Building a Paneth Cell: Exploring Cellular Origin and
Function to Inform Intestinal Host-Microbe Interaction and
Disease
by
Benjamin E. Mead
B.S. Chemical & Biological Engineering University of Colorado Boulder, 2013 Submitted to the Signature MASSACHUSETTS INSTITUTE OF TECHNOLOGY
JUN 1 1
2018
LIBRARIES
ARCH
IVES
Harvard-MIT Program in Health Sciences and Technology, in PartialFulfillment of the Requirements for the Degree of
Doctor of Philosophy in Medical Engineering and Medical Physics at the
MASSACHUSETTS INSTITUTE OF TECHNOLOGY
June 2018
2018 Massachusetts Institute of Technology. All rights reserved.
Signature redacted
of A uthor ... ...
Harvard-MIT Program in Health Sciences and Technology May 09, 2018
Signature redacted
C ertified By ... ...
Jeffrey M. Karp, PhD Professor of Medicine Brigham & Women's Hospital, Harvard Medical School Thesis Supervisor
Signature redacted
A e BEmery N. Brown, MD, PhD
Di or, Harvard-MIT Program in Health Sciences and Technology Professor of Computational Neuroscience and Heath Sciences and Technology
Building a Paneth Cell: Exploring Cellular Origin and Function to Inform Intestinal Host-Microbe Interaction and Disease
by
Benjamin E. Mead
Submitted to the Harvard-MIT Program in Health Sciences and Technology on May 09,
2018 in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in
Medical Engineering and Medical Physics
ABSTRACT
Understanding how our bodies interact with our resident gut microbes may unlock new therapies for multiple diseases. However, we presently lack representative models to study such interactions, limiting therapeutic development. Here, we advance methods of cellular bioengineering to create high-fidelity models of intestinal cell types, in particular the microbe-interfacing antimicrobial Paneth cell. With this model, we study at scale molecular interventions that direct Paneth cell development and function as a means to modulate our gut microbes in health and disease.
Multiple diseases are linked to alterations in Paneth cell function and the composition of the gut bacteria, including inflammatory bowel disease. Past studies of Paneth cells have relied on complex and poorly scaled animal models, or limited in vitro models, including stem cell-derived organoids. The extent to which in vitro models of Paneth cells reproduce
in vivo biology is an unanswered question central to their utility in studying host-microbe
interaction for therapeutic development.
We first present a generalizable approach using single-cell RNA sequencing to compare the identity of in vivo Paneth cells to those of in vitro organoids and based on lineage-defining differences between the two, nominate small molecule interventions to improve model representation. We then validate our improved Paneth cell model through rigorous characterization and a demonstration of functional (antimicrobial activity, niche support) improvements in Paneth cell physiology following our intervention.
With this high-fidelity Paneth cell model, we built a scalable platform to study interventions which may enhance cell function and development. As a proof-of-concept screen, we use a well-defined and clinically-relevant set of small molecules to identify drugs that enhance Paneth cell differentiation and antimicrobial function. We validate the most potent drugs with additional analyses, revealing multiple molecular targets that may serve as therapeutic candidates to restore Paneth cells in disease or act as a new approach to therapeutically shape the gut microbiota.
Thesis Supervisor: Jeffrey M. Karp, PhD
Title: Professor of Medicine
ACKNOWLEDGEMENTS
A thesis is not only a record of scientific and academic pursuits, but proof of the
innumerable provisions of a community. To my community, I owe the greatest thanks for your guidance, support, and steadfast belief in me.
To my many undergraduate advisors and mentors, especially Dr. Diana Carlone, Dr. Joe Magee, and Prof. Stephanie Bryant, whose relentless encouragement led me from the waitlist of a summer research program long ago on to graduate school.
To Prof. Jeff Karp, for your unconditional and unrelenting conviction in maximizing impact, and unwavering belief in my success. Thank you for guiding me to possibilities and opportunities which I could have never imagined. To my thesis committee, it has been an honor and a privilege to learn from you. Prof.'s Jim Collins, Alex Shalek, and Scott Snapper, thank you each for your invaluable recommendations, guidance, and perspective.
To "Team PC", for believing in our work and embarking on this occasionally misguided adventure. Your many efforts have made this thesis all that it is. Allie Braun, Dustin Ammendolia, Sammy Zheng, Lauren Levy, Andy Yang, and Daphne Sze, thank you for your incredible capacity to learn, your patience, commitment, and friendship.
To the Karp and Langer Labs, specifically Prof. Robert Langer, Prof. Xiaolei Yin, Dr. Oren Levy, Dr. Nitin Joshi, Dr. Yuhan Lee, Dr. Kelvin Ng, Peter Jones and the many other lab mates whose advice, perspectives, and camaraderie has been invaluable.
To my many amazing collaborators, and in particular Dr. Jose Ordovas-Montanes, Dr. Prerna Bhargava, Dr. Jaime Cheah, and Christian Soule for your guidance, assistance, and patience through our many 'iterations' of experiments.
To the Kenneth Rainin Foundation, the National Science Foundation, Brigham & Women's Hospital, and the Harvard-MIT program in Health Sciences & Technology, for your generous support of my education and research.
To my many peers and friends in the HST and MIT community, thank you for sharing this experience with me, I am continually inspired by all of your amazing accomplishments and genuine humanity. From our early morning bus rides across the river to HMS lectures to our even earlier mornings in the wards of Mt. Auburn Hospital, your fellowship has made the past five years extraordinary.
To my best friend and fianc6e, Katy, thank you for being there through thick and thin, I couldn't have said it better, "My greatest support would be my future wifey... She is amazing". To my parents, thank you for always embracing and fostering my interests in science and medicine and to my brothers, thank you for always keeping me humble and grounded in reality - I may not be the most athletic, but show me a multiple choice test I can't win!
TABLE OF CONTENTS
ABSTRACT ... 3 ACKNOW LEDGEMENTS... 5 TABLE OF CONTENTS ... 7 LIST OF TABLES ... 11 LIST OF FIGURES ... 111. HARNESSING HOST-MICROBE INTERACTION IN HUMAN HEALTH ... 13
1 .1 P re fa c e ... 13
1.2 Introduction ... 13
1.3 How did we get here?... 13
1.3.1 A brief history of the human-microbe relationship ... 13
1.3.2 Government investment, venture-backing, and industry collaboration power microbiome-inspired therapeutic development ... 15
1.3.3 Contending with complexity ... 15
1.3.4 Emerging approaches to microbiome-inspired therapeutic development...17
1.4 Bugs as drugs: the FMT, FMT-derived approaches, and probiotics...18
1.4.1 The support for bugs as drugs ... 18
1.4.2 FMT's pioneering success supports a wave of microbiota-inspired approaches. 19 1.4.3 Probiotics to complement FMT...20
1.4.4 Probiotics via a GI-axis, for autoimmune, malignant, and metabolic disease...21
1.4.5 Emerging probiotic approaches in malignancy, food allergy, and early life...23
1.4.6 Probiotics for women's health ... 24
1.4.7 Probiotics for the skin... 24
1.4.8 A path forward for bugs as drugs ... 24
1.5 Drugs for bugs: a plethora of prebiotics and next-gen 'antibiotic' approaches .25 1.5.1 Prebiotics in metabolic syndrome... 26
1.5.2 Prebiotics for food allergy... 27
1.5.3 Targeted elimination of problematic bacteria via phages, CRISPR, small molecules, and peptides... 28
1.5.4 Xenobiotics as unintentional drugs for bugs...29
1.5.5 A path forward for drugs for bugs... 29
1.6 Drugs from bugs... 30
1.6.1 SCFAs for asthma...31
1.6.2 Micro biota-derived agents in malignancy ... 31
1.6.3 Searching for microbe-derived bioactive small molecules ... 31
1.6.4 A path forward for drugs from bugs...32
1.7 Thinking outside the bug - the frontier of microbiome research... 33
1.7.1 Autoimmune disease ... 33
1.7.2 The gut-brain axis...34
1.8 Securing the microbiota's impact on health... 35
1.9 Acknowledgements... 36
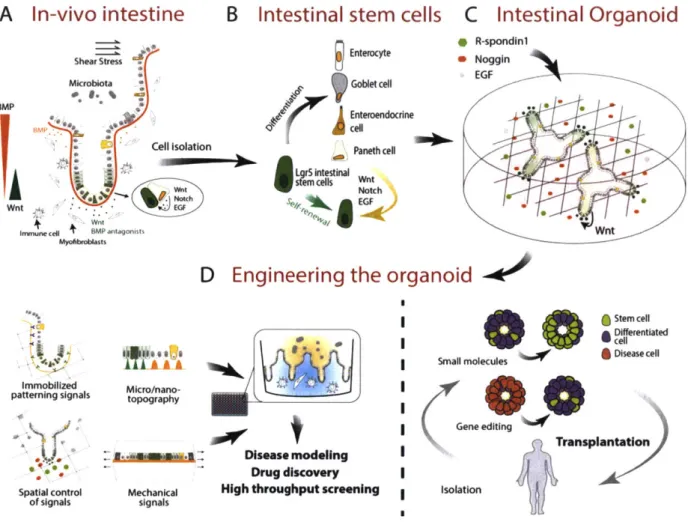
2.1 Biological model systems... 37
2.2 Organoids: self-organizing systems of stem cells & progeny ... 39
2.3 Intestinal Organoids... 41
2.4 Bioengineering organoid systems ... 43
2.5 Engineering the niche ... 44
2.5.1 Customized biomimetic scaffolds ... 44
2.5.2 Adaptability to cell-induced modifications... 46
2.5.3 Spatio-temporal control... 47
2.5.4 Bio-printing and bottom up approaches...47
2.5.5 Monitoring systems in vitro... 48
2.6 Conclusions...49
3. IMPROVING PHYSIOLOGICAL FIDELITY OF ORGANOID-DERIVED PANETH CELLS... 51
3.1 Introduction ... 51
3.1.1 Paneth cells are critical mediators of host-microbe interaction ... 52
3.2 R es u lts ... 53
3.2.1 Benchmarking the Paneth cell of conventional organoids ... 53
3.2.2 Chemical modulation of Wnt and Notch drives Paneth cell enrichment... 57
3.2.3 The proteome as a global measure of Paneth cell representation... 61
3.2.4 Profiling heterogeneity & in vivo representation after chemical induction ... 65
3.2.5 Assessing gains in physiological function after chemical-induction ... 71
3.3 Discussion & conclusions ... 74
3.4 Materials & methods... 77
3.4.1 Crypt culture, enrichment, differentiation... 77
3.4.2 RNA extraction & qRT-PCR ... 78
3.4.3 Confocal imaging of whole cell clusters... 78
3.4.4 High-resolution single-cell imaging... 79
3.4.5 Flow cytometry... 79
3.4.6 Lysozyme functional secretion assay... 79
3.4.7 Quantification of cell viability, apoptosis, cytotoxicity... 80
3.4.8 Bacteria co-culture... 80
3.4.9 Mass spectrometry proteomics preparation & quantification ... 81
3.4.10 Quality control of mass spectrometry performance and data generated... 82
3.4.11 Protein and peptide identification and quantification... 82
3.4.12 Proteome pathway and network analysis... 83
3.4.13 Single-cell RNA-sequencing ... 84
3.4.14 Single-cell RNA-sequencing computational pipelines and analysis... 84
3.4.15 Transcriptional scoring... 85
3.4.16 Quantification and statistical analysis... 86
3.5 Acknowledgements... 86
4. SCALABLE SCREENING OF IN VITRO PANETH CELLS INFORMS FUNCTION, DEVELOPMENT, & SURVIVAL ... 88
4.1 Introduction ... 88
4.1.1 The cellular model bottleneck in drug discovery... 89
4.1.2 Limitations to scaling and screening organoid cultures ... 90
4 .2 R es u lts ... 90
4.2.2 Primary small molecule screen for molecular targets to enhance Paneth cell d e ve lo p m e nt...9 6
4.2.3 Secondary screening validates potent targets to selectively enhance Paneth cell d iffe re ntia tio n in vitro ... 10 1
4.3 Discussion & conclusion...106
4.4 Materials & methods...107
4.4.1 Crypt isolation and organoid culture...107
4.4.2 High-throughput screening: 96-well format...108
4.4.3 High-throughput screening: 384-well plating and assays...108
4.4.4 High-throughput screening: 384-well primary screen format and analysis...109
4.4.5 High-throughput screening: 384-well secondary screen format and early vs. full tre a tm e nt a n a lysis ... 1 1 1 4.4.6 Flow cytometry profiling of organoids in "2.5-D"...112
4.5 Acknowledgements...112
5. PERSPECTIVE & FUTURE DIRECTIONS... 114
5.1 Therapeutic development in the microbiome requires new models - organoids may provide a platform...114
5.2 Contributions to high-fidelity ex vivo model development...115
5.2.1 Rational model design using in vivo benchmarking...115
5.2.2 Rapid model prototyping with scalable chemical induction...116
5.3 Future directions ... 117
5.3.1 Harnessing methods of model development in new systems ... 118
5.3.2 Advancing therapeutic candidates for inflammatory bowel disease...118
LIST OF TABLES
Table 1.1: Companies advancing 'bugs as drugs' therapeutics ... 25
Table 1.2: Companies advancing 'drugs for bugs' approaches... 30
Table 1.3: Companies advancing 'drugs from bugs' approaches... 32
Table 3.1: TaqMan gene expression assays used for qRT-PCR ... 78
LIST OF FIGURES
Figure 1.1: Approaches for microbiome-inspired therapeutics... 17Figure 2.1: Model systems in the life sciences... 38
Figure 2.2: O rganoid developm ent... 40
Figure 2.3: Intestinal organoids... 42
Figure 2.4: Bioengineering approaches to advance organoid-based research and th e ra p y ... 5 0 Figure 3.1: Transcriptional benchmarking of in vitro Paneth cells to in vivo... 56
Figure 3.2: Establishing chemically-induced PC-enriched cultures... 59
Figure 3.3: Image analysis of cell clusters and flow cytometry... 60
Figure 3.4: Characterizing the in vitro PC proteome ... 63
Figure 3.5: Proteomic pipeline and sample-to-sample comparison... 64
Figure 3.6: Structural and functional insights from the in vitro PC proteome... 65
Figure 3.7: Quality metrics for single-cell RNA sequencing ... 66
Figure 3.8: Single-cell RNA-sequencing reveals cellular composition across treatments and origins of proteom ic data... 67
Figure 3.9: Transcriptional identity of Paneth cells within conditions and related to in v iv o ... 6 9 Figure 3.10: Signaling pathways and processes associated with in vitro PC enrichment ... ... ... ... ... . . ... ... ... 7 0 Figure 3.11: CI-PCs have superior function to conventional organoids ... 72
Figure 3.12: CI-PCs reveal putative function of Nupri transcription factor in PC survival ... ... .... ... ... .... ... ... ... ... ... . . . 7 4 Figure 4.1: Scaling Paneth cell culture for secretory measurements ... 92
Figure 4.2: Screening for short-term secretory agents... 94
Figure 4.3: Assessing physiologic-agent effect on in vitro Paneth cell function ... 95
Figure 4.4: Primary screen for small molecule modulators of in vitro PC differentiation 97 Figure 4.5: Quality control metrics for primary screen... 99
Figure 4.6: Dose-response curves for primary screen hits... 100
Figure 4.7: Secondary screening validates set of small molecule PC differentiation e n h a n c e rs ... 1 0 2 Figure 4.8: Effect of early versus full treatments of select agents on PC differentiation ... ... .... ... 1 0 3 Figure 4.9: Cross screening primary hit small molecules in conventional organoids.. 105 Figure 4.10: PC phenotypes for select small molecules in PC-enriched and conventional o rg a n o id s ... 1 0 5
1. HARNESSING HOST-MICROBE INTERACTION IN HUMAN
HEALTH
1.1 Preface
Developing new therapies for complex disease - those of overlapping genetic and environmental origin - may be greatly expedited if highly representative (high-fidelity) model systems with which to study, identify, and validate therapeutic targets were available. Specifically, the emergence of the human microbiota's role in complex disease (including autoimmune, metabolic, and neurologic), has demonstrated the importance of the complex interactions between our tissues and our resident microbes. Such exchanges begin at the intestinal epithelium, where interactions with the gut microbiota are believed to precipitate many types of complex disease, including inflammatory bowel disease. This thesis seeks to advance new approaches to model a specialized cell of the intestinal epithelium, the Paneth cell, which has been clearly linked to complex disease through genetic susceptibilities that alter host-microbe interaction.
As an introduction to the scope of microbiota-associated disease and the breadth of emerging therapeutic approaches, this chapter reviews the present knowledge of microbe interaction in disease as well as emerging therapeutic approaches via host-microbe interaction. This introductory chapter serves to motivate the opportunity for improved models of 'host' systems to expedite development of new therapeutics for complex disease.
1.2 Introduction
The past two decades have produced remarkable breakthroughs in our understanding of the microbes that live within and around us. The human microbiota - the total microbial population inhabiting the human being - exists in near equal number to our own cells'. Some consider our microbiota to be as important as an organ of the human body, having significant correlative and causal associations with human health3. Human microbial communities are implicated in a wide spectrum of disease including obesity, diabetes, autoimmune disease, and infection4-6. As such, the development of microbiota-derived
and -targeted therapeutics is a rapidly emerging business. While this industry has been profiled from both regulatory and commercial perspectives 7-10, a concise understanding
of the scientific breakthroughs across disease areas and technical and biological challenges remaining on the path to commercial success is lacking. Here, we seek to address how scientific advances have driven the present business of microbiome-inspired therapeutics, where the science suggests this industry's impact on disease is most
promising, and the key unknowns which may validate therapeutic opportunities.
1.3 How did we get here?
Since 7000 B.C.E., humans have used microbes in the creation of fermented foods and traditional medicine1 1 - Ancient Egyptians used moldy bread on wounds to prevent
infections1 2 1 3. Yet, it was not until the advent of microscopy in the late
17th century by
Antonie van Leeuwenhoek that we had the first recorded observations of microorganisms. van Leeuwenhoek's observations of stool revealed microbes within the body - a finding that was confirmed 130 years later by the physicians Theodor Frerichs and Theodor Billroth, who observed microorganisms in the intestinal tract14. In the 1860's, the medical
community began the search for tools of microbial destruction, stemming from Louis Pasteur's germ theory, which proposed that infectious diseases are caused by microorganisms, and the work of Joseph Lister, who pioneered the use of antiseptics in surgery to prevent infection15. Most notable was Alexander Fleming's discovery of penicillin in 1928, which jumpstarted the antibiotic era. The development of antibiotics and advances in public health initiatives resulted in a nearly 14-fold reduction in infectious disease deaths in the 20th century16.
However, the antibiotics renaissance entered a twilight in the 2 1st century, in part due to
an incomplete understanding of microbial ecology and poor clinical practices that contributed to the rise of antibiotic resistance17-19. To combat antibiotic resistance,
medicine has embraced changing practices and developing therapies that were once deemed 'fringe' microbiology - including phage therapy, fecal microbiota transplant (FMT) to control pathogenic microbes in the body20,2 1. Specifically, phage therapy and FMT are
gaining traction due to their increased specificity and reduced risk of developing resistance10' 22
The 2 1st century has also brought a shift in perspective towards microbes as supportive
of good health, not just as pathogens. Dietary trends embracing natural foods and products have provided a platform for probiotics-ingestible or topical 'beneficial' bacteria2 3-to emerge as supplements and clinically-validated therapeutics. It is projected
that by the year 2020, the global market for probiotics as dietary supplements will be over
$50 billion2 4.
In addition to the changing clinical needs and desires of present, major technological and scientific advances have brought the microbiome, and it's role in health to prominence. Advances during the Human Genome Project (HGP)2
5-27 directly enabled investigators to
probe the genetic identities of microbial ecosystems28. Individuals and consortia, including the Human Intestinal Metagenome Initiative and the European MetaHIT project, established the techniques and bioinformatic tools to enable metagenomic and
metatranscriptomic studies that have provided a quantitative description of the human microbiome29
-3 3. As sequencing-based techniques and analyses of human microbiota
revealed new insights into the relationship between community variation and microbial load (a factor in both health and disease3 4), potential therapeutic approaches have
emerged. To step beyond profiles of the microbiota, developments in genetic engineering and advances in microbial culturing protocols 35, aseptic facilities, and animal models have provided methods for functional preclinical hypothesis testing, enabling academic research and commercial ventures to expand into the microbiota-inspired drug space.
1.3.2 Government investment, venture-backing, and industry
collaboration power microbiome-inspired therapeutic development
The U.S. National Institutes of Health (NIH) was one of the earliest supporters of microbiota research. The Human Microbiome Project (HMP), established in 2008, provided a major impetus for microbiome research27
,30,37. The NIH contributed $173
million towards the consortium's effort and, from its success, now supports microbiome research across 16 of its 27 centers38. In spring 2016, the United States White House
Office of Science and Technology Policy announced the National Microbiome Initiative
(NMI), a one-year, multi-agency, $120 million research initiative. This announcement
coincided with over 100 external institutions announcing new efforts in microbiome research and translation, including the Gates Foundation ($100 million) and JDRF ($10 million )39.
Accordingly, the number of companies advancing microbiome-based therapeutic candidates has increased exponentially. This growth coincides with unprecedented investment in biotechnology. Many early microbiome-inspired ventures have inked deals through institutional investors, largely venture capital. Notably, Flagship Pioneering founded Seres Therapeutics Inc.-the first microbiome venture to go public (2015). PureTech Health has also emerged as key player in creating and funding microbiome-derived immune modulators with the aid of several global industry leaders. PureTech supports Vedanta's microbiome work targeting C. difficile, inflammatory bowel disease (IBD), and immuno-oncology as well as Commense's preclinical investigation for pediatric asthma, allergy, and other pediatric autoimmune disorders.
In addition to institutional investors, at least three other models have emerged to engage translational efforts in the microbiome. Partnerships or formal collaborations, such as those between start-ups Enterome, Finch Therapeutics, Vedanta Biosciences, and Caelus Health backed by Takeda and Johnson and Johnson's Janssen Pharmaceuticals. These partnerships include modest internal effort from the larger company, an infusion of venture funding, and often academic collaborations. Another approach involves established pharmaceuticals, including Novartis and Merck, committing appreciable internal efforts and working through collaborations with academic centers (e.g. the Novartis-Foundry Sequence-to-Molecule Pipeline collaboration with UCSF, the Broad Institute, MIT, and Harvard University). Finally, several large pharmaceuticals have carved out standalone groups, providing substantial internal investments to build microbiota-inspired pipelines, e.g. J&J's Janssen Human Microbiome Institute. Furthermore, these established collaborations provide the new ventures expertise and guidance through the complicated FDA clinical trial and approval process.
1.3.3 Contending with complexity
Given the number of human diseases associated with the microbiome and the wide variety of approaches under therapeutic investigation, the field is vast and complex. How microbiota-based or -inspired therapeutics are defined, how each 'definition' is characterized, and how each product is reproduced with high fidelity are questions that
must be addressed. These points are especially true of probiotics and FMT approaches. The regulatory line between probiotics as dietary supplements and probiotics as medication is ill-defined. As the field moves forward, defining "supplement," which is not
FDA regulated, versus clinical product, which is FDA regulated, may be a major driver of
product value. Similarly, questions around patentability of microbiota-derived therapeutics will likely steer commercial development. As something that exists in nature, bacteria are not patentable, although engineered or synthetic derivatives may be.
Further, a therapeutic cannot exist without a defined medical issue to treat. Microbiota-based therapeutics are being developed for microbiome-associated disease, but what qualifies as a microbe-associated disease? Researchers and clinicians must be careful to demonstrate microbiota changes are causal for a disease rather than a byproduct of disease. Additionally, if microbiome-associated disease develops over time from environmental exposures, when should therapeutic interventions be employed? Will predisposed individuals need intervention prior to the emergence of symptoms, and further, can an intervention on the microbiota re-establish a healthy system following a long established dysbiosis? Showing superior efficacy in relation to current drugs and lifestyle changes will be a high bar to meet, but critical to validate the field. Finally, given the limitations to clinical trials with infants and children, despite substantial research highlighting early-life exposure as one of the most significant contributors to microbiota-associated disease, ideal therapeutic windows or patient populations may never be investigated clinically.
As well, given differences in patients' microbiomes with the same diagnosis, can a generalized microbiome therapeutic be effective, or is a population-specific formulation required to advance to the clinic? Further, misdiagnosis and mis-selection of patients will hinder clinical translation, a potentially daunting complexity considering the variations within the human microbiome. Short-term temporal variability due to environment provides an additional confounding factor that may elicit a misdiagnosis of microbiota-associated disease phenotypes40. These issues highlight the need for increased
characterization of the microbiota to individual patient and strain-specific levels, including standardized analytic approaches to ensure interstudy reproducibility41. However, there may also be "one size fits most" approaches as in the case of FMT for C. difficile infection
(CDI). Indeed, while there is significant variance in the composition of 'settled' microbial
populations within adults, the gut microbiota is relatively conserved at the species-level across human populations. This suggests that we all possess a limited set of symbiotic passengers rather than a continuum of varying microbiota42. Therefore, more generalized, rather than personalized microbiota-derived and -targeted therapeutic products may provide benefits across patient populations and allow for if not, 'one-size-fits-all', then at least 'some-sizes-fit-most' approaches.
Consideration of these questions will be critical as companies advance their microbiota-based therapeutics through regulatory agencies and test their competitiveness on the market. As with all new avenues of therapeutic development, complexity can be offset by unprecedented efficacy, if microbiota-inspired therapeutics can be shown to provide
solutions to daunting medical dilemmas, overcoming regulatory and economic concerns becomes much easier to stomach.
1.3.4 Emerging approaches to microbiome-inspired therapeutic
development
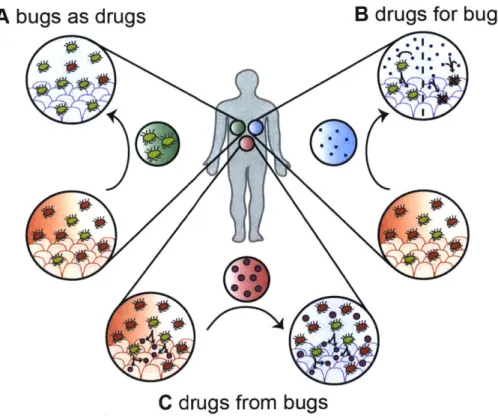
To clarify these translational efforts, we've grouped therapeutic approaches by broad concepts- bugs (bacteria) as drugs, drugs for bugs, and drugs from bugs. We have chosen to divide approaches along these three axes as they best correspond to the spectrum of clinical development in the field, and share many of the same scientific, clinical, and regulatory complexities. 'Bugs as drugs' defines therapeutics that deliver a single bacterium or set of bacteria to a body site of interest-the drug is the bacteria-such as in FMT and probiotics. 'Drugs for bugs' refers to natural or synthetic small molecules or biologics that are delivered to host bacteria (broadly or in a species-specific manner) to elicit therapeutic changes on those targeted bacterial populations, e.g. prebiotics and bacteriophages. 'Drugs from bugs' consists of small molecules or biologics which are derived from or are altered by host bacteria-notably bacterial metabolic byproducts-which effect other members of the host microbiota or host to alter disease (Figure 1.1).
A bugs as drugs
B drugs for bugs
C drugs from bugs
Figure 1.1: Approaches for microbiome-inspired therapeutics
A) Bugs (bacteria) as drugs: delivery of live or spore bacteria (from single isolate to whole community)
to a microbiome alters resident microbiome ecology and provides therapeutic effect.
B) Drugs for bugs (bacteria): delivery of agents to either selectively promote the expansion of desired symbiotic bacteria (including prebiotics) or selectively eliminate pathogens or dysbionts (including phage approaches) within a microbiome to provide therapeutic effect.
C) Drugs from bugs (bacteria): delivery of agents identified to originate from the microbiota and provide
1.4 Bugs as drugs: the FMT, FMT-derived approaches, and probiotics
'Bugs as drugs' constitute isolated bacteria (from various sources) which are delivered as the active component-the "drug", engraft in a microbiota and shift microbial composition to provide therapeutic benefit. We break this category into two approaches: FMT and FMT-derived therapeutics, and probiotics. By in large, FMT and FMT-derived therapeutics utilize enema or oral delivery of donor-derived (or inspired) bacteria into the colon. Probiotic approaches (of typically a single or few bacteria) also utilize oral delivery to the
GI tract or topical delivery to the vaginal canal or skin microbiotas. While proposed
mechanism of action is similar for both approaches-bacteria engrafting and altering local ecosystems-they differ in terms of product development for clinical translation, highlighting two paths with distinct challenges and advantages. FMT-based approaches grew out of a defined FDA pathway beginning with Investigational New Drug (IND) status for FMT in CDI, while probiotics have developed from unregulated dietary supplements to under limited regulation.
1.4.1 The support for bugs as drugs
In 1989, David Strachan proposed the hygiene hypothesis, positing that a developed society's efforts to reduce exposure to infectious agents through improved hygiene (e.g. antibiotics, clean water, household cleaners) limits exposure to microbes important for training immune responses, which in turn increases the rates of autoimmune and allergic disease4 3. As a measure of the importance of this environmental interaction, Brodin et al. have determined from a cohort of 210 twins that a majority of our individual immunological variations stem from non-heritable, environmental, factors, to which the microbiota contributes44. Additional investigations have begun to support this hypothesis and
detangle the complex web of interactions between human genetics, microbial communities, and the environment45,46.
Early-life exposure to the "right" microbiota is central to healthy development and reduces lifetime risk of autoimmune disease. The human microbiota is established at birth: the infant is first inoculated by the mother's vaginal canal, then by breast-milk and environmental exposures47. For those infants born by Cesarean section, however, the neonatal microbiome reflects that of the mother's skin microbiome, rather than that of the proposed 'friendlier microbe'-containing vaginal canal48. By the end of the first year of life,
the infant microbiome has generally reached a steady-state that persists relatively unchanged through adulthood49,50. Multiple studies with germ-free and conventional animal models, as well as prospective humans studies, have established that the
microbiota shapes the immune system51.
Most emerging approaches for targeting and manipulating the human microbiota for therapeutic effect deliver bacteria (live or dormant) to the gut through an FMT or probiotic approach. Many of these approaches are in trials or on the market for diseases in which
for clinical translation. This includes the role of microbiota in normal development, metabolic diseases, autoimmune or immune-mediated processes, and cancers.
1.4.2 FMT's pioneering success supports a wave of microbiota-inspired approaches
As the first FDA-sanctioned microbiota-based therapeutic, FMT exemplifies how clinical
translation of microbiome research occurs. In FMT, whole-biome fecal samples from healthy donors are transplanted into recipients via enema, colonoscopy, or upper tract endoscopy to 'reset' the recipient's microbial ecosystem. FMT was conditionally granted
IND status by the FDA in 2013 for the treatment of recurrent gastrointestinal CD15 2.
Standard treatment for CDI is antibiotics; however, antibiotic-resistant infection recurs in more than 20% of cases53. FMT has shown remarkable efficacy in treating antibiotic-resistant infection with resolution in 90% of treated patients at least one year after treatment54-56. Due to its IND status allowing for accelerated approval for clinical use, FMT presents as an attractive option for commercialization. Currently, over 200 clinical studies are underway or have been completed using FMT as an intervention (per clinicaltrials.gov).
However, FMT is limited by differences in donor microbiota and our understanding of the mechanisms against recurrent infection. "Next-generation" FMT treatments aim to simplify the collection and enteric delivery of therapeutic material and standardize the processing of fecal material to increase efficacy and safety. Indeed these steps to standardize and define the FMT process have been codified by a revision of the FDA's initial IND status regarding FMT for CD157. The new guidelines require that stool banks,
but not physicians nor hospital-based labs, must submit an IND to obtain and distribute donor stool for FMT. This reduces the barriers for physician access to stool for FMT, while increasing regulatory (donor screening and characterization) measures for stool banks. The hope is that the new guidelines do not discourage investment into stool banks, but rather encourage advancement of safe and effective microbiota-derived therapeutics. OpenBiome has emerged as a leader in development and distribution of FMT technology. The non-profit organization runs the first public stool bank, collecting stool samples from rigorously screened donors and providing the material to clinics for CDI treatment and in-house research. Recently, the company has made G3 capsules for oral delivery of concentrated fecal-derived bacteria available. In October 2015, OpenBiome announced a personal stool biobanking program called "Personalbiome," allowing patients to bank their own healthy stool in preparation for therapies that could negatively impact their microbiota-autologous FMT. Furthermore, OpenBiome has reached into a variety of clinical applications and, through partnership with many hospitals, has begun FMT trials for multiple GI-indications including IBD, IBS, cirrhosis, and others. In the same vein, companies such as Pureflora Inc. are focused on developing systems to simplify collection, storage, and processing of fecal material in order to standardize processing for optimal preservation of anaerobic species and to reduce labor costs 58.
In an attempt to clarify therapeutic mechanisms and simplify the therapeutic product, multiple startups have begun to identify, derive, and assess the specific components of FMT. Symbiotic Health has developed a non-invasive and scalable FMT approach (their proprietary enteric biologics delivery platform). In a preliminary human trial, their orally-administered healthy fecal-derived capsule demonstrated 68% single-treatment efficacy and overall 89% success in clearing recurrent CDI, a similar rate as achieved with traditional FMT 58. In that study, however, the gut microbiota of donors was not profiled,
limiting the claims the company can make as to why their capsule is better than traditional FMT. Rebiotix is also advancing a next-gen FMT product known as Microbiota Restoration Therapy (MRT) that delivers a cocktail of live human-derived bacteria to the gut. Their primary candidate RBX2660 for recurrent CDI demonstrated 87.1% efficacy during phase 2 clinical trials in clearing the infection 59. Feasibility trials are underway for
MRT's application to prevent CDI, IBD, hepatic encephalopathy, and multi-drug resistant organism treatment.
Seres Therapeutics Inc. is arguably the highest-profile venture undertaking the clinical translation of FMT-inspired therapeutics. Their approach orally delivers combinations of
50 species of fecal-derived bacterial spores to the gut. Seres claims fecal-derived spores
are more effective than fecal-derived bacteria and have improved safety in patients because the ethanol isolation of spores eliminates potential pathogens such as vegetative bacteria, fungi, and viruses. In a phase lb clinical trial, their lead candidate SER109 showed promising results in treating recurrent CDI 60. However, the subsequent phase 2 study did not show significant improvement versus placebo. After review, the company believes that the unexpected results may be due to suboptimal dosing and misdiagnosis of patients in the trial. Seres discussed its findings with the FDA and has moved into a phase 3 clinical study with SER-109 for patients with recurrent CDI.
Overall, FMT and FMT-inspired therapies face the challenge of disease target. Beyond
CDI, where host-microbe interaction is of paramount importance, the role of the
microbiota independently modulating the host is not clear. Indeed, IBD-one of the most-studied and well-established diseases linked to the microbiota- yet it lags in terms of microbiota-inspired targets under development. The etiology of the disease is still being resolved and clear mechanisms often required for clinical translation have yet to be identified61-63. However, applications in complex disease are often a natural transition for
ventures already in the GI arena, as they can capitalize on parallel use for intellectual property and existing in-house expertise built around the gut microbiome and infectious disease. Despite the challenges, the early success in CDI and the exponential growth of this subfield suggests that the FMT momentum will continue to yield refinements in therapeutic products going forward.
1.4.3 Probiotics to complement FMT
Rather than repopulate the gut via FMT or distill the key components of FMT, many companies are pursuing the often less complicated path of probiotics - using only select strains of naturally occurring or genetically modified bacteria to alter the composition of microbiota. Outlined in the 2007 Food, Drug, and Cosmetic Act64, probiotics may be
marketed with limited clinical evidence of efficacy upfront should the company express intent to use generated revenue to support clinical trials required for FDA approval as a therapeutic. Furthermore, probiotic products fall along a continuum of regulatory classifications by the FDA with varying requirements for clinical research, IND status, and market access. Those marketed as therapeutics are regulated as Live Biotherapeutic Products (LBPs) under the FDA's Center for Biologics Evaluation and Research65.
Conversely, live bacteria probiotics sold and marketed as natural health products and supplements have minimal regulation. Many of the later probiotics are prone to issues of shelf-life stability or poor delivery to site of interest (often the intestine), resulting in the majority of bacteria being dead-on-arrival, raising questions of efficacy66. Indeed, in a
randomized controlled trial the probiotic combination of Lactobacillus and Bifidobacterium strains failed to show any improvement in eczema, which has been associated with changes in the microbiota compared to a placebo control6 7. However, a meta-analysis of
probiotics given for multiple gastrointestinal ailments shows that the use of probiotics is generally beneficial with a few specific exceptions66. Mechanistically, the question of probiotic efficacy appears to be linked to the colonization of the probiotic in question, suggesting that the efficacy of probiotics must be assessed in highly-controlled and individualized settings. For example, the engraftment of Bifidobacterium longum has been shown to be dependent on the individual features of the recipients microbiota6 8;
presumably, in cases of disease, the microbiota would only be amenable to certain flora. Ultimately, for probiotics to gain traction as legitimate therapeutics, the community will need to prove efficacy through mechanism-based, context-dependent study. With stricter regulatory guidelines, probiotics for clinical use-not 'over-the-counter' equivalents-should avoid the pitfalls detailed above.
1.4.4 Probiotics via a GI-axis, for autoimmune, malignant, and metabolic disease
Orally-administered probiotics which engraft into the GI-tract are being explored in a wide range of disease and include both naturally-occurring or rationally-identified species and bacteria engineered to provide not-found-in-nature therapeutic abilities. Vedanta Biosciences is one such venture developing "rationally defined" probiotics from isolated bacterial strains to treat autoimmune disease via GI-axis. Their preclinical product VE-202 is an oral formulation of live Clostridium bacteria that activates Treg cells to secrete IL-10 and reduce inflammation in IBD69. Some companies are taking probiotics one step
further with genetic engineering. Actogenix is developing genetically engineered L. lactis
"Actobiotics", which produce anti-inflammatory proteins for oral mucositis, IBD, and diabetes. ViThera is also engineering L. lactis in yogurt to treat IBD. Both companies have not reported advancing beyond early product development and translational studies. One question that arises is whether the genetically engineered L. lactis will engraft and colonize the intestine, a key hurdle facing most, if not all, probiotic products.
Investigators have identified oral and gut dysbioses in cancer patients receiving PD-1-based immunotherapy for melanoma or epithelial tumors, suggesting the microbiota influences response to immunotherapy70. Responders exhibit greater diversity and abundance of RuminococcaceaelFaecalibacterium correlating with enhanced systemic
and anti-tumor immune responses mediated by increased antigen presentation and effector T cell function in the tumor microenvironment. FMT from cancer patients who respond to immune checkpoint inhibitors into germ-free and antibiotic-treated mice ameliorates PD-1 blockade-mediated anti-tumor effects71. Specifically, A. muciniphilia
restores anti-cancer effects of PD-1 inhibition, although the bacterium's immunomodulatory mechanism of action is still unknown. Similarly, promotion of anti-tumor immune suppression has been observed in patients with pancreatic ductal adenocarcinomas (PDA), though gut bacteria which translocate to the pancreatic tumors72. Treatment with antibiotics reversed the anti-tumor immune suppression and
increased tumor-infiltrating T-cell activation. Strain-specific microbe population shifts can also alter the progression of colorectal cancer though an immune-mediated response7 3.
Additional pioneering studies have established the use of specific gut bacteria to supplement cancer immunotherapy by enhancing the anti-tumor effect74
,75.
These clinically relevant relationships between gut microbiota and immune response, have spurred microbiota-inspired therapeutcs for immuno-oncology. Indeed, Vedanta has partnered with Japanese JSR Corporation to advance microbiome-derived LBPs that induce CD8+ T cells, the predominant effectors in cancer immunotherapy. They plan to file for IND for their lead immune-oncology candidate in 2018.
Another major partnership is that between Seres, MD Anderson Cancer Center, and the Parker Institute for Cancer Immunotherapy. SER-401 containing rationally-defined live bacteria as oral microbe-based immunotherapy is still in preclinical stages for advanced metastatic melanoma. SER-401 is designed to be used in combination with anti-PD-1 checkpoint inhibitors to better understand the safety and efficacy of utilizing microbes to modulate the microbiota during traditional cancer treatment for other cancers beyond melanoma70. The partnership announced in November 2017 the launch of SER-401 in an advanced metastatic melanoma trial comparing the efficacy of the LBP-immunotherapy drug (PD-1 targeting) compared to FMT- and placebo-immunotherapy drug combinations. Other companies posit that the use of live bacteria themselves will be more therapeutically effective than current small molecule drugs for addressing specific sub-diseases of metabolic syndrome. Caelus Health's candidate probiotic CP-001, an oral formulation of live E. halli, is proposed to reduce insulin resistance and prevent type 2 diabetes (T2D) (phase 1 trial). MicroPharma Ltd has patented their probiotic 'superstrain'
Lactobacillus reuteri NCIMB 30242, which claims to support healthy cholesterol levels by
reducing LDL-C absorption7 6
,77. AOBiome is treating hypertension with its ammonia
oxidizing bacteria (AOB) B244 platform (phase 2).
Modulation of gut microbiota TMAO production has been identified as a possible pre-/probiotic route due to associations between TMAOs and cardiovascular and other metabolic diseases. TMAOs have been associated with increased development of atherosclerotic plaques and CVD, although a causal mechanism of action has yet to be determined 78-80. Furthermore, mice receiving microbiota transplants enriched in TMAOs
develop an obesity phenotype81. Multiple microbiota-based interventions, however, have demonstrated that the pro-disease effect can be mitigated 78-80. For example, A.
muciniphila protects mice from low-grade inflammation and obesity associated with
metabolic syndrome 82. To the best of our knowledge, no company has developed a
probiotic therapeutic utilizing A. muciniphila for metabolic syndrome.
1.4.5 Emerging probiotic approaches in malignancy, food allergy, and early life
In addition to the products presently under development, the potential utility of orally-administered probiotics for both cancer and food allergy is an active area of research. A group at the National University of Singapore and Singapore General Hospital has engineered E. coli Nissle 1917 to express a binding site for the heparin sulfate proteoglycan on colorectal cancer tumor cell surfaces and secrete murosinase, which converts glucosinolte, found in cruciferous plants, to its active form sulforaphane, a known
G2/M cellular arrester. Preliminary studies with murine colorectal cancer models showed
enhanced tumor clearance long-term with treatments of the engineered bacteria in parallel with a high cruciferous diet8 3. This proof-of-concept investigation highlights a strategy to safely engraft probiotic bacteria that are therapeutically-effective in a
combination chemoprotective therapy (probiotics and personalized nutrition).
Food allergen-sensitive or allergic individuals are known to have different microbiota population dynamics, i.e. a relative abundance of Bacteroides, Propionibacterium,
Klebsiella within the first months of life versus non-allergic individuals, who are enriched
for Acinetobacter, Clostridium, Proteobacteria (excluding genus Klebsiella)84. As well,
antibiotic and acid-suppression treatments within the first six months of life have been associated with significantly increased prevalence of cow's milk and egg allergies during early childhood85. Notably, Clostridia have been shown to protect against food allergen sensitization by regulating innate lymphoid cell function, Treg cell induction, and intestinal
epithelial permeability86,8 7. As well, germ-free mice have been shown to be more sensitive to food allergens, such as peanuts, than conventional or C/ostridia-colonized mice86. While a Clostridia-based probiotic has yet to be publicly tested, administration of a
Lactobacillus rhamnosus probiotic with peanut oral immunotherapy in peanut-allergic
children induced sustained (although not permanent) unresponsiveness post-treatment88, implying a probiotic-treatable axis between the microbiota and food allergies.
Given the importance of early-life microbial exposure, multiple investigators are pursing interventions to set the 'right' initial microbial seeds. This approach is supported by a studies in newborn mice, where the induction of colonic Treg cells by oral inoculation with a mix of rationally-isolated Clostridium strains reduced the risk of colitis and systemic allergic responses in adulthood 69,87. Alternatively, low-dose antibiotics given to young
mice significantly alter the gut microbiota and promote lasting metabolic consequences, including increased propensity for obesity on a high-fat diet 89. Clinical studies of potential
early-life microbiome interventions are few, but promising. Of note, Dominguez-Bello et al. recently demonstrated that swabbing newborns delivered by Caesarian section with bacteria from the mother's vaginal canal can populate the newborn's microbiota in a manner similar to conventional delivery, though any lasting health consequences from such intervention remain to be determined90. In addition, a 4,557 patient randomized
'synbiotic' (combination of the probiotic L. plantarum and facultative prebiotic fructooligosaccharide), trial in India in healthy term or late-term infants showed a protective effect of a single-strain probiotic treatment to prevent sepsis 91. Based on these investigations, we propose that FMT-inspired or probiotic treatments early in life for individuals identified to be at risk for autoimmune disorders (e.g. genetic predisposition) should be a space in which companies consider development.
1.4.6 Probiotics for women's health
With 580 reference genomes, the urogenital tract is the second-most characterized body region in the HMP (top, unsurprisingly, is the GI tract). In women's health, priority has been placed on identifying selected bacterial strains and understanding the links between perturbations of the urogenital microbiome and infection. Novel therapeutics to target bacterial vaginosis, HIV, and other STDs as well as improved pregnancy outcomes are of particular focus 92,93.
Osel Inc. was one of the first companies to be FDA approved for use of live bacteria as therapeutics. Their two LBPs, LACTIN-V 94 and CBM588 94, have progressed to phase 2 clinical trials for treating recurrent bacterial vaginosis/urinary tract infections and antibiotic-associated diarrhea, respectively. The company is also pursuing use of genetically-engineered bacteria to treat disease-associated dysbiosis such as IBD. Their MucoCept platform using modified Lactobacillus is in preclinical development for prevention of HIV by neutralizing HIV passing through mucosal surfaces 95. Despite the
notable unmet therapeutic needs in this specific facet of women's health, aside from Osel, there is a relative lack of commercial translation in microbiome-inspired ventures targeting women's health, making it ripe for future innovation.
1.4.7 Probiotics for the skin
Several ventures have stepped up to begin positioning topical probiotics to treat the increasing prevalence of microbe-associated skin conditions (e.g. acne, atopic dermatitis
(AD)) and antibiotic-resistant skin infection through restoration and protection of a more
natural flora. MatriSys Biosciences is pursuing commercialization of live bacteria therapies for AD. Their S. hominis A9-based product (MSB-01), which began phase 2a clinical trial in 2016, seeks to increase the presence of the commensal S. hominis on the skin to reduce susceptibility to pathogens, specifically AD-linked S. aureus96. AOBiome
is also pursuing topical microbiota-based therapeutics for autoimmune skin disease with their B244 platform (phase 1). Early successes in probiotic trials for skin disease will likely shape the way for further intervention in microbiome-associated skin disease.
1.4.8 A path forward for bugs as drugs
Continuing development of FMT, FMT-inspired, and probiotic approaches will require the clear demonstrations of efficacy to both prove the scientific validity and meet regulatory standards. There is still only a limited understanding for the mechanisms of these potential therapeutics-a scientific limitation that has led to clear constraints on
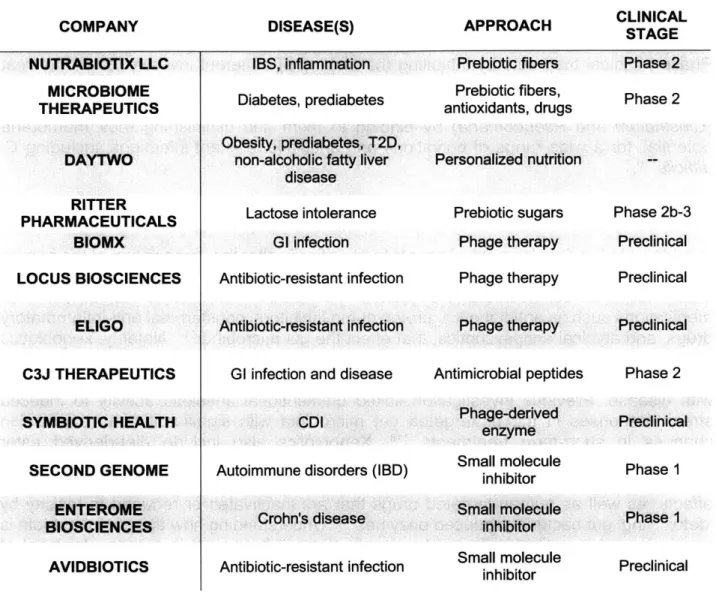
therapeutic translation. Companies are largely pursuing 'me too' approaches, as the risks for developing FMT-inspired approaches beyond what is already known to work is appreciable, and, due to the inherent complexities of these systems, clear mechanisms of action are elusive. As development is being driven from clinical evidence, it is difficult to extrapolate FMT platforms beyond CDI and generate significant investment for other promising microbiota-associated diseases. In total, it appears that for bugs as drugs approaches-and, more broadly, microbiota-inspired therapeutics-to meet clinical needs, industry and academia will need to clearly define therapeutic mechanisms and prove causation, moving away from the large correlative studies that have largely driven interest in the field. Proving causation ties in with demonstrating efficacy, as both require the correct patient population and diagnoses to advance-a challenge facing companies moving into phases 2 and 3 clinical trials (Table 1.1).
Table 1.1: Companies advancing 'bugs as drugs' therapeutics
DISEASE(S) APPROACH CLINICAL STAGE
OPENBIOME SYMBIOTIC HEALTH REBIOTIX SERES THERAPEUTICS INC. VEDANTA BIOSCIENCES ACTOGENIX VITHERA OSEL INC. CAELUS HEALTH MICROPHARMA LTD AOBIOME MATRISYS BIOSCIENCES
CDI, IBD, IBS, cirrhosis
Recurrent CDI
Recurrent CDI, IBD, hepatic encephalopathy, multi-drug resistant organism treatment
CDI; advanced metastatic
melanoma
Autoimmune disease; cancer Oral mucositis, IBD, diabetes
IBD
Recurrent bacterial vaginosis/UTI, antibiotic-associated diarrhea; IBD;
HIV prevention T2D
LDL cholesterol absorption Hypertension; autoimmune skin
disease
Atopic dermatitis-associated antibiotic-resistant skin infection
FMT FMT FMT-inspired FMT-inspired Probiotic Engineered probiotic Engineered probiotic Probiotic; Engineered probiotic Probiotic Probiotic Probiotic Probiotic Early Phase 1-2 Phase 1 Early Phase 1-2 Phase 3; preclinical Preclinical Phase 1-2a Preclinical Phase 2; preclinical Phase I Phase 1-2 Phase 2; phase 1 Phase 2a
1.5
Drugs for bugs: a plethora of prebiotics
approaches
and next-gen 'antibiotic'
Prebiotics and targeted agents of microbial elimination are an important focus of microbiota-inspired therapeutic development. Delivering an agent to a microbial community which elicits a population change is not a novel concept-think, antibiotics.
However, the many new approaches and the emphasis on specificity has expanded therapeutic opportunities around the concept. This is especially true of agents that promote growth and survival of specific symbionts or commensal species of the microbiota, known as prebiotics. 'Drugs for bugs' approaches span the stages of clincal development, with frontrunners in this space working on prebiotics and personalized nutrition, and engineered bacteriophages among other microbe-specific antibiotics emerging as intriguing preclinical approaches. Further, we are beginning to appreciate the unintentional consequences of many common dietary and medicinal agents on our microbial ecosystems, known as xenobiotics.
Prebiotics, substances that promote growth of beneficial bacteria, such as Lactobicilli and
Bifidobacteria, primarily consist of non-digestible fiber, which is fermented by intestinal
bacteria as a source of nutrition97,98. Like probiotics, prebiotics have been sold as natural health supplements for years without FDA regulation. They have the advantages of ease of delivery, a longer shelf-life than LBPs, and often consist of inexpensive compounds that are typically on the FDA's list of generally regarded as safe (GRAS) materials. Despite the prevalence of un-regulated prebiotics, there are multiple ventures seeking clinical approval for their products. A notable example of this is Nutrabiotix LLC's prebiotic fiber, which is currently in phase 2 trials for IBS and inflammation. Other early ventures are using small molecules to specifically target gut microbes, including CDI and species implicated in IBD as a supplement to current drug therapies. One of the limiting factors to progress in this area is the significant inter-individual differences in the response to these interventions, likely due to individual microbiome and genetic variation- such challenges may be better addressed by personalized approaches 99.
In contrast to prebiotics, phage therapy, CRISPR systems, and small molecule or peptide-based antibiotic approaches seek to specifically eliminate certain bacteria with key roles in disease pathophysiology. These approaches circumvent some of the major issues facing current antibiotics-too broad antimicrobial activity, systemic host effects, susceptibility to antibiotic resistance-with more strain-specific targeting and mechanisms of action. Several companies are advancing these 'next-gen' antimicrobial therapeutics through preclinical to Phase 2 clinical trials for eliminating pathogenic bacteria that cause infection themselves or are a key component of disease-associated dysbiotic conditions.
1.5.1 Prebiotics in metabolic syndrome
Recent evidence suggests that the gut microbiota proportionally changes in response to perturbations in diet, a known driver of the development of metabolic syndrome (obesity, elevated blood pressure, and high blood lipid and glucose levels associated with increased risk of cardiovascular disease (CVD) and T2D)100. Most notably, consumption of artificial sweeteners and common dietary emulsifiers (e.g. carboxymethylcellulose and polysorbate-80) leads to functional alterations in the gut microbiome that induce glucose intolerance,101 low-grade inflammation, and metabolic syndrome102. Recently, high salt
commensal Lactobacillus murinus, which modulates TH17 cell-induction of hypertension103.
Preclinical studies investigating the potential of prebiotics or personalized nutrition to alter host microbiota and reverse phenotype in T2D have shown promise 04, and companies
are advancing pre- and probiotics for diabetes and metabolic syndrome. MicroBiome Therapeutics prebiotic therapeutic candidate, NM504, consists of antioxidants and prebiotic GRAS fibers for diabetes and prediabetes, and is designed to shift the gut microflora to improve blood glucose regulation. Notably, NM504 is the first "microbiome modulator" to receive U.S. patent allowance (October 2016). Another candidate, NM505 consists of a reformulation of the common diabetes drug metformin along with the prebiotics in NM504. A phase 2 clinical trial with the latter formulation has indicated that the addition of prebiotics reduces the common GI side effects associated with metformin1 05. While neither formulation is designed to address the glucose resistance to insulin in T2D nor the incidence of the disease, the use of prebiotics as an adjunctive therapeutic to traditional insulin poses an interesting approach for those individuals for whom may be predisposed to prediabetes/diabetes genetically or due to family history. Alternatively, recent research suggests that lincreased consumption of dietary fibers increased the abundance of 15 bacterial strains that actively produce short-chain fatty acids (SCFAs) and are correlated with improved clinical outcomes in T2D patients106.
Expanding on the idea of dietary modulation of the microbiota, personalized nutrition is an emerging approach for prebiotic modulation on the gut microbiome. In a cohort following nearly 1,000 individuals' glycemic responses to food, significant variability was noted with respect to individual glycemic responses on identical diets, raising concern for outcomes of 'one size fits all' dietary interventions 07. To understanding individual
responses to diet, advanced machine learning algorithms have been employed to achieve individual predictions of the glycemic responses to foods, resulting in improved individual blood glucose control. To achieve individualized predictability of glycemic responses, predictive algorithms use a combination of clinical and microbiome features, highlighting the contribution of personalized microbiome features. Similar microbiome-based predictive capabilities were shown with respect to discrete food components such as non-caloric artificial sweeteners 101 and bread 108. In all, microbiome features are a critical component to enabling individual diets to shape clinical responses.
DayTwo, an Israel-based company, is developing personalized nutrition approaches aimed at tailoring diet to the individual as means of improving blood sugar control in obesity, pre-diabetes, T2D, and non-alcoholic fatty liver disease. Long-term interventional studies involving this approach are currently under way, aimed at directly comparing the 'gold standard' nutritional approaches to person-tailored nutrition in individuals predisposed to develop features of metabolic syndrome.
1.5.2 Prebiotics for food allergy
For years, food allergies have been addressed through 'personalized nutrition' approach-avoiding the food that one is allergic to-and through conventional drugs, e.g.