM007287 is also acknowledged. O.J received support from the Koch Institute Frontier Grant and the Prostate Cancer Foundation. JRM acknowledges support from grant F30CA183474 from the NCI and T32GM007753 from NIGMS. MAK and GS acknowledge support from NIH grants 1R01DK075850-01 and 1R01CA160458-01A1. M.G.V.H acknowledges support from the Lustgarten Foundation, the Ludwig Center at MIT, the Broad Institute SPARC program, the Burroughs Wellcome Fund, SU2C, and the NIH (P30CA1405141, R01CA168653).
References
1. Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013; 49:388–398. [PubMed: 23395269]
2. Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem Sci. 2015; 40:130–140. [PubMed: 25639751]
3. White E. Exploiting the bad eating habits of Ras-driven cancers. Genes & Development. 2013; 27:2065–2071. [PubMed: 24115766]
4. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009; 324:1029–1033. [PubMed: 19460998]
5. Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer. 2013; 13:572–583. [PubMed: 23822983]
6. Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010; 35:427–433. [PubMed: 20570523]
7. Kamphorst JJ, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015; 75:544–553. [PubMed: 25644265]
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
8. Commisso C, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013; 497:633–637. [PubMed: 23665962]
9. Schilling U, et al. Design of compounds having enhanced tumour uptake, using serum albumin as a carrier—part II. In vivostudies. Int J Rad Appl Instrum B. 1992; 19:685–695. [PubMed: 1522023] 10. Steinfeld JL. I131 albumin degradation in patients with neoplastic diseases. Cancer. 1960
11. Rothschild MA, Oratz M, Schreiber SS. Regulation of albumin metabolism. Annu Rev Med. 1975; 26:91–104. [PubMed: 1096784]
12. Gupta D, Lis CG. Pretreatment serum albumin as a predictor of cancer survival: a systematic review of the epidemiological literature. Nutr J. 2010; 9:69–69. [PubMed: 21176210] 13. Andersson C, Iresjö BM, Lundholm K. Identification of tissue sites for increased albumin
degradation in sarcoma-bearing mice. J Surg Res. 1991; 50:156–162. [PubMed: 1990221] 14. Brenner DA, Buck M, Feitelberg SP, Chojkier M. Tumor necrosis factor-alpha inhibits albumin
gene expression in a murine model of cachexia. J Clin Invest. 1990; 85:248–255. [PubMed: 2295699]
15. Fearon KC, et al. Albumin synthesis rates are not decreased in hypoalbuminemic cachectic cancer patients with an ongoing acute-phase protein response. Ann Surg. 1998; 227:249–254. [PubMed: 9488524]
16. Jewell WR, Krishnan EC, Schloerb PR. Apparent Cellular Ingress of Albumin in Walker 256 Tumor and Rat Muscle. Cancer Res. 1975; 35:405–408. [PubMed: 1109805]
17. Swanson JA, Watts C. Macropinocytosis. Trends in Cell Biology. 1995; 5:424–428. [PubMed: 14732047]
18. Jonas O, et al. An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci Transl Med. 2015; 7:284ra257.
19. Daly R, Hearn MTW. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit. 2005; 18:119–138. [PubMed: 15565717]
20. Tolner B, Smith L, Begent RHJ, Chester KA. Production of recombinant protein in Pichia pastoris by fermentation. Nat Protoc. 2006; 1:1006–1021. [PubMed: 17406338]
21. Merlot AM, Kalinowski DS, Richardson DR. Unraveling the mysteries of serum albumin-more than just a serum protein. Front Physiol. 2014; 5:299. [PubMed: 25161624]
22. Hou HW, et al. Deformability based cell margination—A simple microfluidic design for malaria-infected erythrocyte separation. Lab Chip. 2010; 10:2605–2609. [PubMed: 20689864]
23. Hou HW, et al. A microfluidics approach towards high-throughput pathogen removal from blood using margination. Biomicrofluidics. 2012; 6:24115–2411513. [PubMed: 22655023]
24. Mayers JR, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014; 20:1193–1198. [PubMed: 25261994] 25. Racoosin EL, Swanson JA. Macropinosome maturation and fusion with tubular lysosomes in
macrophages. J Cell Biol. 1993; 121:1011–1020. [PubMed: 8099075]
26. Klionsky DJ. Autophagy in mammalian systems, Part B. Preface. Meth Enzymol. 2009; 452:xxi– xxii. [PubMed: 19200871]
27. Koivusalo M, et al. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J Cell Biol. 2010; 188:547–563. [PubMed: 20156964] 28. Commisso C, Flinn RJ, Bar-Sagi D. Determining the macropinocytic index of cells through a
quantitative image-based assay. Nat Protoc. 2014; 9:182–192. [PubMed: 24385148] 29. Bardeesy N, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of
pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006; 103:5947–5952. [PubMed: 16585505]
30. Whatcott CJ, et al. Desmoplasia in Primary Tumors and Metastatic Lesions of Pancreatic Cancer. Clin Cancer Res. 2015; 21:3561–3568. [PubMed: 25695692]
31. Mahadevan D, Von Hoff DD. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2007; 6:1186–1197. [PubMed: 17406031]
32. Manic G, Obrist F, Kroemer G, Vitale I, Galluzzi L. Chloroquine and hydroxychloroquine for cancer therapy. Molecular & Cellular Oncology. 2014; 1:e29911–29912. [PubMed: 27308318]
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
33. Palm W, et al. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell. 2015; 162:259–270. [PubMed: 26144316]
34. Kamphorst JJ, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. in. Cancer Res. 2015; 75:544–553. [PubMed: 25644265] 35. Andersson C, Lönnroth C, Moldawer LL, Ternell M, Lundholm K. Increased degradation of
albumin in cancer is not due to conformational or chemical modifications in the albumin molecule. J Surg Res. 1990; 49:23–29. [PubMed: 2359290]
36. Von Hoff DD, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013; 369:1691–1703. [PubMed: 24131140]
37. Wosikowski K, et al. In vitro and in vivo antitumor activity of methotrexate conjugated to human serum albumin in human cancer cells. Clin Cancer Res. 2003; 9:1917–1926. [PubMed: 12738750] 38. Cregg JM, Cereghino JL, Shi J, Higgins DR. Recombinant protein expression in Pichia pastoris.
Mol Biotechnol. 2000; 16:23–52. [PubMed: 11098467]
39. Wang W, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002; 62:6278–6288. [PubMed: 12414658]
40. Wyckoff J, Gligorijevic B, Entenberg D, Segall J, Condeelis J. High-resolution multiphoton imaging of tumors in vivo. Cold Spring Harb Protoc. 2011; 2011:1167–1184. [PubMed: 21969629]
41. Sahai E, et al. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnology. 2005; 5:14. [PubMed: 15910685]
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
Figure 1. Albumin-derived amino acids are found in pancreatic tumors
a. Representative albumin enrichment in the plasma of WT and KP mice following plasma exchange of [15N]-labeled mouse serum albumin ([15N]MSA) for endogenous albumin as assessed by analysis of labeled albumin peptides. The %[15N]MSA shown was obtained by analysis of the albumin peptide LVQEVTDFAK in plasma by LC-MS/MS over time during and after plasma exchange (n = 5 per genotype). The time indicated by the double dagger corresponds to the 30-minute period of plasma exchange.
b. Labeled amino acids in the plasma of WT and KP mice following plasma exchange of [15N]-labeled MSA for endogenous MSA. Plasma was sampled at the time of tissue collection (~12 hours post-exchange). The presence of [15N]-labeled amino acids was determined by GC-MS, (n = 5 per genotype).
c. Following plasma exchange of [15N]-labeled MSA for endogenous MSA in WT and KP mice, the presence of [15N]MSA in tissue was determined by analysis of multiple labeled peptides from normal pancreas (WT) or pancreatic tumors (KP) by LC-MS/MS, (n = 5 per genotype).
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
d. The presence of labeled amino acids in the pancreas (WT) or tumor (KP) from WT and KP mice respectively following plasma exchange of [15N]-labeled MSA for endogenous MSA. Tissues were collected ~12 hours post-exchange and labeled amino acids determined by GC-MS (n = 5 per genotype).
e. Following plasma exchange of [15N]-labeled MSA for endogenous MSA in WT and KP mice, the presence of [15N]MSA in tissue was determined by analysis of multiple labeled peptides from livers of animals with pancreatic tumors (KP) or without pancreatic tumors (WT) by LC-MS/MS, (n = 5 per genotype).
f. The presence of labeled amino acids in the livers of WT or KP mice ~12 hours following plasma exchange of [15N]-labeled albumin for endogenous albumin. Labeled amino acids were determined by GC-MS.
For all panels, (Ala = alanine; Asp = aspartate; Cys = cysteine; Gln = glutamine; Glu = glutamate; Gly = glycine; Ile = isoleucine; Leu = leucine; Lys = lysine; Met = methionine; Phe = phenylalanine; Ser = serine; Thr = threonine; Val = valine), significance differences are noted as P * < 0.05; ** < 0.01; ***<0.001; n.s. difference not significant, by unpaired t-test.
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
Figure 2. Direct assessment of macropinocytosis and albumin catabolism in tumors
a. Schematic depicting device implanted directly into the tumor to release specified contents from reservoirs into distinct tissue regions. Radially outward diffusion of compounds from reservoirs is shown in 3-dimensional (left) and cross-sectional views (right). The orientation of the schematic on the right matches the orientation of device placement relative to tissue in panels b, c, and d.
b. Combined delivery of 70 kDa rhodamine-dextran (Rh-Dextran) and self-quenched, BODIPY-labeled bovine serine albumin (DQ-BSA). Both compounds are released from a single reservoir in MIA PaCa-2 tumors. Radial transport of compound is ~150 μm from the device into tissue after a 24h incubation period. Scale bar, 200 μm. (images are
representative of n = 6 from mice per cell line per condition with triplicate reservoirs). c. Delivery of 70 kDa Rh-Dextran from the device into MIA PaCa-2 and BxPC-3 tumor tissue at 48h. Unlike MIA PaCa-2 tumors, BxPC-3 tumors have been shown previously not to engage in macropinocytosis8. Tissue penetration up to ~550 μm is observed in each tumor. Co-delivery of 70kDa Rh-Dextran with the macropinocytosis inhibitor EIPA leads to reduced Rh-Dextran uptake in MIA PaCa-2 tumor tissue as shown. Scale bar, 150 μm. (images are representative of n = 6 from two mice per cell line per condition with triplicate reservoirs).
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
d. Comparison of albumin breakdown in MIA PaCa-2 and BxPC-3 tumor sections. Devices containing DQ-BSA were implanted into MIA PaCa-2 and BxPC-3 tumors as indicated. Degradation of BSA, indicated by fluorescence signal from DQ-BSA, increases over time in MIA PaCa-2 tumors a depth of 200μm from the device after 1 day, and up to 350μm from the device after 2 days. Scale bar, 200 μm. (images are representative of n = 6 from two mice per cell line per condition with triplicate reservoirs).
e. Examination of albumin and dextran uptake in MIA PaCa-2 and BxPC-3 tumors. Multiphoton images of live tumors implanted with devices containing 70kDa Rh-Dextran and DQ-BSA. Fluorescence from DQ-BSA is observed in individual cells in MIA PaCa-2 tumors by 4h after device implantation, and high concordance is observed between cells that display Rh-Dextran uptake and DQ-BSA fluorescence. Scale bar, 20 μm. (images are representative of n = 2 mice per cell line per condition with duplicate reservoirs). f. Quantification of relative macropinocytic index in MIA PaCa-2 and BxPC-3 xenograft tumors based on DQ-BSA fluorescence. Macropinocytic index is a ratio of the total area of all macropinosomes in a field of designated area (n=5 distinct fields per cell line). For all panels, white arrows indicate direction of delivery of DQ-BSA, Rh-Dextran, or EIPA. (data from n = 2 mice per cell line per condition with duplicate reservoirs used to calculate macropinocytic index). Significance values indicated as P ** < 0.01, by unpaired t-test.
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
Figure 3. Albumin catabolism and fibronectin internalization in autochthonous KrasG12D-driven pancreatic tumors catabolize albumin and internalize fibronectin
a. Differential albumin breakdown in tumor compared to normal pancreatic tissue. Devices containing self-quenched, BODIPY-labeled bovine serine albumin (DQ-BSA) were implanted directly into the pancreas of KP mice with pancreatic tumors. Tissue sections visualized by bright-field (left) and fluorescence (right) microscopy are shown. Arrows indicate reservoir position that corresponds to the site of DQ-BSA delivery. DQ-BSA fluorescence is observed only in areas where the device contacts tumor tissue (top), and not when the device is adjacent to non-malignant pancreatic tissue (bottom). (images are representative of n = 2 mice per cell line per condition with triplicate reservoirs).
b. Cellular fate of albumin catabolism. Multiphoton microscopy images of pancreatic tumors in live KP mice that also harbor a tdTomatoLSL allele (KPT) to mark cancer cells with red
fluorescence. Images were obtained 4h post device implantation to deliver DQ-BSA. Green
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
fluorescence corresponding to catabolism of DQ-BSA localizes with tumor cells marked by red fluorescence. Low (top) and high power (bottom) images are shown. Scale bar, 20 μm for both low and high power images. (images are representative of n = 4 mice per cell line per condition with duplicate reservoirs).
c. Multiphoton images of pancreatic tumors in live KPT mice showing increasing fluorescence from DQ-BSA over time. Red fluorescence marking tumor cells (top) and green fluorescence from DQ-BSA (bottom) are shown at 10- and 90-minutes post device implantation. Scale bar, 50 μm. (images are representative of n = 2 mice per genotype per condition with duplicate reservoirs).
d. Quantification of DQ-BSA fluorescence from the experiment in (C) reveals greater than 3-fold increase in fluorescence from 10- to 90-minutes. tdTomato fluorescence is also shown and changes minimally over the same 90-minute period. (n = 2 mice per genotype per condition with duplicate reservoirs).
e. Multiphoton images of normal pancreas (WT) and pancreatic tumors (KP) in live mice. Second harmonic generation is used to detect fibrillar collagen and displayed to demonstrate background auto-fluorescence in the TRITC channel. Images were obtained 30min post device implantation to deliver TRITC-Fibronectin (yellow). Cellularity of internalized TRITC-fibronectin was visualized and observed only in KP mice. White dashed outline indicates area of magnification to demonstrate cellular distribution of TRITC-fibronectin. Scale bar, 20μM. (Images are representative of n = 2 mice per genotype per condition with duplicate reservoirs).
f. Devices containing self-quenched, BODIPY-labeled bovine serine albumin (DQ-BSA) with vehicle or the lysosomal inhibitor hydroxychloroquine (HCQ) were implanted directly into the pancreas of KP mice with pancreatic tumors. Tissue sections visualized by bright-field (left) and fluorescence (right) microscopy are shown. White arrows indicate reservoir position that corresponds to the site of DQ-BSA delivery. A strong decrease in DQ-BSA is observed in the presence of HCQ (images are representative of n = 5 distinct regions of tumor per condition).
g. Quantification of fluorescent intensity of DQ-BSA in the presence and absence of HCQ (n = 5 distinct regions of tumor per condition). Significance values indicated as P ** < 0.01, by unpaired t-test.
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
Figure 4. Local depletion of amino acids following inhibition of macropinocytosis in KrasG12D -driven pancreatic tumors in vivo
a. Devices containing EIPA were implanted directly into pancreatic tumors of KP mice. 24 hours after device implantation, serial sections from the tumor were analyzed by matrix assisted laser desorption ionization coupled to imaging mass-spectrometry (MALDI-IMS). Positive ion mode was used to detect the macropinocytosis inhibitor, EIPA (top panels; EIPA diffused approximately 400–500μM). Negative ion mode used to detect the amino acids aspartate and glutamate (middle and bottom panels respectively). A scale bar representing the intensity range of ions detected for each image is included in the lower right of each panel and the length of this bar corresponds to 1mm. The left panels (MS) show the mass spectrometry signal only, the center panels (BF, MS) show an overlay of the mass spectrometry signal with a bright field image of an adjacent tissue section, and the right panels (H&E, MS) show an overlay of the mass spectrometry signal and an H&E stain of an adjacent tissue section. Schematic demonstrating device placement is included in the upper right as a white circle with an arrow indicating direction of EIPA release. A blue line indicates the extent of the EIPA signal in the H&E, MS images. Images are representative of n = 2 KP mice with triplicate reservoirs.
b. Qualitative signal for EIPA (blue) and signal from the amino acids aspartate (top) and glutamate (bottom) (red) are shown as separate and overlaid images as indicated. Also
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
A
uthor Man
uscr
ipt
shown is an outline (blue line) of the extent of EIPA signal overlaid on the aspartate and glutamate signal as indicated. White scale bar corresponds to 1mm.
c. Devices containing vehicle control (polyethylene glycol) were implanted directly into pancreatic tumors of KP mice. 24 hours after device implantation, serial sections from the tumor were analyzed by matrix-assisted laser desorption ionization coupled to imaging mass-spectrometry (MALDI-IMS). Positive ion mode was used to demonstrate the absence of the macropinocytosis inhibitor, EIPA (top panels; No EIPA observed, vehicle). Negative ion mode used to detect the amino acids aspartate and glutamate (middle and bottom panels respectively). A scale bar representing the intensity range of ions detected for each image is included in the lower right of each panel and the length of this bar corresponds to 1mm. The left panels (MS) show the mass spectrometry signal only, the center panels (BF, MS) show an overlay of the mass spectrometry signal with a bright field image of an adjacent tissue section, and the right panels (H&E, MS) show an overlay of the mass spectrometry signal and an H&E stain of an adjacent tissue section. Schematic demonstrating device placement is included in the upper right as a white circle with an arrow indicating direction of vehicle release. A blue dashed line indicates the approximate extent of diffusion based on diffusion of EIPA in A. Images are representative of n = 2 KP mice with triplicate reservoirs.
d. Qualitative signal for EIPA (blue) and signal from the amino acids aspartate (top) and glutamate (bottom) (red) are shown as separate and overlaid images as indicated. Also shown is an outline (blue dashed line) of the expected extent of EIPA diffusion if it were included in the reservoir. White scale bar corresponds to 1mm.