International Immunology, Vol. 9, No. 1, pp. 179–187 © 1997 Oxford University Press
Thymocytes control the CD4 gene differently
from mature T lymphocytes
Yasushi Uematsu, Alena Donda
and
Gennaro De Libero
Experimental Immunology, Department of Research, University Hospital, Hebelstrasse 20, 4031 Basel, Switzerland
Keywords: enhancer, gene regulation, silencer, thymic development, transgenic mouse
Abstract
We analyzed the activity of the enhancer, the promoter and the silencer of the human CD4 gene during T cell development using transgenic mice. Immunofluorescence studies on thymic populations of mice carrying transgenes in various combinations of these regulatory DNA elements revealed that thymocytes control the CD4 gene in a different manner than mature peripheral T lymphocytes. The 59-positive regulatory unit, consisting of the promoter and the 59 enhancer, is already active at the CD4–CD8–double-negative (DN) stage of development. However,
its activity becomes lower in the double-positive and a fraction of the CD4FCD8int/–cell population,
indicating that an additional enhancer, located in either the first or the third intron of the CD4 gene, is required for CD4 gene expression in this population. The other studied regulatory element is the minimal CD4 silencer which inhibits CD4 gene expression in peripheral CD8 T lymphocytes. This silencer is inactive in the most immature DN thymocytes, which probably use a distinct silencer mechanism to down-regulate CD4 gene expression. Unexpectedly, the CD4 silencer is also active in CD4FCD8int/–cells of the thymus, implying that an anti-silencer may be required to resume CD4
expression in this cell population. Altogether, the CD4 gene is regulated by several positive and negative regulatory mechanisms which come into play in a developmentally coordinated manner.
Introduction
αβT lymphocytes are activated when the TCR interacts with express neither CD4 nor CD8. Upon progression to the next differentiation step, these double-negative (DN) cells turn on antigens associated with self-MHC molecules. This
inter-action involves engagement of CD4 or CD8 glycoproteins, both the CD4 and the CD8 genes and become
double-positive (DP) cells. DP cells start to express TCR on the cell known as co-receptor molecules. The CD4 or CD8 molecules
expressed on the T cell bind to class II or class I MHC surface and undergo thymic selection, which confers self-MHC restriction and self-tolerance to the T cell repertoire molecules on the antigen-presenting cells respectively,
stabilizing the interaction between the TCR and antigen– (reviewed in 3,4,6). After this process, DP cells lose one of the co-receptors and become either CD4 single-positive (SP) MHC complex (reviewed in 1) and facilitating transmission of
activation signals through the tyrosine kinaselck (2). or CD8 SP cells and migrate into the periphery as mature T lymphocytes. These findings point out the importance of CD4 CD4 and CD8 molecules are co-expressed on the majority
of thymocytes, while they are mutually exclusively expressed and CD8 in both reactivity and development of the T cell. Furthermore, they suggest that co-receptor expression may on mature peripheral T cells. During thymic development, the
class of the MHC molecule interacting with the TCR on be regulated in a programmed and coordinated manner. Little is known about the molecular mechanisms controlling co-immature T cells determines the co-receptor phenotype at
the mature stage (reviewed in 3,4). Furthermore, most CD41 receptor expression during development as well as those which maintain the stable CD4 or CD8 SP phenotype in cells acquire helper function, while CD81 cells frequently
show cytotoxic activity (5). The biological meaning of the mature cells.
During the last few years, the CD4 promoter and enhancer concordance between the T cell functions and the co-receptor
phenotype remains unclear. were identified (7–10), but turned out to be not responsible
for lineage-specific CD4 gene expression. More recently, The co-receptor phenotype changes as T lymphocytes
develop in the thymus. The most immature thymocytes an intronic DNA element, characterized as a silencer, has
Corresponding author: Y. Uematsu Received3 July 1996,accepted3 October 1996
use distinct molecular mechanisms for both positive and negative regulation of CD4 expression from those used by mature T lymphocytes.
Methods
Transgenic mice and DNA constructs
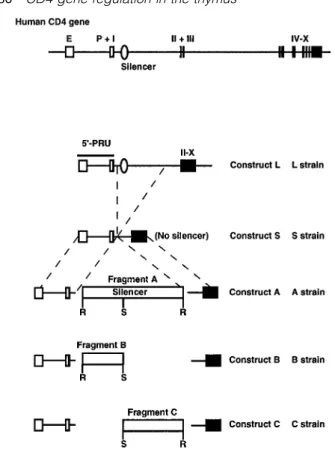
The transgenic mouse strains L, S, A, B and C were previously described, and are named according to the introduced construct (11). Briefly, the L strains were established by introducing the promoter, the enhancer, the first exon followed by the entire first intron and cDNA sequence encoding the rest of the exons of the human CD4 gene. This construct also contains a 2.5 kb sequence derived from the 59 part of the third intron. Construct S consists of the same elements except that the third intron and ~8 kb of the first intron have been deleted. Constructs A, B and C were made by inserting fragment A, B and C into construct S respectively (Fig. 1). Fragment A is the 484 bpRsaI fragment, containing the CD4 silencer isolated from the first intron of the human CD4 gene.
Fragments B and C were yielded by SacII digestion of
fragment A. Non-transgenic C57BL/6J mice (Biological Research Laboratories, Fu¨llinsdorf, Switzerland) were used as a negative control.
Immunofluorescence analysis
Fig. 1. Genomic organization of the human CD4 gene and the The transgenic and control mice were analyzed between 6
transgene constructs. E, enhancer; P, promoter. The exons (closed and 15 weeks of age. Cell suspensions from mouse thymi boxes) are numbered with Roman numerals. The 484 bp RsaI
and lymph nodes were kept at 4°C and washed with PBS
fragment (fragment A) and its subfragments (fragments B and C)
supplemented with 1% BSA and 0.02% sodium azide. First,
are indicated as open boxes and are drawn in a different scale from
the cells were incubated with 1% normal mouse serum.
the other genomic elements. Details of the transgene construction
have been described (11). Subsequently the cell preparations were stained with the
following mAb directly labeled with FITC, phycoerythrin (PE), Red613 or biotin: mouse anti-hCD4 (B-B14; Serotec, Oxford, UK), rat anti-mouse CD4 (GK1.5; Becton Dickinson, Mountain been found to be important for lineage-specific expression of View, CA), rat anti-mouse CD8 (H53.6.7, Becton Dickinson), the CD4 gene in mature T lymphocytes of humans and mice hamster anti-mouse CD3ε(500A2; PharMingen, San Diego, (11–13). However, in these studies the activities of these CA), rat anti-mouse CD69 (PharMingen, San Diego, CA) and positive (promoter and enhancer) and negative (silencer) rat anti-mouse heat-stable antigen (HSA, CD24) (J11d, kindly elements have been examined mainly in mature T lympho- provided by U. Staerz). Biotinylated antibodies were detected
cytes, but not during thymic development. by adding streptavidin–TriColor (Caltag, South San Francisco,
In the present study, in order to address the question of CA) or streptavidin–allophycocyanin (Southern Biotechnology whether the human CD4 silencer elements control transgene Associates, Birmingham, AL). In addition, unlabeled GK1.5 expression in thymic populations in the same manner as in and H53.6.7 in combination with PE-conjugated goat anti-rat peripheral T cells, we measured human CD4 (hCD4) reporter Ig anti-serum (Southern Biotechnology Associates) were used gene expression in the thymus using the transgenic strains, to gate CD4–CD8–DN cells by excluding PE-positive cells.
which we had used for the previous study (11). The activity Three-color and four-color flow cytometric analyses were
of the human promoter and enhancer elements, which we performed on a FACScan and a FACS Vantage (Becton
introduced into the transgenic constructs, has not well been Dickinson) respectively. Acquired data were analyzed with characterized in immature T lymphocytes, and hence we the Lysys II and CELLQuest programs (Becton Dickinson). included analysis of promoter/enhancer activity of the human
CD4 gene in this study. We studied activity of the human CD4
Results
promoter in combination with the human CD4 enhancer as the 59-positive regulatory unit (PRU) in the thymus. We then
DN CD3–thymocytes use different silencing mechanisms from analyzed the repressive effect of the human CD4 silencer on
those of peripheral CD8 SP cells reporter gene expression driven by the 59-PRU. Transgene
expression in different thymic populations according to TCR In a previous study, we localized the human homolog of the mouse CD4 silencer to the 484 bpRsaI fragment in the first and co-receptor expression was measured by multi-color
Table 1. Expression level of the transgene in different transgene was expressed at various levels in both CD31and
transgenic lines CD3–cells, indicating that in these cells, construct L is unable
to control the expression of the transgene properly. In the S strains, transgene expression was detected at high levels in
Construct Line Copy numbera Expression levelb
the majority of CD3– cells. However, ~20% of CD31 cells Control 0 1.9 express no or low levels of hCD4 in these strains. Introduction
L #70 ~40 75.0
of fragment A, containing the silencer element active in mature
#72 ~40 70.5
cells (11), repressed the expression of hCD4 in the great
S #29 ~50 137.0
#31 ~50 130.8 majority of CD31cells but only in 75% of CD3–cells. Strikingly,
#47 ~40 81.0 fragment B, which also contains the minimal silencer active
A #10 ~20 65.5
in mature CD8 SP cells, failed to down-regulate the
trans-#19 ~20 53.3
gene in CD3–cells, while it efficiently silenced hCD4
expres-B #11 1 4.4
#50 ~5 24.3 sion in CD31cells. Fragment C showed no apparent effects
C #8 ~100 291.6 in transgene expression.
#16 1 4.0
High-level transgene expression in DP thymocytes requires
aEstimated by Southern hybridization (data not shown).
bMedian of fluorescence intensity of human CD4 in mouse CD4 an intronic sequence of the hCD4 gene
SP lymph node cells using FITC-conjugated B-B14.
We next analyzed CD41CD81DP cells, which are the most abundant cells in the thymus and represent the inter-mediate precursors of mature T lymphocytes. In the L strains, fragment, to which we refer as fragment A (Fig. 1), is functional the transgene was expressed in DP thymocytes at similarly in a CD8 lineage-specific manner. Furthermore, the lineage- high levels as in peripheral CD4 SP T cells (Tables 1 and 2, specific silencing activity was confined within the 190 bp and Fig. 3). On the contrary, the S strains showed reduced RsaI–SacII subfragment, referred to as fragment B (Fig. 1). transgene expression (Tables 1 and 2, and Fig. 3). DP
Fragment C (Fig. 1), the other 296 bp SacII–RsaI sub- thymocytes from the A, B and C strains also expressed
fragment, did not show the silencing activity (11). To the transgene at lower levels than peripheral CD4 SP cells address the question of whether the human CD4 regulatory (Fig. 3). These observations suggest that the 59-PRU alone is elements control transgene expression in thymic populations insufficient for high-level expression of the CD4 gene in DP in the same manner as in peripheral T cells, we measured cells and that these immature thymocytes require a positive the reporter gene expression in the thymus using the same regulatory sequence, distinct from the 59-enhancer, to main-transgenic strains. Because the transgene expression tain CD4 expression at high levels. These data also suggest depends largely on the copy number integrated in the that this DNA element may be located either in the 59 part of genome (Table 1), we evaluated the transcriptional activity of the third intron, which is included in construct L, or in the first the different constructs in thymic populations by comparing
intron but outside of fragment A. the expression of hCD4 in thymocytes with that in CD4 SP
lymph node cells of the same animal.
Full activation of the silencer and the 59-PRU in CD4int/–CD81
We first analyzed thymic DN cells, which include both CD3–
thymocytes cells, representing the most immature T cell precursors, and
In thymic CD4int/–CD81 cells, a weak transgene expression
mature CD31 cells (either TCRαβ1 or γδ1). To distinguish
was detected in the L, A and B strains (Fig. 4), suggesting between these two DN populations, thymocytes were stained
that the silencer is already active when the surface CD4 with anti-CD4 and anti-CD8 mAb with one color, and the
protein starts to decrease. Mice with constructs S and C, negative cells were studied for expression of CD3 and hCD4
lacking the silencer sequence, expressed hCD4 in CD4int/–
(Fig. 2 and Table 2). In the L strains, which bear a construct
with the whole first intron and a part of the third intron, the CD81 cells as strongly as in peripheral T lymphocytes (Fig.
Fig. 2.Two-dimensional dot-plot for CD3 and human CD4 in DN gated thymocytes. The quadrants discriminate positive and negative cells. DN cells were detected by a combination of anti-mouse CD4 and anti-mouse CD8 mAb as described. Numbers in quadrants indicate the percentage of gated cells.
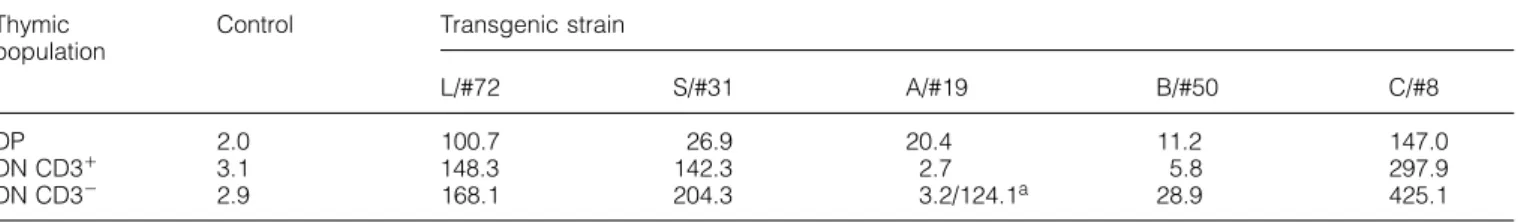
Table 2. Expression levels of transgene in the thymic DP, DN CD31 and DN CD32 (triple-negative) cells in different transgenic strains
Thymic Control Transgenic strain population
L/#72 S/#31 A/#19 B/#50 C/#8
DP 2.0 100.7 26.9 20.4 11.2 147.0
DN CD31 3.1 148.3 142.3 2.7 5.8 297.9
DN CD32 2.9 168.1 204.3 3.2/124.1a 28.9 425.1
Median of fluorescence intensity measured with FITC-conjugated B-B14.
aThis population shows two peaks (Fig. 2). The value of each peak is separately calculated.
Fig. 3.Comparison of transgene expression in the DP thymic population (thick lines) and peripheral CD4 SP cells (thin lines). The gates used for these analyses are indicated by the square in the dot-plot.
Fig. 4.Comparison of transgene expression in the CD4int/–CD81thymic population (thick lines) and peripheral CD4 SP cells (thin lines). The
staining profile of a normal littermate of the same population is superimposed with dotted lines.
4), indicating that the 59-PRU is sufficient for full expression located in the first or in the 59 part of the third intron. The other 70% of this population expresses the transgene at the in this cell population.
same high levels as peripheral CD4 SP cells. Unexpectedly, High-level transgene expression in a fraction of CD41CD8int/–
in the A and B strains, we detected only a smaller number of thymocytes requires an intronic sequence of the hCD4 gene cells with high levels of hCD4 (Fig. 5), implying that the negative regulatory element in fragment B may also be active Thymic CD41CD8int/– cells of the L strains expressed the
transgene at high levels, as expected (Fig. 5). Surprisingly, in the thymic CD41CD8int/–population.
In order to estimate developmental stage of the hCD4lo
the S and C strains showed low transgene expression (hCD4lo)
in ~30% of this population. This observation again suggests cells of the S strain, thymocytes with the CD41CD8int and CD41CD8–phenotype were separately analyzed. The
propor-that, like in DP thymocytes, the expression of the CD4 gene
Fig. 5.Comparison of transgene expression in the CD41CD8int/–thymic population (thick lines) and peripheral CD4 SP cells (thin lines).
or in the 59 part of the third intron, is required for CD4 expression at a high level in the DP and a fraction of CD41CD8int/–populations. They also imply that triple-negative thymocytes may use a silencing mechanism distinct from that of mature CD8 SP lymphocytes. The results presented in this study are consistently obtained from independent lines carrying the same transgenes, indicating that the pattern of reporter gene expression is likely due to intrinsic properties of the transgenic constructs but not to effects of integration sites of the transgenes.
Discussion
In this study we assessed the activity of the positive and negative regulatory elements of the human CD4 gene during T cell development using transgenic mice. For gene activation,
Fig. 6.Comparison of transgene expression in the CD41CD8intthymic
two factors must be considered: (i) expression of activation
population and in the CD41CD8–thymic population in the S strain.
The gates used for these analyses are indicated by the squares in molecule(s) that bind to the regulatory element and (ii) the
the dot-plot. chromatin structure, which determines accessibility of the
regulatory element and is controlled by a locus control region. The 59-PRU, consisting of the promoter and the 59 enhancer, is able to transcribe the reporter gene in construct S in in the CD41CD8– fraction (Fig 6). We further analyzed the
phenotype of the hCD4locells with mAb against other differen- early thymic populations with the DN CD3–phenotype. This indicates that the transcription factors interacting with the 5 9-tiation markers. This cell subset expresses CD3 at similar
levels as the hCD4hicells of the same population (Fig. 7A). PRU are already available in early thymocytes. These results also show that the 59-PRU is accessible in this thymic popula-They also express high levels of CD69 (Fig. 7B) and have a
small cell volume (Fig. 7C). Furthermore, hCD4locells belong tion. However, we cannot distinguish whether this is asso-ciated with the transgene itself or with its particular integration to the HSAhipopulation (Fig. 7D). These findings suggest that
the hCD4lo cells are immature cells, but in a late stage of site. We believe that the latter case is unlikely, because we observed the same phenotype in three independent differentiation, most likely immediately preceding the mature
CD4 SP cells. transgenic S strains. It is still possible that chromatin structure
affects in a variable manner the accessibility of the 59-PRU In conclusion, the analyses of the transgenic thymi showed
that: (i) transcription factors for the 59-PRU are already avail- in different developmental stages.
This transcriptional potential of the 59-PRU is not constantly able in the DN CD3–stage, (ii) the 59-PRU alone is capable
of expressing the transgene only in reduced levels in a maintained throughout the thymic development. Thymic DP cells and a fraction of CD41CD8int/–cells from the S strains fraction of DN CD31, the majority of DP and a fraction of
CD41CD8int/–cells, (iii) the silencer mapped in fragment B is express reduced levels of the transgene (Figs 3 and 5).
CD41CD8int/– hCD4lo cells are CD31 and CD691 (Fig. 7A active in DN CD31, but inactive in triple-negative thymocytes,
(iv) this silencer is already active in CD8 committed late and B), indicating that they have recently been activated, possibly already undergone positive selection (14). On the thymocytes, when the CD4 protein starts to decrease, and
(v) this silencer can also repress transgene expression in other hand, they appear small in size (Fig. 7C) and express high levels of HSA (Fig. 7D), thus displaying an immature CD41CD8int/–cells. These observations suggest that an
Fig. 7.Four-color analyses of CD41CD8int/–cells in the S strain. (A) Left panel, a dot-plot for CD4 and CD8 showing the gate used in this analysis; right panel, a dot-plot for hCD4 and CD3 in the CD41CD8int/–gated thymocytes. (B) A dot-plot for hCD4 and CD69 in the thymocytes
gated as in panel (A). (C) Histograms of forward scatter of total (left panel) and hCD4locells (right panel) of the CD41CD8int/–population. The gates were shown in (A). The vertical bars discriminate small and large cells. (D) Two-dimensional dot-plot for hCD4 and HSA in total thymocytes. The quadrants discriminate positive and negative cells.
59-PRU is decreased from the DP to a stage immediately in most of the thymocytes (12). This discrepancy might be due to the different extent of deletion of the first intron of the preceding to mature CD4 SP thymocytes and suggest that
constructs used in the two studies. Indeed, a sequence of 8 an additional enhancer-like element is required for normal
kb is removed from the human CD4 gene in our construct S, levels of CD4 gene expression in these cell populations.
while a sequence of 6 kb is deleted from the mouse CD4 These results are consistent with the recent study which
gene in the other study and therefore still containing a long showed that transgenic mice carrying the human CD4 cDNA
stretch of the first intron of the mouse CD4 gene. We suggest driven by human CD4 promoter, without intron sequences,
that the difference of the two studies might be explained with also express lower levels of the human CD4 protein in
the deletion, in our construct S, of a region containing an immature thymocytes (15). In both studies, the activity of
important enhancer-like element, which instead is included in the 59-PRU was assayed by measuring surface expression
the construct used in the other study. Another explanation, of the transgene products. Although it has already been
which we believe is unlikely but cannot be excluded, is that demonstrated that CD4 expression is controlled at the
tran-an inefficient interaction occurs between humtran-an regulatory scriptional level in mature T lymphocytes (13), it still remains
elements and mouse DNA binding proteins. This possibility to be determined whether, in thymocytes, the transgene
is not supported by the finding that our transgenic L strains, expression is also repressed at the transcriptional level.
which have the complete first intron of the human CD4 gene, Interestingly, the CD41CD8int/– hCD4lo cells share several
express normal levels of the transgene.
characteristics with CD41 thymocytes which express CD3 The activity of the 59-PRU is also decreased in a fraction and are detected in class II MHC-deficient mice, which lack of DN CD31cells. By analyzing the S and C strains, which mature CD4 SP cells. In these animals the thymic CD4 SP carry the transgenes lacking a large part of the first intron, cells remain immature, as a consequence of the absence of ~20% of DN CD31cells express decreased levels of hCD4,
the MHC molecules required for further maturation. Indeed, as seen in the CD41CD8int/–population. Thus, this population
they appear small and express decreased levels of CD8 and may represent cells at the stage equivalent to the
CD3, but high levels of HSA (16), thus representing a late CD41CD8int/– hCD4lopopulation, being direct precursors of
immature population. Further studies are required to deter- more mature DN CD31T cells.
mine whether both types of cells belong to the same popu- Curiously, in the same strains, the CD4int/–CD81population
lation. lacks hCD4locells. This observation may indicate that
differen-In apparent contrast with our results, transgenic mice tiation stages of CD8 committed cells are different from CD4 carrying a construct similar to our construct S, but with committed cells, as already suggested by several groups
(17–20). elements of mouse origin, express high levels of the transgene
Altogether, our study shows that the transcriptional activity of the 59-PRU is fully operative at early stages of thymic development, when thymic precursor cells are still triple-negative, and is reduced from the beginning of the DP stage until the cells become SP. The period with reduced 59-PRU activity includes the stage when T cells undergo positive selection and overlaps the period when DP cells down-regulate CD8 expression. It remains to be determined whether the change of the 59-PRU activity is instrumental for lineage commitment and whether it is due to the use of different transcription factors binding to the 59-PRU or to a modification of the same factors.
The activity of the CD4 silencer on transgene expression driven by the 59-PRU was then investigated in transgenic thymus carrying constructs A, B or C. We observed that in the DN thymic CD3–and CD31cells this silencer behaves in
different ways. In triple-negative thymocytes, fragment A was partially inhibitory, while fragments B and C were inactive. In DN CD31 cells, fragments A and B were inhibitory, while fragment C was inactive. Thus, in the triple-negative popula-tion, which contains the most immature cells, the CD4 gene is repressed with different mechanisms from those used in CD8 SP cells. In contrast, more mature DN CD31cells control the CD4 gene in a manner similar to CD8 SP cells. This is also the case with thymicγδDN cells, which do not express
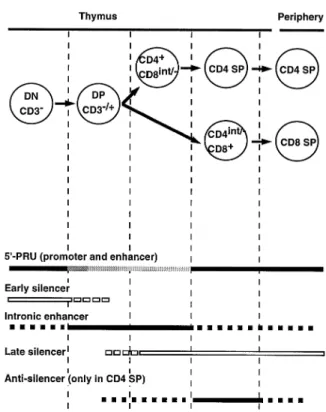
Fig. 8. Regulatory model for CD4 gene expression during T cell
the transgene in the A and B strains (data not shown).
development. Closed bars indicate positive regulatory activity. Open
The inactivity of the intronic silencer in the most immature
bars indicate silencing activity. The reduced enhancing activity of
thymocytes is also shown by the finding that in the L strains the 59-PRU is displayed as gray shading. Broken bars indicate where hCD4 is partially repressed in the DN population, while it is enhancing or silencing activity is not determined.
fully inhibited in mature CD8 SP cells. We propose the following hypothesis to explain these results. A regulatory sequence, present in construct L but outside of fragment A,
attenuates the effect of the CD4 silencer in DN, but not in (11). Our findings suggest that in this latter population, an antisilencing mechanism restores CD4 gene expression. CD8 SP cells. In DN thymocytes, complete repression of the
CD4 gene is mediated by an additional negative regulatory The present study has shown that the combination of
multiple regulatory proteins controls the CD4 gene in a tissue-element, excluded from construct L. Thus, two additional
elements, with opposite regulatory functions and different and stage-specific manner. This type of gene regulation is not peculiar to the CD4 gene. For example, lineage-specific locations in the CD4 locus, would be involved in the CD4
gene regulation of immature thymocytes. Alternatively, it is expression of the TCR γ gene seems to be established by enhancer–silencer interaction (21). Furthermore, YY1, a possible that the copy number of the transgene might affect
the silencing activity in the thymic DN population, because transcription factor which was originally cloned and charac-terized as a repressor protein (22), can be converted into an the L strains carry higher copy numbers of the transgene
than the A and B strains (Table 1). enhancer factor depending on the flanking sequence of the
binding sequence (23,24). Thus the YY1 protein controls An additional unexpected finding of this study is that the
CD4 silencer located in the first intron is not active in a CD8 genes with YY1 binding motifs in a tissue-specific manner. A third example is the regulation of Ig genes driven by the lineage-specific manner, but represses CD4 expression also
in CD4 committed cells. Indeed, in both the A and B strains, octamer motif, which interacts with a panel of octamer binding proteins with various types of tissue-specificity. Oct-1, a which carry two constructs with this silencer element, the
hCD4 transgene is repressed in CD4int/–CD81 as well as ubiquitously expressed octamer binding protein, confers B cell specificity to Ig gene expression when it also binds to a in CD41CD8int/– populations. Therefore, it is necessary to
postulate the presence of an antisilencer, capable of counter- B cell-specific co-activator protein (25,26). Thus, including CD4, a number of genes have been found to be regulated acting the activity of the silencer in the CD4 committed
thymocyte. As a normal expression of the transgene is not by a simple on–off mechanism, but by complex interactions between several regulatory proteins.
observed in the L strains, we conclude that this antisilencer
element is located in the first intron, outside of fragment A. According to our results, we propose the following model for CD4 gene regulation during T lymphocyte differentiation. In conclusion, at least two distinct silencing mechanisms
control CD4 expression during development of SP T lympho- This model consists of six different regulatory elements, which come into play at different stages of differentiation (Fig. 8). cytes in the thymus: (i) an early silencer for DN CD3–
thymo-cytes, and (ii) the late silencer active in CD8 SP thymocytes Among these six elements, the promoter and the enhancer, which constitute the 59-PRU, and the late silencer have been and peripheral CD8 SP T cells as well as CD4 SP thymocytes
PRU positive regulatory unit
characterized (7–13,27,28). The other three are an early
SP single positive
silencer, an intronic enhancer and an antisilencer, which have not been identified so far. In the most immature thymocytes (triple-negative cells), the 59-PRU is already capable of
activat-References
ing the CD4 gene. An early CD4 silencer is active at this stage,
repressing CD4 expression in immature DN thymocytes. Upon 1 Schwartz, R. H. 1985. T-lymphocyte recognition of antigen in association with gene products of the major histocompatibility
progression to the DP stage, the repressive effect of the early
complex.Annu. Rev. Immunol.3:237.
silencer is decreased by down-regulation of the silencer factor
2 Veillette, A., Bookman, M. A., Horak, E. M. and Bolen, J. B. 1988.
or by activation of a competing positive regulatory element. The CD4 and CD8 T cell surface antigens are associated with Activity of the 59-PRU is also decreased and CD4 expression the internal membrane tyrosine-protein kinase p56lck.Cell55:301.
3 Kisielow, P. and von Boehmer, H. 1995. Development and
requires additional enhancement from the postulated
selection of T cells: facts and puzzles.Adv. Immunol.58:87.
enhancer element, likely present in the first intron. In more
4 von Boehmer, H. 1994. Positive selection of lymphocytes. Cell
mature thymocytes, the late silencer, located in fragment A, 76:219.
is activated irrespectively of lineage commitment and, thus, 5 Bendelac, A. and Schwartz, R. H. 1991. CD41and CD81T cells
acquire specific lymphokine secretion potentials during thymic
the late silencer is not per se responsible for the
lineage-maturation.Nature353:68.
specific CD4 gene expression. In CD4int/–CD81 thymocytes
6 Nossal, G. J. 1994. Negative selection of lymphocytes. Cell
and peripheral CD8 SP cells, the CD4 gene is down-regulated
76:229.
as a result of activation of the late silencer. On the other hand, 7 Sawada, S. and Littman, D. R. 1991. Identification and in CD41CD8int/– thymocytes, an additional enhancer or an characterization of a T-cell-specific enhancer adjacent to the
murine CD4 gene.Mol. Cell. Biol.11:5506.
antisilencer is activated, thus conferring the lineage specificity
8 Siu, G., Wurster, A. L., Lipsick, J. S. and Hedrick, S. M. 1992.
to CD4 expression in T cells.
Expression of the CD4 gene requires a Myb transcription factor.
The transgenic mice analyzed in this study express human Mol. Cell. Biol.12:1592.
CD4 from the early stages of thymic development. Since 9 Sawada, S. and Littman, D. R. 1993. A heterodimer of HEB and
an E12-related protein interacts with the CD4 enhancer and
human CD4 has been shown to be able to support T
lympho-regulates its activity in T-cell lines.Mol. Cell. Biol.13:5620.
cyte development (29), it is of immunological interest whether
10 Salmon, P., Giovane, A., Wasylyk, B. and Klatzmann, D. 1993.
human CD4 affects T cell development, including thymic
Characterization of the human CD4 gene promoter: transcription
selection. In all transgenic animals we studied, the proportion from the CD4 gene core promoter is tissue-specific and is of thymic populations (DN, DP and SP) as well as the total activated by Ets proteins.Proc. Natl Acad. Sci. USA90:7739.
11 Donda, A., Schulz, M., Bu¨rki, K., De Libero, G. and Uematsu, Y.
number of thymocytes appeared normal (data not shown).
1996. Identification and characterization of a human CD4 silencer.
However, it still remains possible that T cell repertoire
forma-Eur. J. Immunol.26:493.
tion might be affected by the early expression of hCD4. The 12 Sawada, S., Scarborough, J. D., Killeen, N. and Littman, D. R. transgenic lines we have produced may be a useful tool to 1994. A lineage-specific transcriptional silencer regulates CD4
gene expression during T lymphocyte development.Cell77:917.
address this question.
13 Siu, G., Wurster, A. L., Duncan, D. D., Soliman, T. M. and Hedrick,
In conclusion, the present study analyzing the known
S. M. 1994. A transcriptional silencer controls the developmental
regulatory elements of the human CD4 gene has shown that
expression of the CD4 gene.EMBO J.13:3570.
they control CD4 gene expression only in some thymic 14 Swat, W., Dessing, M., von Boehmer, H. and Kisielow, P. 1993. subpopulations. Furthermore, the results also suggest the CD69 expression during selection and maturation of CD4181
thymocytes.Eur. J. Immunol.23:739.
presence of additional, not yet identified regulatory regions,
15 Salmon, P., Boyer, O., Lore`s, P., Jami, J. and Klatzmann, D. 1996.
which are necessary for the proper CD4 gene control in all
Characterization of an intronless CD4 minigene expressed in
the analyzed thymocyte populations. The complete under- mature CD4 and CD8 T cells, but not expressed in immature standing of CD4 gene regulation during thymic development thymocytes.J. Immunol.156:1873.
16 Cosgrove, D., Gray, D., Dierich, A., Kaufman, J., Lemeur, M.,
requires identification and characterization of these postulated
Benoist, C. and Mathis, D. 1991. Mice lacking MHC class II
DNA elements.
molecules.Cell66:1051.
17 Marodon, G. and Rocha, B. 1994. Generation of mature T cell populations in the thymus: CD4 or CD8 down-regulation occurs Acknowledgements
at different stages of thymocyte differentiation.Eur. J. Immunol. 24:196.
We gratefully acknowledge A. Braun and M. Dessing for technical
18 Lundberg, K., Heath, W., Kontgen, F., Carbone, F. R. and assistance, and A. Livingston, U. Staerz and S. Takeda for the
Shortman, K. 1995. Intermediate steps in positive selection: generous gifts of some of mAb. We also thank P. Kisielow and
differentiation of CD418int TCRint thymocytes into CD4–81TCRhi
L. Mori for useful discussions, and M Bu¨rk for careful reading of the
thymocytes.J. Exp. Med.181:1643. manuscript. This work is supported by Swiss National Science
19 Suzuki, H., Punt, J. A., Granger, L. G. and Singer, A. 1995. Foundation (NF31-36686.92) to Y. U., Stiftung der Emilia
Guggenheim-Asymmetric signaling requirements for thymocyte commitment to Schnurr to Y. U. and Margarete und Walter Lichtenstein-Stiftung to
the CD41 versus CD81 T cell lineages: a new perspective on G. D. L.
thymic commitment and selection.Immunity2:413.
20 Benveniste, P., Knowles, G. and Cohen, A. 1996. CD8/CD4 lineage commitment occurs by an instructional/default process Abbreviations
followed by positive selection.Eur. J. Immunol.26:461.
21 Lefranc, M. P. and Alexandre, D. 1995. γδ lineage-specific DN double negative
DP double positive transcription of human T cell receptorγgenes by a combination of a non-lineage-specific enhancer and silencers. Eur. J. hCD4 human CD4
HSA heat-stable antigen Immunol.25:617.
22 Shi, Y., Seto, E., Chang, L. S. and Shenk, T. 1991. Transcriptional PE phycoerythrin
repression by YY1, a human GLI-Kru¨ppel-related protein, and novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding relief of repression by adenovirus E1A protein.Cell67:377.
23 Natesan, S. and Gilman, M. 1995. YY1 facilitates the association proteins.Cell80:497.
27 Hanna, Z., Simard, C., Laperriere, A. and Jolicoeur, P. 1994. of serum response factor with the c-fos serum response element.
Mol. Cell. Biol.15:5975. Specific expression of the human CD4 gene in mature CD41
CD8–and immature CD41CD81T cells and in macrophages of
24 Bauknecht, T., Jundt, F., Herr, I., Oehler, T., Delius, H., Shi, Y.,
Angel, P. and Zur-Hausen, H. 1995. A switch region determines transgenic mice.Mol. Cell. Biol.14:1084.
28 Blum, M. D., Wong, G. T., Higgins, K. M., Sunshine, M. J. and the cell type-specific positive or negative action of YY1 on
the activity of the human papillomavirus type 18 promoter.J. Lacy, E. 1993. Reconstitution of the subclass-specific expression of CD4 in thymocytes and peripheral T cells of transgenic mice: Virol.69:1.
25 Gstaiger, M., Knoepfel, L., Georgiev, O., Schaffner, W. and identification of a human CD4 enhancer.J. Exp. Med.177:1343. 29 Killeen, N., Sawada, S. and Littman, D. R. 1993. Regulated Hovens, C. M. 1995. A B-cell coactivator of octamer-binding
transcription factors.Nature373:360. expression of human CD4 rescues helper T cell development in mice lacking expression of endogenous CD4.EMBO J.12:1547. 26 Strubin, M., Newell, J. W. and Matthias, P. 1995. OBF-1, a