HAL Id: hal-03115234
https://hal.archives-ouvertes.fr/hal-03115234
Submitted on 19 Jan 2021HAL is a multi-disciplinary open access archive for the deposit and dissemination of sci-entific research documents, whether they are pub-lished or not. The documents may come from teaching and research institutions in France or abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est destinée au dépôt et à la diffusion de documents scientifiques de niveau recherche, publiés ou non, émanant des établissements d’enseignement et de recherche français ou étrangers, des laboratoires publics ou privés.
Mambalgin-1 pain-relieving peptide locks the hinge
between α4 and α5 helices to inhibit rat Acid-Sensing
Ion Channel 1a
Miguel Salinas, Pascal Kessler, Dominique Douguet, Daad Sarraf, Nicolo
Tonali, Robert Thai, Denis Servent, Eric Lingueglia
To cite this version:
Miguel Salinas, Pascal Kessler, Dominique Douguet, Daad Sarraf, Nicolo Tonali, et al.. Mambalgin-1 pain-relieving peptide locks the hinge between α4 and α5 helices to inhibit rat Acid-Sensing Ion Channel 1a. Neuropharmacology, Elsevier, 2021, pp.108453. �10.1016/j.neuropharm.2021.108453�. �hal-03115234�
Mambalgin-1 pain-relieving peptide locks the hinge between α4 and α5
helices to inhibit rat Acid-Sensing Ion Channel 1a
Miguel Salinas1*, Pascal Kessler2, Dominique Douguet1, Daad Sarraf2, Nicolo Tonali2,3, Robert Thai2, Denis Servent2*, Eric Lingueglia1*
1
Université Côte d'Azur, CNRS, IPMC, LabEx ICST, FHU InovPain, France 2
Université Paris Saclay, CEA, Département Médicaments et Technologies pour la Santé (DMTS), SIMoS, 91191 Gif-sur-Yvette, France
3
CNRS, BioCIS, Université Paris-Saclay, 92290 Châtenay-Malabry, France.
*Address for correspondence
Miguel SALINAS and Eric LINGUEGLIA: Université Côte d'Azur, CNRS, Institut de Pharmacologie Moléculaire et Cellulaire, UMR7275, 06560 Valbonne, France. Phone: 33 4 93 95 34 23; Fax: 33 4 93 95 77 08; E-mail: salinas@ipmc.cnrs.fr and lingueglia@ipmc.cnrs.fr
And,
Denis SERVENT: Université Paris Saclay, CEA, Département Médicaments et Technologies pour la Santé (DMTS), SIMoS, 91191 Gif-sur-Yvette, France. Phone: 33 1 69 08 52 02; Fax: 33 1 69 08 90 71; Email: denis.servent@cea.fr
Graphical Abstract
Highlights
• Fingers I and II of Mamb-1 are required for interaction with the rASIC1a thumb domain • K8 in Mamb-1 finger I interacts with Y358 in rASIC1a thumb domain
• No apparent contact between the toxin core and the lower part of the thumb domain • Mamb-1 does not act directly on the pH sensor but on the thumb domain
Abstract
Acid-sensing ion channels (ASICs) are proton-gated cationic channels involved in pain and other processes, underscoring the potential therapeutic value of specific inhibitors such as the three-finger toxin mambalgin-1 (Mamb-1) from snake venom. A low-resolution structure of the human-ASIC1a/Mamb-1 complex obtained by cryo-electron microscopy has been recently reported, implementing the structure of the chicken-ASIC1/Mamb-1 complex previously published. Here we combine structure-activity relationship of both the rat ASIC1a channel and the Mamb-1 toxin with a molecular dynamics simulation to obtain a detailed picture at the level of side-chain interactions of the binding of Mamb-1 on rat ASIC1a channels and of its inhibition mechanism. Fingers I and II of Mamb-1 but not the core of the toxin are required for interaction with the thumb domain of ASIC1a, and Lys-8 of finger I potentially interacts with Tyr-358 in the thumb domain. Mamb-1 does not interfere directly with the pH sensor as previously suggested, but locks by several contacts a key hinge between α4 and α5 helices in the thumb domain of ASIC1a to prevent channel opening. Our results provide an improved model of inhibition of mammalian ASIC1a channels by Mamb-1 and clues for further development of optimized ASIC blockers.
Keywords: acid-sensing ion channel; sodium channel; pain; mambalgin; toxin; inhibition
mechanism
Chemical compounds studied in this article: Mambalgin-1 (PubChem CID: 121513904).
Abbreviations: The abbreviations used are: ASICs, acid-sensing ion channels; rASIC1a, rat
acid-sensing ion channel-1a; cASIC1, chicken acid-sensing ion channel-1; hASIC1a, human acid-sensing ion channel-1a; Mamb-1, mambalgin-1; SAR, structure−activity relationship; PDB, Protein Data Bank; SEM, standard error of the mean.
1. Introduction
Mambalgins are three-finger toxins isolated from mamba venoms (Baron et al., 2013; Diochot et al., 2012) that are accessible to solid-phase chemical synthesis (Mourier et al., 2016; Schroeder et al., 2014). They act as pain-relieving peptides in rodent (Diochot et al., 2016; Diochot et al., 2012; Verkest et al., 2018) and can be as potent as morphine but with fewer unwanted side effects and through an opioid-independent pathway involving the specific inhibition of ASIC1a- and ASIC1b-containing channels, a set of Acid-Sensing Ion Channels (ASICs) widely expressed throughout the pain pathways. ASICs (Waldmann et al., 1997) are amiloride-sensitive and voltage-independent cation channels that predominantly conduct Na+ ions and are activated by extracellular acidification (ASIC1-3) and lipids (ASIC3) (Marra et al., 2016). They are widely expressed in the peripheral and the central nervous systems, where they are involved in physiological and pathophysiological processes ranging from synaptic plasticity and neuronal injury to nociception and mechanoperception (Deval and Lingueglia, 2015; Wemmie et al., 2013). In addition to their role in pain (Deval et al., 2008; Diochot et al., 2012), ASICs also emerge as interesting targets with potential clinical applications in psychiatric disorders (Wemmie et al., 2013), stroke and neurodegenerative diseases (Friese et al., 2007; Xiong et al., 2004), and cancer (Berdiev et al., 2003; Liu et al., 2016) illustrating the potential therapeutic value of specific inhibitors like mambalgins (Bychkov et al., 2020; Diochot et al., 2016; Verkest et al., 2018).
Understanding in detail how mambalgins bind and inhibit mammalian ASIC channels is therefore an essential step for further engineering and development of optimized ASIC blockers. Mamb-1 inhibits rat ASIC1a by acting as a gating modifier that stabilizes the closed state of the channel, suggesting a preferential binding to this state (Diochot et al., 2012). Binding of Mamb-1 to the closed state of ASIC1a has been confirmed by the cryo-EM low
resolution structure of Mamb-1 in complex with chicken ASIC1 (Sun et al., 2018) and very recently with human ASIC1a (Sun et al., 2020).
Both structures are very similar, indicating no major differences between the cASIC1/Mamb-1 and hASIC1a/Mamb 1 complexes despite some differences regarding the pharmacological effect of Mamb-1 on chicken and human channels. The structures suggest that the toxin binds to the upper part of the thumb domain, but finger II does not penetrate deeply into the acidic pocket as proposed in the initial model combining docking of the toxin on rASIC1a and structure−activity relationships (SAR) (Mourier et al., 2016; Salinas et al., 2014). However, the cryo-EM structures, even at the latest 3.9 Å resolution of the hASIC1a/Mamb-1 complex (Sun et al., 2020), did not allow to build a very precise view of the peptide/channel complex. However, the authors proposed a model where Mamb-1 inhibits hASIC1a by a closed state trapping mechanism through a direct interaction with the pH sensor of the channel to prevent the collapse of the acidic pocket necessary for channel opening (Sun et al., 2020).
We combine here a complementary set of approaches including structure-activity relationship of both rASIC1a and Mamb-1, and molecular dynamics simulation of the interaction of Mamb-1 with the closed state of rASIC1a modeled from the crystal structure of the closed state of cASIC1 solved in 2018 (Yoder et al., 2018), to obtain a detailed picture at the level of side-chain interactions, of peptide binding and inhibition of mammalian ASIC1a channels. Data fit well with the recent cryo-EM low resolution structure of hASIC1a/Mamb-1, but the better resolution of our model at the interface between the channel and the peptide allows to propose a new interpretation of the inhibition mechanism where Mamb-1 does not directly act on the pH sensor.
2. Materials and methods
2.1 Plasmid constructions and mutagenesis
The coding sequences of rat ASIC1a and chicken ASIC1 (GenBank Accession Number #U94403 and # NM_001040467, respectively) and their related mutants were subcloned into the NheI/NotI restriction sites of the pCI vector (Promega). Mutants of rat ASIC1a and chicken ASIC1 were obtained by recombinant PCR strategies as previously described (Salinas et al., 2006).
2.2 Xenopus oocyte preparation, DNA injection and electrophysiology
Animal handling and experiments fully conformed to French regulations and were approved by local governmental veterinary services (authorization number E06-152-5 delivered by the Ministère de l’Agriculture, Direction des Services Vétérinaires). Animals were anesthetized by exposure for 20 min to a 0.1% solution of 3-aminobenzoic acid ethyl ester (MS-222) (Sigma) buffered at pH 7.4. Oocytes were surgically removed and dissociated with collagenase type IA (Sigma) in the presence of trypsin inhibitor (Sigma), then injected into the nucleus in the animal hemisphere with 18-23 nl of ratASIC1a (1-2 ng/µl) or pCI-cASIC1 (1 ng/µl) plasmids, or with their mutants at the same concentrations. Oocytes were kept at 19 °C in ND96 solution containing: 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 2 mM MgCl2, and 5 mM HEPES (pH 7.4 with NaOH) with penicillin (6 µg/ml) and streptomycin (5 µg/ml). ASIC currents were recorded 1–2 days after injection using the two-electrode voltage-clamp technique with two standard glass microelectrodes (0.5–2.5 mega-ohms) filled with 3 M KCl, and a manual setup (Dagan TEV 200 amplifier, Dagan Corp.). The electrodes
were always implanted in the oocyte animal hemisphere, then oocytes were clamped at -50 mV, and ASIC currents were activated by rapid changes in extracellular pH induced by a microperfusion system. HEPES was replaced by MES (5 mM) in experiments performed using low pH (pH 6.5-4.0). Stimulation, data acquisition, and analysis for manual recordings were performed using pCLAMP 9.2 software (Axon Instruments). All experiments were performed at 19°C in ND96 solution supplemented with 0.05% fatty acid- and globulin-free-bovine serum albumin (Sigma) to prevent nonspecific adsorption of the peptides to tubing and containers. Synthetic Mamb-1 and its variants were applied 30 s at pH 7.47 before the acid stimulation.
2.3 Synthesis of Mamb-1 and its variants
Peptide synthesis of Mamb-1 (PubChem CID: 121513904) and alanine variants was performed on a Protein Technologies, Inc Prelude synthesizer at a 25 μmoles scale using a tenfold excess of Fmoc-amino acid relative to the preloaded Fmoc-Lys(Boc)-wang-LLresin (0.33 mmol/g). Fmoc-protected amino acids were used with the following side chain protections: ter-butyl ester (Glu, Asp), ter-butyl ether (Ser, Thr, Tyr), Trityl (Cys, His, Asn, Gln), ter-butoxycarbonyl (Lys), 2,2,5,7,8-pentamethyl-chromane-6-sulfonyl (Arg).
Amino acids were coupled twice for 10 min using 1:1:2 amino acid/HCTU/NMM in NMP. Pseudoproline dipeptides (Fmoc-L-Val-L-Thr[PSI(Me,Me)Pro]-OH, Fmoc-L-Asn(Trt)-L-Thr[PSI(Me,Me)Pro]-OH, Fmoc-L-Ser(tBu)-L-Ser[PSI(Me,Me)Pro]-OH) where coupled once for 1 h. After incorporation of each residue, the resin was acetylated for 5 min using a 50-fold excess of a mixture of acetic anhydride and NMM in NMP. Fmoc deprotection was performed twice for 2 min using 20% piperidine in NMP, and 30 sec NMP top washes were performed between deprotection and coupling steps. Following chain assembly, the
peptidyl-resin was treated with a mixture of TFA/TIPS/phenol/water (87.5:2.5:5:5) for 2 h. The crude peptide was obtained after precipitation and washed in cold ethyl ether. The different peptides were purified by reverse phase HPLC using a Vydac C4 semi-preparative column (Waters, USA; 250 x 10 mm; 4 ml.min-1; solvent A, H2O/TFA 0.1%; solvent B, acetonitrile; gradient, 20 - 40% solvent B in 40 min), and checked by mass spectrometry using ESI-MS (Bruker, Germany).
Disulfide bond formation and characterization. The purified peptides were dissolved in
guanidine-HCl 6 M in 0.1 M Tris-HCl pH 8 and diluted (1/100) in degassed 0.1 M Tris-HCl pH 8 buffer in the presence of reduced (GSH) and oxidized (GSSG) glutathione (molar ratio of 1/10/100 peptide/GSSG/GSH at a peptide concentration of 10 µM) and then incubated for 24 h at room temperature. After acidification, purification of the refolded toxins was performed on a Vydac semi-preparative column (Waters, USA; 250 x 10 mm; 4 ml.min-1; solvent A, H2O/TFA 0.1%; solvent B, acetonitrile; gradient, 20 - 40% solvent B in 40 min). After analytical reverse phase HPLC on a Vydac C4 analytical colum (Waters, USA; 250 x 4.5 mm; 1 ml.min-1; solvent A, H2O/TFA 0.1%; solvent B, acetonitrile; gradient, 20 - 40% solvent B in 40 min) (Supplemental Figure 1.1), each toxin (Table 1) was checked by mass spectrometry using ESI mass spectra (Bruker, Germany) (Supplemental Figure 1.2). Their circular dichroïsm spectra revealed a typical β-sheet signature with a minimum signal close to 213 nm, similar to that of Mamb-1, thus confirming a correct pairing of the four disulphide bridges (Supplemental Figure 1.3).
2.4 Bioinformatics
Homology modelling. The structural model of rat ASIC1a (UniProt P55926; 526 amino
identity of 90%. The comparative protein structure modeling program Modeller 9v8 (Sali and Blundell, 1993) was used to perform the homology modeling of the closed form of the trimer complex based on the experimental PDB structure 5WKV (Yoder et al., 2018). The N-termini [1-40] and C-termini [458-526] of rASIC1a were removed as no reliable coordinates are available for these regions.
Molecular dynamics simulations (MD). The stability of the complex between the rASIC1
homology model and the Mamb-1 crystal structure (PDB ID code 5DU1; (Mourier et al., 2016) was evaluated with the use of molecular dynamics simulations. The complex configuration published previously by Sun et al. was used for initializing MD simulations (Sun et al., 2018). The rASIC1a/Mamb-1 complex was prepared using the leap module in AmberTools-1.4. Hydrogen atoms were added and ionizable residues were set to predicted protonation states at pH 7.0. TIP3 water molecules and Na+ counterions were added to the system. All simulations and system equilibrations were performed using Amber 11 software (forcefields parm99.dat and frcmod.ff03). During simulations, positional restraints were applied to the transmembrane helices. Energy minimization was performed in a two-step process, first to relax water molecules and then to minimize the entire system. Then, the solvated system was equilibrated with two steps. First, the system was heated to 300 K and equilibrated for 10 ps under a constant volume ensemble (NVT) followed by a 10 ps constant pressure simulation (NPT). An additional 100 ps simulation under the NPT ensemble was run prior to production simulations to allow for further temperature and pressure equilibration. Production runs were then performed for 1 ns. The resulting trajectories were analyzed using the Ptraj module of the AMBER 11 package and a snapshot was collected for every 20 ps. Results of the molecular simulation show a stabilization of the complex without major displacement from the initial configuration and without major changes in the conformation of Mamb-1 or rASIC1a. The RMSD of the backbone of the conformation of Mamb-1 after 1 ns
when compared to the initial structure was only 1.11 Å. The RMSD of the backbone of the conformation of the rASIC1a trimer after 1 ns when compared to the initial structure was 2.36 Å.
3. Theory/calculation
3.1 Calculation of the pH0.5 of activation and inactivation (pH0.5act and pH0.5inact), and of the half maximal inhibitory concentration (IC50) of Mamb-1 and its variants
The pH-dependent curves of activation and steady-state inactivation were fitted to a sigmoidal dose-response curve with variable slope using the Hill equation: I = Imax/[1 + (10−pH0.5/10−pH)nH] where Imax is the maximal current, pH0.5 is the pH at which half of the maximal current is measured, and nH is the Hill coefficient.
The dose-response curves of mambagin-1 (and its variants) on rat ASIC1a (and its mutants) were fitted to a sigmoidal dose-response curve using the Hill equation: I=Bottom + (Imax-Bottom)/ [1 + (10-pIC50/10-p[Toxin])nH] where I
max is the maximal current, IC50 is the half maximal inhibitory concentration of Mamb-1 or its variants, pIC50 is -logIC50, [Toxin] is the variable concentration of toxin being tested and nH is the Hill coefficient.
3.2 Double-mutant cycle analysis
The difference in binding energy caused by a single mutation is calculated through ∆∆Gwt1,wt2→mut1,wt2 = ∆Gmut1,wt2 - ∆Gwt1,wt2 = RT.ln(Kdw1t,wt2/ Kdmut1,wt2), with R = 1.99 cal/mol/K and T = 293 K. When the ∆∆G associated with a couple of modifications (i.e., on the toxin and on the channel) was significantly different from the sum of the ∆∆G values
associated with each single modification, the two modified residues were considered to be in proximity and possibly in interaction (Hidalgo and MacKinnon, 1995). The coupling energy (∆∆Ginteraction or∆∆Gint), which reflects the interaction energy for the two modified residues, was calculated from ∆∆Gint = RT.ln[(Kwt1,wt2 . Kmut1,mut2) / (Kwt1,mut2 . Kmut1,wt2)], where K is Kd or Ki, wt1 is wild-type channel, mut1 is mutant channel, wt2 is wild-type Mamb-1, mut2 is variant Mamb-1. According to the equation Ki = [T](I/Io)/(1-(I/Io)) (Hidalgo and MacKinnon, 1995) where Io is the control current level and I is the current in presence of toxin concentration [T], we can deduce that Ki = IC50 when [T] = IC50. Thus, the free energy of
interaction can also be defined as follows (Mourier et al., 2016):
∆∆Gint = RT.ln[(IC50wt1,wt2 . IC50mut1,mut2) / (IC50wt1,mut2 . IC50mut1,wt2)] (Equation 1) To understand the graphical meaning of the dose-response curves shifts, it is possible to rewrite equation 1 as follows:
Because lnX = ln10.logX, the equation 1 can be written:
∆∆Gint = RT.ln10.log [(IC50wt1,wt2 . IC50mut1,mut2)/(IC50mut1,wt2 . IC50wt1,mut2)] ∆∆Gint = RT.ln10.[(logIC50wt1,wt2 + logIC50mut1,mut2) - (logIC50wt1,mut2 + logIC50mut1,wt2)] Because-log IC50 = pIC50
∆∆Gint = RT.ln10.[(pIC50wt1,mut2 + pIC50mut1,wt2) - (pIC50wt1,wt2 + pIC50mut1,mut2)] By consequence ∆∆Gint = 0 when:
pIC50wt1,wt2 + pIC50mut1,mut2 = pIC50wt1,mut2 + pIC50mut1,wt2 This equality can be rewritten as follows:
pIC50mut1,mut2 - pIC50wt1,wt2 =pIC50wt1,mut2 - pIC50wt1,wt2 + pIC50mut1,wt2 - pIC50wt1,wt2
or
Thus, the additive effect of two mutations (∆∆Gint = 0 kcal.mol-1; meaning that the two residues probably do not interact) can be visualized graphically when the curve shift induced by the double mutation is equal to the sum of the curve shifts induced independently by the two single mutations (Figure 4).
On the other way, a ∆∆Gint less than 0, i.e., when the effect of a couple of mutations is smaller than the sum of the effects associated with each single mutation (Figure 4D), strongly suggests that both residues are in contact. It can even be possible that the effect of the double mutation could be similar to the strongest single mutant effect, the weakest effect being included in the strongest effect (Figure 4C).
∆pIC50 wt1,wt2> mut1,mut2 < ∆pIC50 wt1,wt2> wt1,mut2 +∆pIC50 wt1,wt2>mut1,wt2
3.3 Statistical analysis
Data analysis was performed using GraphPad Prism 8 software. Statistical analysis of the half-maximal pH of activation and inactivation and of the IC50s was performed from the sigmoidal fits of each recording. Data are presented as Mean ± SEM or Mean with 95% Confidence Interval in brackets. Significances were calculated with one-way analysis of variance followed by a Dunnett-test: ns P > 0.05, *P < 0.05, **P < 0.01 ***P < 0.001; when compared to the control condition. Statistical reporting also includes the degrees of freedom (DF), and the exact p value. For example, an ANOVA is reported as F(DFn,DFd) = x.xx; P = 0.xxxx, and a t-test as t(DF) = x.xx; P = 0.xxxx. Individual data points have been indicated where possible.
The standard error associated with ∆∆Gint was calculated as: RT.ln10.[ δpIC50wt,mut+δpIC50mut,wt+δpIC50wt,wt+δpIC50mut,mut ], where the different δpIC50 are the standard error associated with pIC50 measurement.
4. Results
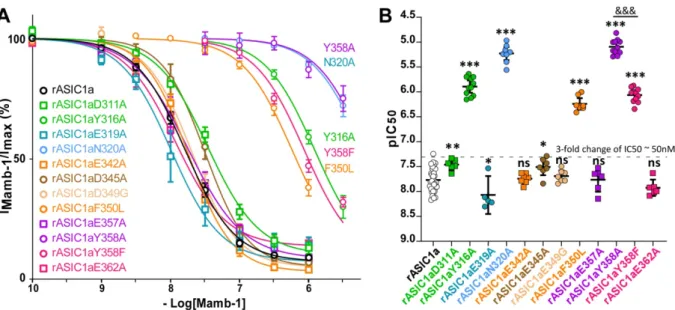
4.1 Comprehensive mapping of Mamb-1 residues potentially facing the rASIC1a channel The cASIC1/Mamb-1 and hASIC1a/Mamb-1 cryo-EM low resolution structures (5.4 and 3.9 Å, respectively) show direct contact of fingers I and II of the toxin with the thumb domain of the channel, and possibly contact of the core of Mamb-1 with the lower part of the thumb (Sun et al., 2020; Sun et al., 2018). We have performed an alanine-scanning analysis to further explore the possible involvement of finger I, finger II and the core of the toxin (Figure 1A). The dose-response curves measuring inhibition of rASIC1a by Mamb-1 and its variants were all performed under the same conditions with peptides applied at pH 7.47 before acid stimulation at pH 5.0. A role of Met-25 in finger II was suggested based on the NMR structure of Mamb-1 (Schroeder et al., 2014) and on the cryo-EM low resolution structures of cASIC1/Mamb1 and hASIC1a/Mamb-1 complexes (Sun et al., 2020; Sun et al., 2018), with possible contribution to a hydrophobic patch composed of Met-25, Pro-26, Phe-27, Leu-30 and Leu-32. However, the M25A variant did not affect the IC50 value of Mamb-1 on rASIC1a (Figure 1B and 1D and Table 1), which does not support a direct role for Met-25 in the interaction with the channel. On the other hand, the N22A modification in finger II has a significant impact on the Mamb-1 IC50 (Figure 1B and 1D, Supplemental Figure 1.4A and 1.4D and Table 1) suggesting a possible involvement of Asn-22. In addition, the IC50 values of Q5A, H6A and K8A variants at the tip of finger I were decreased respectively by 8-, 120- and 6-fold as compared to wild-type Mamb-1 (Figure 1B and 1D, Supplemental Figure 1.4A and 1.4D and Table 1), highlighting the important role of this region in the interaction of Mamb-1 with rASIC1a.
Val-10, Thr-11, His-13, Arg-14 and Met-16 located in the core of the toxin (Figure 1A) appear to face the cASIC1 and hASIC1a channels in the cryo-EM low resolution structures and seem therefore good candidates for participating in the interface with the channel. In rASIC1a, the V10A and T11A variants significantly affected IC50, but the magnitude of these effects was very limited (less than 3-fold) and in the same range as residue variants not exposed at the interface (Figure 1C-D, Supplemental Figure 1.4A and 1.4D and Table 1; (Mourier et al., 2016). The other three variations targeting the same region, i.e., H13A, R14A and M16A, did not have any statistically significant impact on the IC50 (Figure 1C-D, Supplemental Figure 1.4A and 1.4D and Table 1).
Mamb-1 and some of its variants have been also tested at one concentration (10-6.0 M) on cASIC1 (Supplemental Figure 1.4B-C). This concentration was chosen to visualize possible shifts of the dose-response curves by more than ½ log based on the IC50 of Mamb-1 on cASIC1 (~120 nM) (Sun et al., 2018). The T11A, H13A, R14A and M16A variants were not significantly different from wild-type Mamb-1, with a shift of the dose-response curve estimated to be lower than or around ½ log (note that a shift of the dose response curve by ½ log is equivalent to a 3-fold change of the IC50) (Supplemental Figure 1.4B-C).
Replacement of Phe-27 or Arg-28 residues at the tip of finger II with alanine has been shown to significantly alter the inhibitory effect of Mamb-1 on rASIC1a (Mourier et al., 2016), cASIC1 (Sun et al., 2018) and hASIC1a (Sun et al., 2020). However, we show here that these two positions accept other residues without affecting the inhibition mechanism. The R28K variant, which behaved like wild-type Mamb-1 (Figure 1E-F), suggests that the guanidinium group of Arg-28 can be replaced by an amino group without affecting the interaction at all. Similarly, the lack of effect of the F27Y variant suggests that a hydroxyl group can be added at this position without inducing steric hindrance or electrostatic repulsion (Figure 1E-F).
These data identify an important role of His-6 at the tip of toxin finger I in the Mamb-1/rASIC1a interaction, together with a significant contribution of Gln-5 and Lys-8 in the same region, and of Asn-22 in finger II. The core of Mamb-1 does not appear to be important for its interaction with rat ASIC1a or chicken ASIC1.
4.2 A cluster of four residues in rASIC1a is central for the effect of Mamb-1
A mapping by alanine-scanning mutagenesis of the surface of the thumb domain in rASIC1a was next performed to identify residues potentially facing Mamb-1 (Figure 2A-B). It was based on the cryo-EM low resolution structures of ASIC1/Mamb-1 complexes (Sun et al., 2020; Sun et al., 2018) and on a modeled structure of the closed state of rASIC1a that we built by homology modeling with the crystal structure of cASIC1 in the closed state (Yoder et al., 2018). Cysteine residues forming disulfide bridges have been excluded from this screening to avoid disruption of the structure of the thumb domain, as well as Pro-346, which cannot be mutated without affecting the overall structure of the α5 helix (Figure 1C-D) (Jasti et al., 2007). Inhibition by Mamb-1 at two concentrations (10-7.0 M and 10-7.5 M) was first evaluated (Figure 2A-B and Table 2). Only mutations of Tyr-316, Asn-320, Phe-350 and Tyr-358 had a strong impact, while all the other mutants had either significant but limited impact (IC50 < 50 nM, which is too small to be formally distinguished from a possible indirect effect), or no significant effect (Figure 2A-B and Table 2). Three residues (Tyr-316, Asn-320, and Tyr-358) appeared therefore to be important for inhibition by the toxin in addition to Phe-350 (Figure 2A-B and Table 2) that we had previously identified (Mourier et al., 2016; Salinas et al., 2014) (Figure 2C-D). The Y316F and Y358F mutants had also been tested to quantify the involvement of the hydroxyl group of both Tyr-316 and Tyr-358 (Figure 2A-B and Table 2). The Y358F mutation strongly affected the inhibition by Mamb-1 while the
Y316F mutation had no effect, suggesting that the hydroxyl group of Tyr-358, but not of Tyr-316, is involved in the interface with the toxin. Based on the cryo-EM low resolution structures, the Glu-343, Asp-346 and Asp-350 residues in cASIC1, equivalent to Glu-344, Asp-347 and Asp-351 in hASIC1a, and to Glu-342, Asp-345 and Asp-349 in rASIC1a, appear as possible candidates to interact with the Arg-28 residue of Mamb-1. However, inhibition by Mamb-1 was not strongly altered in the E342A, D345A or D349G mutants of rASIC1a (Figure 2A-B, Figure 3A-B and Table 2). The negatively charged residues Asp-311 and Glu-319 also appear as possible candidates to interact with Lys-8 but none of the D311A and E319A mutants strongly altered inhibition by Mamb-1 (Figure 2A-B, Figure 3A-B and Table 2). It is important to note that the effect of some mutants can be statistically significant, especially when tested at a single concentration, but remained however much lower than the effect observed with the most potent mutants. Their IC50 values were under 50 nM (i.e., a less than 3-fold change of the IC50) as observed with mutants acting indirectly for example on the folding of the structure, on the gating, or on the cooperativity between subunits.
Two fixed concentrations of Mamb-1 are clearly not sufficient to accurately estimate the alteration of inhibition by the different rASIC1a mutants, and the IC50 of Mamb-1 was next determined for mutants showing either strong or limited but significant effects on the inhibition by the toxin at 10-7.0 M and 10-7.5 M. Small or not statistically significant effects (i.e., IC50 < 50 nM) were measured for the D311A, E319A, E342A, D345A, D349G, E357A and E362A mutants (Figure 3A-B and Table 2), in good agreement with the data obtained with only two toxin concentrations (Figure 2A-B). Conversely, the strongest effect was observed for the Y358A and N320A mutants, which were both equivalent and, on the other hand, an order of magnitude higher than the effects measured for the Y316A and F350L mutants (Figure 3A-B and Table 2). The impact of the Y358F mutation was an order of magnitude smaller than that of Y358A and equivalent to the one of Y316A (Figure 3A-B and
Table 2). This result strongly suggests that the Tyr-358 aromatic ring and hydroxyl group are both involved in the binding of Mamb-1, unlike Tyr-316 where the suppression of the hydroxyl group by the Y316F mutation had no effect on inhibition by the toxin, suggesting that only its aromatic ring is necessary (Figure 2A-B and Table 2).
Because change in the biophysical properties of rASIC1a can indirectly impact its inhibition by Mamb-1, potential shifts of the pH-dependent activation and inactivation curves of rASIC1a mutants were investigated. No activation at conditioning pH used for toxin application (pH 7.47) was shown (Supplemental Figure 3), demonstrating the absence of destabilization of the closed state of the channel by these mutations, which could have altered the inhibition since Mamb-1 stabilizes the closed state. The Y316A and F350L mutants were even able to decrease the pH-dependent activation (Supplemental Figure 3A, 3C, and 3E), suggesting on the contrary a greater stability of the closed state. In this case, a modification of the biophysical properties of the channel would have improved rather than decreased inhibition. On the other hand, the pH0.5 of activation of the Y316F, N320A, Y358A and Y358F mutants was not significantly different from wild-type rASIC1a (Supplemental Figure 3A, 3C and 3E). In addition, opening of the channel and of its mutants is maximal at pH 5.0 (used to measure the number of uninhibited channels for the dose-response curve) (Supplemental Figure 3A), indicating that all mutants are tested under the same conditions. The strong effect on the inhibition by Mamb-1 observed with these mutants cannot therefore be explained by the very limited changes in pH-dependent activation. Compared to wild-type rASIC1a, the pH-dependent inactivation was equivalent for the Y316F, Y358A and Y358F mutants or was slightly decreased for the Y316A, N320A and F350L mutants (Supplemental Figure 3B, 3D and 3E). However, none of these mutants were inactivated at pH7.47 (Supplemental Figure 3B) indicating that their effect on the inhibition by Mamb-1 cannot be indirectly explained by a change of pH-dependent inactivation. Changes in the initial
biophysical properties of the different rASIC1a mutants are therefore not able to explain the strong impact on the inhibition by Mamb-1, thus excluding an effect due to a change in the pH-dependent gating and rather supporting an alteration of the contact between the mutated channels and the toxin.
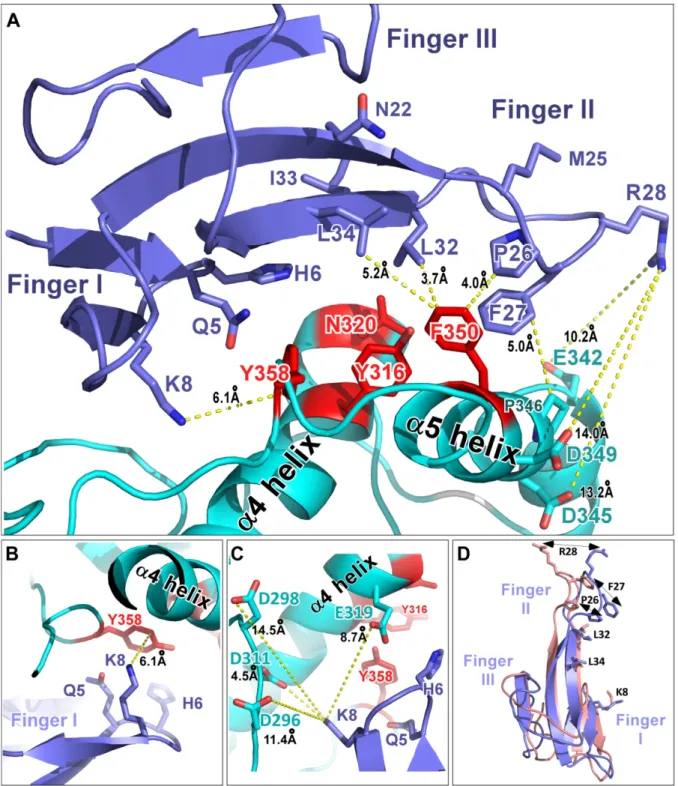
These data reveal that a cluster of four residues (Tyr-316, Asn-320, Phe-350 and Tyr-358), forming a hinge between α4 and α5 helices of the rASIC1a thumb domain (Figure 2D), is crucial for inhibition by Mamb-1 possibly through direct contacts with the toxin. Residues surrounding this cluster, in particular Asp-345 and Asp-349 located in the acidic pocket, do not seem to play a key role.
4.3 Exploration of the contact between finger I of Mamb-1 and the cluster of four residues in the thumb domain of rASIC1a
We showed previously using a double mutant cycle analysis that Phe-350 in rASIC1a could be in contact with Leu-32 in finger II of Mamb-1 (Mourier et al., 2016). We have used the same type of analysis to identify putative contacts between residues in finger I of Mamb-1 and the key residues in the thumb domain of rASIC1a. Tyr-316, Asn-320 and Tyr-358 of rASIC1a are the best candidates for possible binding to finger I, possibly targeting Lys-8. Double mutant cycle analysis of the different combinations between the K8A variant in Mamb-1 and the Y316A or N320A mutation in rASIC1a showed a pure additive effect between the mutants (the free energy of interaction, ∆∆Gint, is close to 0; Figure 4A-B, 4F and Table 3). It means that the effects of the two mutations are independent and therefore does not suggest a contact between Lys-8 and Tyr-316 or Asn-320. On the other hand, the effect induced by the double mutation K8A/Y358A was equivalent to the shift induced by the Y358A mutation alone, revealing a non-additive effect that strongly suggests proximity
between these two residues (Figure 4C, 4F and Table 3). Because of the strong right-shift of the apparent toxin affinity associated with several of these mutants, it was not possible to establish the complete dose-response curves. However, it was possible to extrapolate the curves from a fit of the first points taking into account the margins of error that are reflected in the final calculation of the ∆∆Gint. The Y358F mutation was also non-additive with the K8A variant but had a weaker effect compared to the Ala mutation (Figure 4D, 4F and Table 3), which suggests that the hydroxyl group of Tyr-358 is also involved in the contact in addition to its aromatic ring. On the other hand, the Y358F mutation was purely additive with the Q5A variant (Figure 4E-F and Table 3), suggesting the absence of contact between Gln-5 and Tyr-358.
Overall, our data showed that Lys-8 at the tip of Mamb-1 finger I could be in close proximity with Tyr358 in the thumb domain of rASIC1a. On the contrary, the side chain of Gln-5 does not seem to interact with the side chain of Tyr-358.
4.4 Molecular dynamics simulation of the interaction between Mamb-1 and rASIC1a in its closed state
The model structure of the closed state of rASIC1a (Figure 2C-D), modeled from the crystal structure of the closed state of cASIC1 (Yoder et al., 2018), has been used to carry out a molecular dynamics simulation of the interaction with Mamb-1 independently of the structure-activity approach.
Potential contacts between fingers I and II of Mamb-1, and α4 and α5 helices in the thumb domain of rASIC1a are observed in the model (Figure 5A). The side chain of Gln-5 in finger I is very closed to the backbone of Glu-357 (Figure 5B-C), but no contact is observed with the
side chain of Tyr-358, in agreement with double mutant cycle analysis data. Interestingly, the two key residues Tyr-358 and Asn-320 appear to be close to His-6 (Figure 5A).
The contact between Lys-8 in Mamb-1 finger I and Tyr-358 in rASIC1a determined by double mutant cycle analysis is visible, showing possibly a π:cation bond between the positively charged amino group of the side chain of Lys-8 and the aromatic ring of Tyr-358 (Figure 5A-B). The side chain of Lys-8 adopts a new orientation compared to the one in the crystal structure of the toxin (Figure 5D) and seems far away from Asp-298 and Asp-296 in rASIC1a (Figure 5C). The two negatively charged Asp-311 and Glu-319 in the same region of the channel are closer to Mamb-1 Lys-8 but do not seem to play a role, as suggested by the SAR results with the D311A and E319A rASIC1a mutants (Figure 5C). The molecular dynamics simulation also shows that Leu-32 (and maybe Pro-26) in Mamb-1 finger II are in the right configuration to establish CH:π interaction with the aromatic ring of Phe-350 in rASIC1a α5 helix (Figure 5A), in good agreement with the direct interaction between Leu-32 and Phe-350 that we had previously suggested (Mourier et al., 2016). Tyr-316 in rASIC1a α4 helix does not appear to interact directly with Mamb-1 (Figure 5A and Figure 6A) but is at the interface of the other three key residues, i.e., Asn-320, Phe-350 and Tyr-358 (Figure 6B and Figure 2C-D) as also observed in the X-ray structure of the closed state of cASIC1 (Yoder et al., 2018). The hinge between α4 and α5 helices formed by Tyr-316, Asn-320 and Tyr-358 on one side, and Phe-350 (probably together with Pro-346) on the other side (Figure 2C-D) adopts two different configurations between the closed state stabilized by Mamb-1 and the open/desensitized states (Figure 6A-B). In the rASIC1a/Mamb-1 complex, the closed state conformation seems to be stabilized both by finger I contact with Tyr-358 and possibly Asn-320, which are in close proximity to Tyr-316 in α4 helix, and by the interaction of finger II with Phe-350 and maybe Pro-346 in α5helix (Figure 5A and Figure 6A).
No direct contact with the channel of Asn-22 or Ile-33 in Mamb-1 is found (Figure 5A) despite their significant effect in SAR analysis. It is possible that the N22A and I33A variants affect inhibition by disorganizing the interface between finger II and III, and between finger I and II through disruption of the intramolecular bond, as observed in the X-ray structure of Mamb-1 between side chain of Asn-22 and backbone of Asn-47, and between backbone of Ile-33 and side chain of His-6 (PDB code 5DU1). The model also shows that the side chain of Met-25 is not involved at the interface (Figure 5A), which is consistent with the SAR result with the M25A variant.
The lack of effect of the E342A, D345A and D349G mutants in rASIC1a, which seem to be the only residues that could interact with Arg-28 (Figure 5A) suggests that the significant impact of the R28A variant may be indirect, as can also be inferred from the cryo-EM low resolution structures of the cASIC1/Mamb-1 and hASIC1a/Mamb-1 complexes.
The configuration of the rASIC1a/Mamb-1 complex obtained from molecular dynamics simulation (Supplemental Figure 6) is globally similar to the representation proposed by Sun et al. (Sun et al., 2020) for the cryo-EM low resolution structure of hASIC1a/Mamb-1 and cASIC1/Mamb-1 complexes, respectively (note that these structures are not yet available in the PDB repository). The conformational changes of the α4 and α5 helices are relatively limited between the closed state model of rASIC1a (Figure 2D, left panel) and the model of rASIC1a/Mamb-1 complex generated by molecular dynamics simulation (Figure 6A, left panel).
The model of interaction between Mamb-1 and the closed state of rASIC1a is globally consistent with our mutagenesis data and is coherent with the cryo-EM low resolution structure of the hASIC1a/Mamb-1 complex. However, our model reveals at the level of side chain interactions that fingers I and II of the toxin lock the hinge between α4 and α5 helices in the thumb domain of the channel through several contacts.
5. Discussion
We used mutagenesis and molecular dynamics simulation based on the closed-state structure of rASIC1a modeled from the closed state crystal structure of cASIC1 (Yoder et al., 2018) to further explore the rASIC1a/Mamb-1 complex, revealing a new detailed picture of the toxin/channel interface and providing an improved model of its inhibition mechanism.
5.1 Fingers I and II of Mamb-1, but not the core of the toxin, are required for interacting with rASIC1a
The structure-activity analysis complements previous data from our group as well as from Sun et al. (Mourier et al., 2016; Sun et al., 2020; Sun et al., 2018) to achieve a detailed and exhaustive mapping of the Mamb-1 residues crucial for its inhibitory effect on ASIC1 channels and possibly involved in the channel/toxin interface. Residues essential for the inhibition of rASIC1a by Mamb-1 are identified all along finger II (Leu-32 >> Leu-34, Phe-27, Arg-28, Ile-33, Asn-22) but also at the tip of finger I (His-6 >> Lys-8, Gln-5) (Supplemental Figure 1.4A and 1.4D) in good agreement with the ASIC1/Mamb-1 cryo-EM low resolution structures and mutagenesis data (Sun et al., 2020; Sun et al., 2018), except for the effects of the Q5A and K8A variants, which were not statistically significant in the study of Sun et al..
On the other hand, the domain comprising the core of the toxin, which is positioned close to the lower part of the thumb domain of the channel in the cASIC1/Mamb-1 and hASIC1a/Mamb-1 complex suggesting possible contacts, does not appear to be important for the interface with rASIC1a nor cASIC1. The weak effects associated with mutagenesis of
residues in the lower part of the thumb domain of rASIC1a are consistent with a model where the core of the toxin is not required for interacting with the channel.
5.2 A new interpretation of the Mamb-1 inhibition mechanism of ASIC1a channels
The mapping by mutagenesis of the residues at the surface of the thumb domain of rASIC1a involved in the effect of Mamb-1 performed here reveals, together with the mapping done in a previous study (Salinas et al., 2014), a cluster of four residues in the upper part of this domain (Tyr-358, Asn-320, Tyr-316, Phe-350) crucial for inhibition by the toxin, while residues surrounding this group have a negligible role, including residues located in the acidic pocket like Asp-345 and Asp-349.
Binding of Mamb-1 finger I to the thumb domain of rASIC1a is strongly suggested by the double mutant cycle analysis showing an interaction of Lys-8 at the tip of finger I with Tyr-358 in rASIC1a, a residue closely connected to the α4 helix of the thumb domain through its contact with Tyr-316 (Figure 2C-D, Figure 5B and Figure 6A-B). A contact between Leu-32 in finger II of the toxin with Phe-350 in the α5 helix in the upper thumb domain of rASIC1a was previously identified (Mourier et al., 2016). It appears from the structure of the closed state of chicken ASIC1 that the acidic pocket adopts an enlarged conformation (Yoder et al., 2018) that is different from the collapsed conformation observed in the desensitized/open states. The relative orientation of α4 and α5 helices is indeed different in the closed state and the desensitized/open states of the channel (Yoder et al., 2018). Phe-350, which is necessary for anchoring Mamb-1, undergoes a strong rotation towards the outside of the acidic pocket due to the rotation of the α5 helix in the thumb domain (Rook et al., 2020; Sun et al., 2018; Yoder et al., 2018). As a consequence, Mamb-1 finger II does not need to penetrate deeply into the acidic pocket in the closed state for interacting with Phe-350, as
proposed in our initial model based on the desensitized/open states of rASIC1a (Mourier et al., 2016).
The molecular dynamics simulation of Mamb-1 binding to the modeled structure of the closed state of rASIC1a, which was done independently of the structure-activity experiments, is in good agreement with our experimental mutagenesis data including a potential interaction between Lys-8 and Tyr-358, and between Leu-32 and Phe-350 in the toxin and the channel, respectively. The core of the toxin does not interact with the thumb of the channel although located close to it. In the model (Figure 6 and Supplemental Figure 6), Tyr-316 interacts with Asn-320 (π:CH bond), Phe-350 (parallel aromatic rings; i.e., π:π bond) and Tyr-358 (T shaped structure, i.e., π:CH bond) that are all essential for inhibition by Mamb-1 (Figure 2C-D). Tyr-316 itself seems to be too deeply buried to come into direct contact with the toxin, but may be important for the cohesion of the whole hinge by establishing such a network of interactions. Mamb-1 could therefore affect the stability of the hinge between α4 and α5 helices formed by Tyr-316, Asn-320 and Tyr-358 on one side, and Phe-350 (and possibly Pro-346) on the other side (Figure 2C-D, Figure 6B and Supplemental Figure 6). The upper part of the α4/α5 helices is stiffened by two disulphide bridges contrary to the lower part where the region between Tyr-358 and the α5 helix appears to be less structured and very flexible, as suggested by the molecular dynamics simulation (Figure 2D). The flexibility of the lower part allows the movement of the α5 helix necessary for the collapse by protons of the acidic pocket that triggers channel opening (Yoder et al., 2018). Our model predicts that Mamb-1 targets the hinge between α4 and α5 helices in the thumb domain to lock it, preventing its motion and leading to stabilization of the expanded shape of the acidic pocket and thus to stabilization of the closed state.
This model is different from the one recently published by Sun et al. (Sun et al., 2020). The authors propose that Mamb-1 inhibits hASIC1a through the direct interaction of Arg-28 in the
toxin with both Asp-347 and Asp-351 (corresponding to Asp-345 and Asp-349 in rASIC1a, respectively) located into the pH sensor (i.e., the acidic pocket) of the channel to prevent collapse of the acidic pocket necessary for channel opening. The cryo-EM low resolution structures from chicken and human did not allow to clearly assign the side chain of Arg-28 to Asp-347 and Asp-351, which are 10.5 and 10.2 Å apart, respectively, making interaction through salt bridges unlikely. In addition, the cryo-EM structure of the hASIC1a/Mamb-1 complex is fully compatible with the modified orientation of the Lys-8 side chain seen in our molecular dynamics simulation, but not with the orientation proposed by Sun et al. (Sun et al., 2020). An interaction is nevertheless supported by the D347A and D351G mutation in hASIC1a, which have been described to impact inhibition by the toxin (Sun et al., 2020). However, since the effects were estimated by testing only one concentration of Mamb-1, the real impact on the IC50 is hard to estimate without a complete dose response curve, as discussed before in this paper. Molecular dynamics simulation also shows that Phe-27 in Mamb-1 cannot be stacked further against the thumb domain to bring Arg-28 closer to Asp-345 and Asp-349 in rASIC1a, which is consistent with the lack of effect of the toxin variants F27Y and R28K. In the model proposed by Sun et al. (Sun et al., 2020), the direct interaction of Arg-28 in Mamb-1 with Asp-351 in hASIC1a acidic pocket hinders its interaction with Arg-190 located in the β-ball that is mandatory for the collapse of the acidic pocket (Jasti et al., 2007). However, Mamb-1 is still able to inhibit an ASIC1a mutant where Arg-190, Asp-258 and Gln-259 have been replaced by a Lys, Gln and Glu, respectively (Salinas et al., 2014), which is not supporting a direct implication of Arg-190 in the Mamb-1 inhibition mechanism. In addition, the same ASIC1a mutant is potentiated by the spider toxin PcTx1 (Salinas et al., 2014), which inhibits ASIC1a (Escoubas et al., 2000) by interfering directly with Asp-345, Asp-349 and Arg-190 (rASIC1a numbering) (Baconguis and Gouaux, 2012: Salinas, 2006 #2014) similarly to what has been proposed for Mamb-1 by Sun et al.. This
behavior is again not compatible with a mechanism of inhibition based on a direct interaction of Arg-28 with Asp-345 and/or Asp-349 to shield them from protonation, preventing H-bond with Arg-190. So even if the formation of salt bridges between Arg-28 and Asp-345 and/or Asp349 cannot be completely ruled out, its importance in the inhibition mechanism of Mamb-1 appears to be small compared to locking of the α4/α5 hinge in the thumb domain.
Globally, the inhibition mechanism of Mamb-1 can be compared to the one of neurotoxins from scorpions and sea anemones that uses a voltage-sensor trapping mechanism to alter channel gating (Catterall et al., 2007). Indeed, the voltage-sensors responsible for sodium, potassium and calcium channels activation are positively charged transmembrane segments, inaccessible to the binding of toxins. To alter their gating, these toxins need to act at distance of the sensor by binding to the extracellular S3/S4 loop to prevent their transmembrane movements that are required for channel opening. For example, hanatoxins from tarantula and agatoxins from spiders bind to the S3/S4 loop of voltage dependent potassium and calcium channels, respectively, to inhibit their activation by shifting the voltage dependent activation to more positive membrane potentials (Bourinet et al., 1999; Li-Smerin and Swartz, 2000; Swartz and MacKinnon, 1997), and act by trapping these voltage sensors in their closed states. Similarly, Mamb-1 shifts the pH dependent activation to more acidic pH (Diochot et al., 2012) and does not need to act directly on the negatively charged residues of the pH sensor but on the nearby α4/α5 helices, which are crucial for the overall conformational change of the acidic pocket. This mechanism is very different from the one of the toxin PcTx1, which directly targets the pH sensor (Baconguis and Gouaux, 2012), and could explain the different properties of these two toxins regarding ASIC1a inhibition.
The four residues necessary for inhibition of ASIC1a by Mamb-1 are all shared between ASIC1a and ASC1b, which supports the fact that the toxin is able to selectively inhibit both channels. However, it should be noted that Mamb-1 does not inhibit rASIC1a and rASIC1b
channels in exactly the same way. Rat ASIC1a is almost completely inhibited by the toxin whatever the pH of activation used contrary to rat ASIC1b, but also to human ASIC1a and chicken ASIC1 (sharing together the four residues necessary for inhibition), which show a maximum inhibition at high concentration of Mamb-1 of only 70% (Diochot et al., 2012), 80% (Sun et al., 2020), and 40% (Sun et al., 2018), respectively. Our findings could therefore explain the specificity of Mamb-1 but not the difference observed in the maximal effect of the toxin between rASIC1a and rASIC1b, hASIC1a or cASIC1. Candidate domains to explain such a difference include the palm domain, based on chimeras between ASIC1a and ASIC1b that point to a role for this domain in the inhibition mechanism of Mamb-1 (Salinas et al., 2014), or the N-terminal and first transmembrane domains (TM1), which have been proposed to indirectly influence the effect of Mamb-1 (Salinas et al., 2014) as well as the effect of the small molecules GMQ and amiloride (Besson et al., 2017).
The data presented here give a detailed picture of mambalgin-1 binding on rat ASIC1a and of its inhibition mechanism that is not direct on the pH sensor but indirect through the channel thumb domain, providing clues for the development of optimized ASIC blockers. It is noticeable that the rat and human ASIC1a sequences share 98.1% identical amino acids over their total lengths of 526-528 residues (only 10 differences), and that the key residues identified in this study are all conserved between the two species, supporting the relevance of the data for human ASIC1a.
Acknowledgments
We thank S. Diochot, A. Baron, E. Deval, J. Noël, A. Negm, C. Verkest, M. Chafai, K. Delanoë, L. Pidoux and B. Labrum for discussions and comments, V. Friend and J. Salvi-Leyral for expert technical assistance, and V. Berthieux for secretarial assistance.
Funding
This work was supported by CNRS, INSERM, CEA, Université Côte d’Azur and the Agence Nationale de la Recherche (grant numbers ANR-11-LABX-0015-01 and ANR-17-CE18-0019).
Declarations of interest
None.
Author contributions
MS, Conception and design, acquisition, analysis and interpretation of data. PK, Conception and design of toxin variants.
DD, Conception and design of the molecular dynamics simulation. EL, Conception and design.
DServent, Conception and design.
DSarraf, Synthesis and physico-chemical characterization of Mamb-1 variants. NT, Synthesis and physico-chemical characterization of Mamb-1 variants.
RT, Mass spectra of Mamb-1 variants.
All the authors have contributed to drafting or revising the manuscript. All authors have given approval to the final version of the manuscript.
Author ORCIDs
Miguel Salinas – https://orcid.org/0000-0002-7445-123X Eric Lingueglia – http://orcid.org/0000-0003-3902-3405 Denis Servent – http://orcid.org/0000-0002-0774-1691 Pascal Kessler – http://orcid.org/0000-0001-7976-1811 Dominique Douguet – http://orcid.org/0000-0001-6209-6464 Daad Sarraf – http://orcid.org/0000-0002-3922-6643
Nicolo Tonal – http://orcid.org/0000-0002-1435-5676 Robert Thai – http://orcid.org/0000-0001-9438-4225
Appendix A: Supplementary data
Supplemental Figure legends
Supplemental Figure 1.1. Reverse phase HPLC chromatograms of the purified Mamb-1
alanine variants after refolding.
Supplemental Figure 1.3. Circular dichroïsm analysis of wild-type Mamb-1 and its
alanine variants. The three-finger fold signature is characterized by maxima and minima at
202 nm and 213 nm, respectively, in their circular dichroïsm spectra.
Supplemental Figure 1.4. Alanine-scanning mutagenesis data summarizing residues of
Mamb-1 that affect the inhibition of rASIC1a and cASIC1. A) Mamb-1 and its variants
listed in descending order according to the IC50 value on rASIC1a (boxed data from this study; other data from Mourier et al., 2016. B) Inhibition of cASIC1 by Mamb-1 and its T11A, H13A, R14A and M16A variants located in the core region. Peptide concentrations at 10-6 M for cASIC1 were chosen to be able to detect a rightward shift of the IC50 greater than 1/2 log. C) Inhibition of chicken ASIC1 by wild-type Mamb-1 and its variants shown in (B). The residual current of cASIC1 after inhibition by 10-6 M of Mamb-1 or its variants is expressed as a percentage of control current (IMamb-1/Imax). Data are presented as the mean with 95% confidence interval in brackets, n = 6, one-way analysis of variance followed by a Dunnett-test: ns P > 0.05, F(4,25) = 1.439; P = 0.2504. In cASIC1a, the T11A, H13A, R14A and M16A variants did not differ significantly from wild-type Mamb-1 D) Residue positions on the X-ray structure of Mamb-1 (PDB code 5DU1). Color code illustrates the impact of the alanine mutation on IC50 (yellow for IC50 values up to 50 nM, salmon for IC50 values between 50 nM and 500 nM, and red for IC50 values above 500 nM). The green dashed line indicates the potential pharmacophore (same color code as in Figure 1A).
Supplemental Figure 3. pH dependence of rat ASIC1a and its key mutants. A-B) The pH-dependent curves of activation (A) and inactivation (B) for rASIC1a (black lines; n = 9-7) and its mutants (n = 6-11). Solid lines are fits of the mean values obtained for each pH to a sigmoidal dose-response curve with variable slope. C-D) Statistical analysis of pH0.5 of
activation (C) and inactivation (D) calculated from the sigmoidal fits of each recording performed for determination of the mean pH-dependent curves. Statistical significances were calculated with one-way analysis of variance followed by a Dunnett-test (ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001; when compared to wild-type rASIC1a (n = 9-7) noted in black). F(6,45) = 16.69; P < 0.0001 for “pH0.5 activation” and F(6,45) = 13.27; P < 0.0001 for “pH0.5 inactivation”. E) Values of the half-maximal pH of activation and inactivation shown in A- D with the same color code. Data are presented as the mean with 95% confidence interval in brackets.
Supplemental Figure 6. A structural model of rASIC1a inhibition by Mamb-1 (enlarged
view of Figure 6A in the context of a single channel subunit). Mamb-1 is shown in blue
and rASIC1a in grey except the region covering the α4 and α5 helices that is highlighted in cyan. Side chains of key residues are shown for Mamb-1 and for the hinge between α4 and α5 helices of rASIC1a (in red). Prolines are shown in green.
References
Baconguis, I., Gouaux, E., 2012. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489, 400-405. doi: 10.1038/nature11375.
Baron, A., Diochot, S., Salinas, M., Deval, E., Noel, J., Lingueglia, E., 2013. Venom toxins in the exploration of molecular, physiological and pathophysiological functions of acid-sensing ion channels. Toxicon 75, 187-204. doi: 10.1016/j.toxicon.2013.04.008.
Berdiev, B. K., Xia, J., McLean, L. A., Markert, J. M., Gillespie, G. Y., Mapstone, T. B., Naren, A. P., Jovov, B., Bubien, J. K., Ji, H. L., Fuller, C. M., Kirk, K. L., Benos, D. J., 2003. Acid-sensing ion channels in malignant gliomas. J Biol Chem 278, 15023-15034. doi: 10.1074/jbc.M300991200. Besson, T., Lingueglia, E., Salinas, M., 2017. Pharmacological modulation of Acid-Sensing Ion
Channels 1a and 3 by amiloride and 2-guanidine-4-methylquinazoline (GMQ). Neuropharmacology 125, 429-440. doi: 10.1016/j.neuropharm.2017.08.004.
Bourinet, E., Soong, T. W., Sutton, K., Slaymaker, S., Mathews, E., Monteil, A., Zamponi, G. W., Nargeot, J., Snutch, T. P., 1999. Splicing of alpha 1A subunit gene generates phenotypic variants of P- and Q-type calcium channels. Nat Neurosci 2, 407-415. doi: 10.1038/8070.
Bychkov, M., Shulepko, M., Osmakov, D., Andreev, Y., Sudarikova, A., Vasileva, V., Pavlyukov, M. S., Latyshev, Y. A., Potapov, A. A., Kirpichnikov, M., Shenkarev, Z. O., Lyukmanova, E., 2020. Mambalgin-2 Induces Cell Cycle Arrest and Apoptosis in Glioma Cells via Interaction with ASIC1a. Cancers (Basel) 12. doi: 10.3390/cancers12071837.
Catterall, W. A., Cestele, S., Yarov-Yarovoy, V., Yu, F. H., Konoki, K., Scheuer, T., 2007. Voltage-gated ion channels and gating modifier toxins. Toxicon 49, 124-141. doi: 10.1016/j.toxicon.2006.09.022.
Deval, E., Lingueglia, E., 2015. Acid-Sensing Ion Channels and nociception in the peripheral and central nervous systems. Neuropharmacology 94, 49-57. doi: 10.1016/j.neuropharm.2015.02.009. Deval, E., Noel, J., Lay, N., Alloui, A., Diochot, S., Friend, V., Jodar, M., Lazdunski, M., Lingueglia,
E., 2008. ASIC3, a sensor of acidic and primary inflammatory pain. EMBO J 27, 3047-3055. doi: 10.1038/emboj.2008.213.
Diochot, S., Alloui, A., Rodrigues, P., Dauvois, M., Friend, V., Aissouni, Y., Eschalier, A., Lingueglia, E., Baron, A., 2016. Analgesic effects of mambalgin peptide inhibitors of acid-sensing ion channels in inflammatory and neuropathic pain. Pain 157, 552-559. doi: 10.1097/j.pain.0000000000000397.
Diochot, S., Baron, A., Salinas, M., Douguet, D., Scarzello, S., Dabert-Gay, A. S., Debayle, D., Friend, V., Alloui, A., Lazdunski, M., Lingueglia, E., 2012. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 490, 552-555. doi: 10.1038/nature11494.
Escoubas, P., De Weille, J. R., Lecoq, A., Diochot, S., Waldmann, R., Champigny, G., Moinier, D., Menez, A., Lazdunski, M., 2000. Isolation of a tarantula toxin specific for a class of proton-gated Na+ channels. J Biol Chem 275, 25116-25121. doi: 10.1074/jbc.M003643200.
Friese, M. A., Craner, M. J., Etzensperger, R., Vergo, S., Wemmie, J. A., Welsh, M. J., Vincent, A., Fugger, L., 2007. Acid-sensing ion channel-1 contributes to axonal degeneration in autoimmune inflammation of the central nervous system. Nat Med 13, 1483-1489. doi: 10.1038/nm1668.
Hidalgo, P., MacKinnon, R., 1995. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science 268, 307-310. doi: 10.1126/science.7716527.
Jasti, J., Furukawa, H., Gonzales, E. B., Gouaux, E., 2007. Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH. Nature 449, 316-323. doi: 10.1038/nature06163.
Li-Smerin, Y., Swartz, K. J., 2000. Localization and molecular determinants of the Hanatoxin receptors on the voltage-sensing domains of a K(+) channel. J Gen Physiol 115, 673-684. doi: 10.1085/jgp.115.6.673.
Liu, C., Zhu, L. L., Xu, S. G., Ji, H. L., Li, X. M., 2016. ENaC/DEG in Tumor Development and Progression. J Cancer 7, 1888-1891. doi: 10.7150/jca.15693.
Marra, S., Ferru-Clement, R., Breuil, V., Delaunay, A., Christin, M., Friend, V., Sebille, S., Cognard, C., Ferreira, T., Roux, C., Euller-Ziegler, L., Noel, J., Lingueglia, E., Deval, E., 2016. Non-acidic activation of pain-related Acid-Sensing Ion Channel 3 by lipids. EMBO J 35, 414-428. doi: 10.15252/embj.201592335.
Mourier, G., Salinas, M., Kessler, P., Stura, E. A., Leblanc, M., Tepshi, L., Besson, T., Diochot, S., Baron, A., Douguet, D., Lingueglia, E., Servent, D., 2016. Mambalgin-1 Pain-relieving Peptide, Stepwise Solid-phase Synthesis, Crystal Structure, and Functional Domain for Acid-sensing Ion Channel 1a Inhibition. J Biol Chem 291, 2616-2629. doi: 10.1074/jbc.M115.702373.
Rook, M. L., Williamson, A., Lueck, J. D., Musgaard, M., Maclean, D. M., 2020. beta11-12 linker isomerization governs acid-sensing ion channel desensitization and recovery. Elife 9. doi: 10.7554/eLife.51111.
Sali, A., Blundell, T. L., 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234, 779-815. doi: 10.1006/jmbi.1993.1626.
Salinas, M., Besson, T., Delettre, Q., Diochot, S., Boulakirba, S., Douguet, D., Lingueglia, E., 2014. Binding site and inhibitory mechanism of the mambalgin-2 pain-relieving peptide on acid-sensing ion channel 1a. J Biol Chem 289, 13363-13373. doi: 10.1074/jbc.M114.561076.
Salinas, M., Rash, L. D., Baron, A., Lambeau, G., Escoubas, P., Lazdunski, M., 2006. The receptor site of the spider toxin PcTx1 on the proton-gated cation channel ASIC1a. J Physiol 570, 339-354. doi: 10.1113/jphysiol.2005.095810.
Schroeder, C. I., Rash, L. D., Vila-Farres, X., Rosengren, K. J., Mobli, M., King, G. F., Alewood, P. F., Craik, D. J., Durek, T., 2014. Chemical synthesis, 3D structure, and ASIC binding site of the toxin mambalgin-2. Angew Chem Int Ed Engl 53, 1017-1020. doi: 10.1002/anie.201308898.
Sun, D., Liu, S., Li, S., Zhang, M., Yang, F., Wen, M., Shi, P., Wang, T., Pan, M., Chang, S., Zhang, X., Zhang, L., Tian, C., Liu, L., 2020. Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1. Elife 9. doi: 10.7554/eLife.57096.
Sun, D., Yu, Y., Xue, X., Pan, M., Wen, M., Li, S., Qu, Q., Li, X., Zhang, L., Liu, L., Yang, M., Tian, C., 2018. Cryo-EM structure of the ASIC1a-mambalgin-1 complex reveals that the peptide toxin mambalgin-1 inhibits acid-sensing ion channels through an unusual allosteric effect. Cell Discov 4, 27. doi: 10.1038/s41421-018-0026-1.
Swartz, K. J., MacKinnon, R., 1997. Mapping the receptor site for hanatoxin, a gating modifier of voltage-dependent K+ channels. Neuron 18, 675-682. doi: 10.1016/s0896-6273(00)80307-4.
Verkest, C., Piquet, E., Diochot, S., Dauvois, M., Lanteri-Minet, M., Lingueglia, E., Baron, A., 2018. Effects of systemic inhibitors of acid-sensing ion channels 1 (ASIC1) against acute and chronic mechanical allodynia in a rodent model of migraine. Br J Pharmacol 175, 4154-4166. doi: 10.1111/bph.14462.
Waldmann, R., Champigny, G., Bassilana, F., Heurteaux, C., Lazdunski, M., 1997. A proton-gated cation channel involved in acid-sensing. Nature 386, 173-177. doi: 10.1038/386173a0.
Wemmie, J. A., Taugher, R. J., Kreple, C. J., 2013. Acid-sensing ion channels in pain and disease. Nat Rev Neurosci 14, 461-471. doi: 10.1038/nrn3529.
Xiong, Z. G., Zhu, X. M., Chu, X. P., Minami, M., Hey, J., Wei, W. L., MacDonald, J. F., Wemmie, J. A., Price, M. P., Welsh, M. J., Simon, R. P., 2004. Neuroprotection in ischemia: blocking calcium-permeable acid-sensing ion channels. Cell 118, 687-698. doi: 10.1016/j.cell.2004.08.026.
Yoder, N., Yoshioka, C., Gouaux, E., 2018. Gating mechanisms of acid-sensing ion channels. Nature 555, 397-401. doi: 10.1038/nature25782.
Table 1
Pharmacological Properties of Mamb-1 variants. pIC50 Mean values with 95% confidence intervals in brackets and their calculated IC50 (= 10-pIC50) of the inhibition of rASIC1a current by wild-type and Mamb-1 variants shown in Figure 1D. Color code illustrates the impact of the alanine mutation on IC50 (yellow for IC50 values up to 50 nM, salmon for IC50 values between 50 nM and 500 nM, and red for IC50 values above 500 nM). Asterisks indicate a statistically significant difference compared with wild-type Mamb-1 (indicated in black) done from 4-31 dose-response curves for Mamb-1 and each variant (one-way analysis of variance followed by a Dunnett-test: ns P > 0.05, * P < 0.05, ** P < 0.01, *** P < 0.001, and F(11,87) = 121.9; P < 0.0001). Hill slopes (Mean ± SEM) were calculated from the mean dose-response curves shown in Figure 1B-C. The location of Mamb-1 variants in the three-finger structure is indicated in the left column.
Location Peptides pIC50 IC50 (nM) Hill slope Fold-change
Mamb-1 7.77 (7.68-7.86) 17 -1.2 ± 0.1 1.0 Extremity of finger I Q5A 6.85*** (6.79-6.92) 140 -1.2 ± 0.1 8.2 H6A 5.69*** (5.59-5.79) 2037 -1.9 ± 0.8 119.7 K8A 7.01*** (6.86-7.17) 97 -1.0 ± 0.1 5.7 Core / Finger I V10A 7.40** (7.21-7.59) 40 -1.1 ± 0.1 2.3 T11A 7.38** (6.91-7.85) 42 -0.8 ± 0.1 2.4 H13A 7.64 ns (7.43-7.85) 23 -1.2 ± 0.1 1.4 R14A 7.68 ns (7.41-7.96) 21 -0.9 ± 0.1 1.2 M16A 8.00 ns (7.84-8.16) 10 -0.9 ± 0.1 0.6 Side of finger II N22A 6.66*** (6.52-6.81) 218 -2.2 ± 0.2 12.8 M25A 7.80 ns (7.67-7.93) 16 -1.5 ± 0.3 0.9
Table 2
Inhibition by Mamb-1 of rASIC1a and its mutants targeting the thumb domain.
The residual current at pH 5.0 after inhibition by Mamb-1 at 10-7.0 M (Figure 2A) and 10-7.5 M (Figure 2B) is expressed as a percentage of control current without toxin (IMamb-1/Imax). pIC50 values from Figure 3B and its corresponding calculated IC50 (n.d., not determined). Mutations inducing a strong alteration of inhibition by Mamb-1 (i.e., IC50 > 500 nM; in red) and those having a weak impact (i.e., IC50 < 50 nM; in yellow). IMamb 1/Imax (n = 7-25) and pIC50 (n = 5-31) values are means with 95% confidence intervals in brackets, asterisks indicate a statistically significant difference from the control condition rASIC1a indicated in black (one-way analysis of variance followed by a Dunnett-test: ns P > 0.05, * P < 0.05, ** P < 0.01, *** P < 0.001, with F(19,181) =334.2; P < 0.0001 for “Mamb-1 10-7.0 M” condition, F(19,182) = 88.61; P < 0.0001 for “Mamb-1 10-7.5 M” condition, and F(12,112) = 317.9; P < 0.0001 for IC50 of Mamb-1. Location of the mutants in the secondary structure of the thumb domain is shown in the left column.
Location IMamb-1
/
Imax (%) pIC50 Mamb-1IC50 (nM) rASIC1a mutants Mamb-1 10-7.0 M (100 nM) Mamb-1 10-7.5 M (32 nM) rASIC1a 7.9 (6.7-9.1) 28.6 (24.8-32.4) 7.77 (7.68-7.86) 17
α4
-h
el
ix
D311A 30.5*** (26.5-34.5) 60.3*** (56.6-64.0) 7.46** (7.34-7.58) 35 R315A 21.0*** (17.6-24.4) 41.9* (37.2-46.7) n.d. n.d. Y316A 94.0*** (92.3-95.7) 97.1*** (95.4-98.9) 5.89*** (5.77-6.02) 1282 Y316F 13.7ns (9.2-18.3) 19.7ns (14.7-24.7) n.d. n.d. E319A 14.7** (10.6-18.7) 28.5ns (22.3-34.7) 8.07ns (7.69-8.45) 9 N320A 99.2*** (97.9-100.4) 100.2*** (98.7-101.7) 5.23*** (5.10-5.36) 5902 N322A 10.7ns (7.7-13.7) 37.3ns (33.9-40.7) n.d. n.d.α5
-h
el
ix
E342A 14.1* (10.8-17.4) 42.4*** (35.1-19.8) 7.74 ns (7.65-7.84) 18 D345A 22.1*** (16.4-27.8) 53.9*** (47.6-60.2) 7.51 ns (7.34-7.67) 31 D349G 14.8** (11.5-18.1) 36.6ns (32-41.2) 7.69 ns (7.80-7.58) 20 F350L 87.8*** (84.7-90.8) 96.1*** (94.5-97.7) 6.24*** (6.12-6.36) 574 E353A 4.6ns (3.1-6.0) 16.6* (9.2-24.1) n.d. n.d. K354A 5.9ns (2.6-9.2) 8.6*** (5.7-11.4) n.d. n.d.U
ns
tr
uc
tu
re
d
flex
ibl
e l
oo
p
Q356A D355A 32.7*** (24.0-41.4) 9.7ns 44.9*** (37.5-52.2) n.d. n.d. (3.8-15.5) 34.1ns (19.1-49.2) n.d. n.d. E357A 21.1*** (18.4-23.8) 44.8*** (38.8-50.9) 7.76 ns (7.52-8.00) 17 Y358A 95.5*** (91.7-99.4) 96.7*** (92.4-101.0) 5.09*** (4.99-5.19) 8110 Y358F 94.7*** (91.9-97.6) 99.5*** (95.5-103.6) 6.06*** (5.97-6.16) 863 E362A 22.4*** (19-25.9) 45.2*** (37.8-52.6) 7.93 ns (8.09-7.76) 12Table 3
Double-mutant cycle analysis. Upper panel: pIC50 (Mean ± SEM) calculated for wild-type Mamb-1 and its variants, for their inhibition on wild-type rASIC1a and its mutants from the mean dose-response curves shown in Figure 4 (number of records for plotting the curves given in brackets). Lower panel: Variation in free energy of interaction (∆∆Gint) calculated from the pIC50 values shown in the upper panel, as described in Materials and Methods (Theory/calculation). ∆∆Gint values different from zero are noted in bold.
rASIC1a Y316A N320A Y358A Y358F
Mamb-1 7.79 ± 0.02 (31) 5.91 ± 0.03 (11) 5.26 ± 0.03 (10) 5.17 ± 0.07 (14) 6.07 ± 0.03 (11) K8A 7.08 ± 0.04 (7) 5.15 ± 0.02 (3) 4.76 ± 0.12 (3) 5.08 ± 0.03 (7) 5.64 ± 0.04 (3) Q5A 6.95 ± 0.02 (6) _ _ _ 5.23 ± 0.03 (11)
Y316A/K8A N320A/K8A Y358A/K8A Y358F/K8A Y358F/Q5A
∆∆Gint
(kcal.mol-1)