doi:10.1093/jxb/eru368 Advance Access publication 13 September, 2014
v
ReseaRch PaPeR
Characterization of two distinct subfamilies of SUN-domain
proteins in Arabidopsis and their interactions with the novel
KASH-domain protein AtTIK
Katja Graumann1,*, Emmanuel Vanrobays2,*, Sylvie Tutois2, Aline V. Probst2, David E. Evans1 and
Christophe Tatout2,†
1 Department of Biological and Medical Sciences, Oxford Brookes University, Oxford OX3 0BP, UK
2 UMR CNRS 6293 INSERM U 1103 Clermont Université, GReD, 24 Avenue des Landais, BP80026 63171 Aubière Cedex, France * These authors contributed equally to this work.
† To whom correspondence should be addressed. E-mail: christophe.tatout@univ-bpclermont.fr
Received 24 May 2014; Revised 23 July 2014; Accepted 8 August 2014
Abstract
SUN-domain proteins belong to a gene family including classical Cter-SUN and mid-SUN subfamilies differentiated by the position of the SUN domain within the protein. Although present in animal and plant species, mid-SUN proteins have so far remained poorly described. Here, we used a combination of genetics, yeast two-hybrid and in planta transient expression methods to better characterize the SUN family in Arabidopsis thaliana. First, we validated the mid-SUN protein subfamily as a monophyletic group conserved from yeast to plant. Arabidopsis Cter-SUN (AtSUN1 and AtSUN2) and mid-SUN (AtSUN3 and AtSUN4) proteins expressed as fluorescent protein fusions are membrane-associated and localize to the nuclear envelope (NE) and endoplasmic reticulum. However, only the Cter-SUN subfam-ily is enriched at the NE. We investigated interactions in and between members of the two subfamilies and identified the coiled-coil domain as necessary for mediating interactions. The functional significance of the mid-SUN subfamily was further confirmed in mutant plants as essential for early seed development and involved in nuclear morphology. Finally, we demonstrated that both subfamilies interact with the KASH domain of AtWIP1 and identified a new root-specific KASH-domain protein, AtTIK. AtTIK localizes to the NE and affects nuclear morphology. Our study indicates that Arabidopsis Cter-SUN and mid-SUN proteins are involved in a complex protein network at the nuclear mem-branes, reminiscent of the LInker of Nucleoskeleton and Cytoskeleton (LINC) complex found in other kingdoms. Key words: KASH domain, LINC complex, mid SUN, nuclear envelope, protein interactions, SUN domain.
Introduction
The nuclear envelope (NE) is made up of an inner and outer membrane connected at the nuclear pore complex by the pore domain. In most organisms, protein complexes, comprising proteins of the SUN (Sad1/Unc84)-domain family in the inner nuclear membrane (INM) and proteins of the KASH (Klarsicht, ANC-1, and Syne Homology)-domain family in the outer nuclear membrane (ONM), span the nuclear peri-plasm. This LInker of Nucleoskeleton and Cytoskeleton (LINC) complex (Crisp et al., 2006) is involved in a vari-ety of nuclear and cellular processes including homologous
recombination and movement of nuclei, and appears to be con-served in eukaryotes. Recent studies in plants (Murphy et al., 2010; Graumann et al., 2010; Oda and Fukuda, 2011) have revealed the presence of proteins with a SUN domain at the C-terminus showing significant homology to those in animals and yeast. In addition, the WPP-domain-Interacting Proteins (WIPs) and the SUN-Interacting NE (SINE) proteins were recently described as the first plant KASH-domain proteins (Zhou et al., 2012b, 2014). However, while the C-terminal SUN proteins appear well conserved in eukaryotes, KASH
© The Author 2014. Published by Oxford University Press on behalf of the Society for Experimental Biology. All rights reserved. For permissions, please email: journals.permissions@oup.com
proteins are not. The KASH domain is mainly distinguished by structural features such as a C-terminal transmembrane (TM) domain, which is followed by a short periplasmic tail that mediates interactions with SUN proteins. In animals, the final four amino acids of the KASH domain are com-monly PPPX while the plant-specific WIP and SINE pro-teins contain a conserved xVPT motif (Zhou et al., 2012b, 2014). The N-terminal, cytoplasmic part of the KASH pro-teins is very diverse, as are the functions of KASH propro-teins. In animals and yeast, KASH-domain proteins mainly link to cytoskeletal elements and are involved in nuclear shape, positioning, and movement, amongst other functions (Starr and Fridolfsson, 2010). In plants, the WIP–SUN bridges are also required for maintaining the shape of nuclei but in addition mediate anchorage of RanGAP proteins to the NE (Zhou et al., 2012b). Recent evidence suggests that WIPs are connected to the cytoskeleton, by interacting with WIT pro-teins, which link myosin XI-i to the NE (Tamura et al., 2013). In addition, SINE–SUN interactions link more directly to the actin cytoskeleton by SINE1 binding to actin through its armadillo-repeat domain (Zhou et al., 2014). Both actin linkages are involved in positioning of nuclei (Tamura et al., 2013; Zhou et al., 2014). AtSINE1 is also the first tissue-spe-cific plant KASH protein to be identified, and is expressed predominantly in guard cells (Zhou et al., 2014).
In the ‘classical’ SUN-domain proteins, which are located at the INM, the SUN domain is C-terminal, in the nuclear periplasm, and interacts with the KASH domain of KASH proteins located in the ONM. Two of these ‘classical’ C-terminal SUN-domain proteins (referred below as Cter-SUN) have been described in Arabidopsis thaliana (AtSUN1 and AtSUN2: Graumann et al., 2010; Oda and Fukuda, 2011) and maize (Zea mays) (ZmSUN1 and ZMSUN2: Murphy
et al., 2010). However, searching the genomes of higher plants for SUN-domain proteins, in common with other organisms, reveals the presence of an additional subfamily of proteins containing a central SUN domain. Analysis of these pro-teins in maize (Murphy et al., 2010) has suggested the pres-ence of a subfamily of three, two of which (ZmSUN3 and ZmSUN4) are ubiquitously expressed while one (ZmSUN5) is pollen specific. Antibody staining has indicated a location at the nuclear periphery. Murphy et al. (2010) have used the term Plant-prevalent Mid-SUN 3 TM (PM3) for these pro-teins, referred to hereafter as mid-SUN proteins. The renam-ing from PM3-SUN to mid-SUN is based on the findrenam-ings that mid-SUN proteins are not unique to higher plants; the best studied in mammalian tissues (mouse) is termed osteopoten-tia (Opt) and is resident in the rough endoplasmic reticulum (rER) (Sohaskey et al., 2010). Mutagenesis of Opt results in a range of defects in type I collagen formation resulting from failure of the rER to expand. Sohaskey et al. (2010) suggest that Opt may have a role as a biomechanical adap-tor that positions and stabilizes the rER through interaction with the cytoskeleton. Recently, analysis of mid-SUN protein Sun-Like Protein 1 (SLP1) in yeast (Friederichs et al., 2012) revealed that it also resides in the ER, and is involved in the localization of the Cter-SUN-domain protein MPS3 to the NE. The study showed that SLP1 is localized with EMP65
(ER Membrane Protein of 65 kDa) in cortical and perinu-clear ER but, while it does not appear to interact directly with MPS3, it does affect its localization at the NE. Friederichs
et al. (2012) therefore hypothesize a role for the mid-SUN protein in the maintenance of the MPS3 pool at the NE. Thus both types of SUN-domain protein may be active within the cell, but the functional relationship between Cter-SUN and mid-SUN proteins remains an open question.
The present study provides characterization of the
Arabidopsis mid-SUN proteins in terms of their subcellular
localization, tissue expression, and protein interactions. We found that AtSUN3, AtSUN4, and AtSUN5 interact with each other and with the classical Cter-SUN-domain pro-teins forming protein complexes, which we hypothesize to be involved in anchorage to the INM. Consequently, plants deficient in members of the mid-SUN proteins show altered nuclear morphology. We also provide evidence that the mid-SUN proteins interact with plant KASH proteins, and iden-tify AtTIK, a new plant KASH protein, that controls nuclear morphology in root cells. Taken together, these findings pro-vide novel insights into NE-bridging complexes at the plant NE.
Materials and methods
Membrane yeast two-hybrid systemDetails of the Split-Ubiquitin-based Membrane Yeast Two-Hybrid (MYTH) system have been published previously (Snider
et al., 2010a,b). NMY51 strain MATa, his3Δ200, trp1-901,
leu2-3,112, ade2, LYS2::(lexAop)4-HIS3, ura3::(lexAop)8-lacZ,
ade2::(lexAop)8-ADE2, GAL4, as well as prey plasmid pPR3N (2μ,
TRP1, AmpR), and bait plasmid pBT3N (CEN, LEU2, KanR), were
purchased from DUALSYSTEM Biotech (http://www.dualsystems. com). Yeast media used were a standard Yeast Nitrogen Base (YNB) supplemented with amino acids and bases as required. Interaction efficiencies were recorded using drop tests on Permissive Medium (PM: YNB without Leu and Trp) and Test Medium (TM: YNB without Leu, Trp, Ade, and His) with serial dilutions (100 000, 10 000, 1000, 100, and 10 cells per drop) of a given strain grown in permissive medium and incubated at 30°C.
According to the expected membrane topology of the SUN pro-teins we fused the N-terminal part of the AtSUN propro-teins to the reporters. Constructs were generated by ‘gap-repair’ homologous recombination in vivo in yeast (Oldenburg et al., 1997). cDNAs were amplified using chimeric primer pairs containing 5′ ends with 35 bp of homology to the linearized yeast target plasmid (pBT3N for baits or pPR3N for preys), and 3′ ends matching either the first 18 bases (forward primer) or the reverse complement of the last 18 bases (reverse primer) of the different cDNAs. After digestion by SfI1, plasmids pBT3N or pPR3N and PCR-amplified cDNAs were co-transformed into yeast in the ratio 1:3 (Vector:Insert). Clones were selected on plates containing test medium. All constructs were checked by PCR on colonies and sequenced after recovery of the plasmids. For the list of all primers see Supplementary Table S1. Baits were all subsequently tested for their level of self-activation and validated according to Snider et al. 2010a. All baits satisfied these preliminary requirements. Screens for new interacting proteins were performed using a cDNA library (DUALSYSTEM Biotech) cloned into the prey vector pDSL-Nx (2μ, TRP1, AmpR). This
library includes 6-day-old seedlings, a mixture of dark grown (etio-lated) seedlings and seedlings exposed to blue and far -red light. The vector pOst1–NubI (2μ, TRP1, AmpR) expressing a fusion of the
yeast-resident ER protein Ost1 to the wild-type NubI portion of
yeast ubiquitin (able to self associate to the Cub half of the ubiqui-tin protein) was used as positive control prey.
The correct expression of the preys was tested by western blot-ting using an antibody directed against the HA-tag of the fusion protein. Proteins from total extracts expressing the preys were sepa-rated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were used for Coomassie staining (loading control) or pro-tein transfer onto nitrocellulose membrane (Amersham Hybond-ECL). The preys were detected with a rabbit anti-HA tag (Abcam) diluted 1:4000 followed by enhanced chemiluminescence detection (Pierce-ECL).
Domain predictions
The domain structure for Cter-SUN, mid-SUN, and KASH con-structs were analysed using http://www.ch.embnet.org/software/ COILS_form.html for coiled-coil (CC) domains (Lupas et al., 1991) and http://www.cbs.dtu.dk/services/TMHMM for TM helices (Krogh et al., 2001). The SUN domain was defined first using UniProt data-base annotation and then refined manually from protein alignment including the Cter-SUN protein human SUN2 as a reference. Crystal structure and site directed mutagenesis of the SUN domain from human SUN2 has recently been obtained (Zhou et al., 2012a), and provides a framework for defining the SUN-domain boundaries. The KASH domain was defined first by locating the TM domain and then by protein alignment with other KASH-domain proteins.
Phylogenetic reconstruction
Mid-SUN coding sequences were used for phylogenic reconstruc-tion. Protein sequences were restricted to the SUN domain and were selected from mid-SUN proteins that had already been described. This included A. thaliana (F4I316: ARATH_SUN3, F4I8I0: ARATH_SUN4, and F4JPE8: ARATH_SUN5), Z. mays (AC195254: MAIZE_SUN3, D3KCC3: MAIZE_SUN4, and B6SKI2: MAIZE_SUN5), Schizosaccharomyces pombe (O59729: SCHPO_SLPI), Rattus norvegicus (Q710E6: RAT_SUCO),
Saccharomyces cerevisiae (Q12232: YEAST_SLP1), Homo sapiens
(Q9UBS9: HUMAN_SUCO), Mus musculus (Q8C341: MOUSE_ SUCO) and Dictyostelium discoideum (Q54MI3: DICDI_SUNB). Cter-SUN from A. thaliana (Q9FF75: ARATH_SUN1 and Q9SG79: ARATH_SUN2) and H. sapiens (Q9UH99: HUMAN_SUN2) were used as outgroups. SUN domains were aligned with MUSCLE mul-tiple alignment of protein sequences (Castresana, 2000). Maximum Likelihood (ML) analyses were performed with PhyML with default parameters and bootstrap analysis (Guindon et al., 2010).
T-DNA mutants
T-DNA mutants sun3-1 (Flag_374A03), sun4-1 (Salk_022028),
sun5-1 (Salk_126070), and tik-1 (Salk_205745) were obtained from Arabidopsis stock centres and established as homozygous lines. All
mutants are in the Col genetic background except sun3-1 which is in the Ws genetic background. Because Col and Ws display distinct features in their nuclear morphology, the two genetic backgrounds were considered separately. Double and triple mutants were estab-lished by crossings and identified by PCR genotyping using primers described in Supplementary Table S1. Strong reduction of respec-tive transcripts was demonstrated for each mutant by RT-PCR using standard conditions and actin as an internal control. Percentages of wild-type and aborted seeds were scored in siliques from self-ferti-lization of plants from two distinct genotypes: SUN3 sun4-1 sun5-1 and SUN3/sun3-1 sun4-1 sun5-1. Between 15 and 30 siliques were analysed for each genotype in two independent experiments.
Cloning and fluorescent protein fusions
The coding sequences of AtSUN3, AtSUN4, full-length AtTIK, the TM domain, KASH domain of AtTIK (AtTIK-KASH), and
AtTIK-∆KASH truncation missing the KASH domain were
ampli-fied using the gene-specific primers listed in Supplementary Table S1. Gateway attB-flanking sequences were added to each of the constructs and gateway technology was used for cloning into pDONR207 and afterwards into expression vectors pCambia 1300, pK7CWG2, and pK7WGC2. Vectors containing GFP-calnexin, AtSUN1-YFP, and AtSUN2-YFP were used as described previ-ously (Graumann et al., 2007, 2010). Expression vectors were trans-ferred to Agrobacterium tumefaciens GV3101 strains by heat-shock transformation, and were used for transient expression.
Transient expression and confocal microscopy
Nicotiana tabaccum and N. benthamiana leaves were infiltrated with
agrobacteria carrying expression vectors for transient expression, as described by Sparkes et al. (2006). AtSUN3 and AtSUN4 con-structs were co-infiltrated with the silencing inhibitor p19 to increase expression. The subcellular localization of fluorescent fusion pro-teins in living plant tissue was detected with a Zeiss LSM 510 confo-cal microscope. Images were processed using the Zeiss LSM image browser and Adobe Photoshop.
For ER/NE Fluorescence Intensity (FI) analysis, infiltrated leaf sections were first treated with 25 μM Latrunculin B for 30 min to cause ER sheet formation (Graumann et al., 2007). The pinhole was kept low to image a thin optical section of either the ER or the NE sheet. Up to five equally sized, randomly distributed regions of interest (ROIs; 3 µm2) were measured for each sheet. For each ran-domly selected cell, the average ER FI was divided by the average NE FI to calculate the ER/NE FI ratio. For each sample, the average ratio was calculated. The number of cells analysed for each sample approximated 25.
FRAP was used to investigate the mobility of AtSUN3 and AtSUN4 fluorescent protein fusions in the ER and NE. FRAP was carried out as described by Graumann et al., 2007. Briefly, tran-siently expressing N. benthamiana lower epidermal leaf cells were first treated with Latrunculin B and then imaged with the 514 nm laser to excite the YFP. Scanning transmission was kept low and bleaching was performed at 100% transmission. The fluorescence was monitored in a constant-sized ROI pre- and post-bleach. The raw data was converted to percentage and mobile fractions and half times were calculated as described by Graumann et al., 2007, 2010. Student’s t-test was used for statistical analysis; 30 nuclei per sample were photobleached.
In planta protein interaction studies
Acceptor photobleaching fluorescence resonance energy trans-fer (apFRET) was used to detect in planta protein interactions. ApFRET was performed as described in Graumann et al. (2010) and Graumann (2014). Briefly, transiently expressing
N. benthami-ana leaf tissue was used as described in the previous section. YFP
was excited with 514 nm light and CFP with 458 nm light. The YFP laser transmission was kept low during scanning to avoid pho-tobleaching but was set at 100% during bleaching. Five pre-bleach and five post-bleach scans were carried out in a constant sized ROI. Fluorescence intensity was monitored in the ROI and analysed using Microsoft Excel. For each sample, approximately 35 nuclei were used. Student’s t-test was carried out for statistical analysis.
Analysis of root nuclei
Arabidopsis seedlings were grown on half-strength MS agar. For root
growth analysis, plates were scanned on days 3, 7, 10, and 14 after germination. ImageJ (Schneider et al., 2012) was used for measuring and Excel for data analysis. For analysis of nuclei, 14-day-old seed-lings were removed from the plate and immersed in PBS and eth-idium bromide (50:50) solution for ~5 min. Seedlings were imaged with a Zeiss LSM510 confocal microscope using the ×63 oil immer-sion lens and zoom factor 2. Zeiss LSM image browser was used for
width and length measurements and circularity index was calculated by the following equation:
Circularity index = width/length.
ImageJ was used for nuclear area measurements. All measurement data were analysed using Excel and Student’s t-test was used for sta-tistical analysis. For root-length measurements, 40 plants per sample were used. For analysis of nuclei, at least 15 nuclei from three plants (total of at least 45 nuclei) were used per sample.
Results
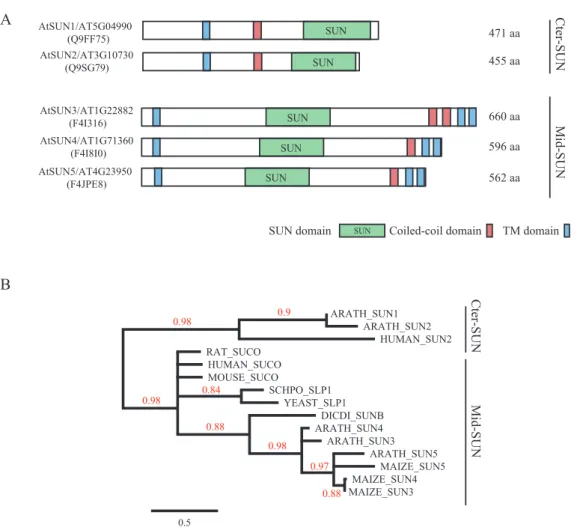
Arabidopsis has five SUN-domain-containing proteins According to the Pfam database at http://pfam.sanger.ac.uk (Punta et al., 2011), the SUN-domain protein family com-prises over 30 different architectures that can be grouped into proteins with an internal SUN domain (mid-SUNs) and proteins containing a SUN domain at their C-terminus (clas-sical or Cter-SUNs). Plant SUN-domain proteins were ini-tially described in Arabidopsis by Graumann et al. (2010) and by Murphy et al. (2010) in maize. The latter also described the existence of both subfamilies of SUN-domain pro-teins in plants. Further analysis of the Arabidopsis genome revealed five AtSUN proteins: the two Cter-SUNs (AtSUN1 and AtSUN2) initially described by Graumann et al. (2010) and three new mid-SUN proteins (AtSUN3, AtSUN4 and AtSUN5) (Fig. 1A). The mid-SUN proteins have three TM domains – one at the N-terminus and two at the C-terminus, with the latter preceded by a CC domain (Fig. 1A).
We analysed the SUN domain of selected plant and animal mid-SUN proteins to generate a phylogenetic tree using the SUN domain of Cter-SUN as an outgroup. The topology of the tree strongly suggests that the SUN domains of the two subfamilies of SUN proteins diverged early during evolution before the emer-gence of the plant and animal kingdoms and are now forming two distinct monophyletic groups (Fig. 1B). Most species display Cter-SUN and mid-SUN proteins, suggesting that both groups have been conserved through evolution (Field et al., 2012).
We then performed an extensive search in the databases for the presence of proteins with a central SUN domain in order to list mid-SUN proteins in dicot species. The SUN domain of mid-SUN proteins is well conserved (Supplementary Figure S1). Within the 13 selected species analysed all mid-SUN proteins contain a single mid-SUN domain except Vitis
vinifera, in which a central and C-terminal SUN domain
are found within the same protein (data not shown). Our analysis revealed that it is difficult to distinguish SUN3 from SUN4 except in Brassicaceae, while the SUN5 proteins form a separate group from SUN3–4 (Supplementary Figure S2). Interestingly, while AtSUN3 and AtSUN4 are expressed at low to medium levels in most tissues (Supplementary Figure S3A), AtSUN5 is more expressed in anther, pollen, and vari-ous endosperm tissues (Supplementary Figure S3B). This resembles the observations made for the maize desynaptic (dy) gene which is proposed to be ZmSUN3 and which is mainly expressed at meiosis (Murphy and Bass, 2012).
The NE localization of AtSUN3 and AtSUN4 was ana-lysed by transient expression of fluorescent protein fusions
in planta. Unfortunately AtSUN5 did not express and
could not be included in the following experiments on mid-SUN protein subcellular localization. YFP-Atmid-SUN3 and AtSUN4-YFP localize to the nuclear periphery (Fig. 2A and 2B). In addition, both fusion proteins also localized to the ER. To determine whether AtSUN3 and AtSUN4 are pre-dominately NE- or ER-localized, we measured ER and NE fluorescence of single, randomly selected cells and compared ER/NE-fluorescent intensity ratios with those of ER marker GFP-calnexin and NE markers AtSUN1-YFP/AtSUN2-YFP (Fig. 2C and Supplementary Figure S4). A ratio below 1 is indicative of NE accumulation, as seen for AtSUN1-YFP (0.45 ± 0.01) and AtSUN2-AtSUN1-YFP (0.56 ± 0.03), while a ratio above 1 is due to ER accumulation, as observed for GFP-calnexin (1.98 ± 0.19). The ER/NE fluorescence inten-sity ratios of 0.91 ± 0.08 for YFP-AtSUN3 and 1.19 ± 0.07 for AtSUN4-YFP are both close to 1, indicating that the two mid-SUN proteins appear evenly distributed in the ER and NE (Fig. 2C). Thus, while Cter-SUN proteins are mainly localized at the NE, mid-SUN proteins also accumulate at the ER, reminiscent of yeast and mammalian mid-SUN localiza-tion (Sohaskey et al., 2010; Friederichs et al., 2012).
To investigate whether AtSUN3 and AtSUN4 are function-ally associated with the ER and NE membranes, we carried out FRAP experiments. Both the mobile portion of a protein population and the rate of protein movement (as indicated by the half time) can be assessed by FRAP and are indicators of protein interactions. Previously, Graumann et al., (2007) showed that non-functional proteins like Lamin B Receptor (LBR) are highly mobile (above 90% mobile fraction) in the ER and NE, while functional NE proteins involved in pro-tein–protein interactions within membranes are immobilized (Graumann et al., 2010). Both YFP-AtSUN3 and AtSUN4-YFP appear highly immobilized in both ER and NE mem-branes (Fig. 2D), indicating that they are involved in protein interactions and therefore functional in these locations. Interestingly, SUN3 was more mobile than SUN4 (Fig. 2D;
P < 0.01). While SUN4 mobility seemed similar in both ER
and NE membranes (P > 0.05), SUN3 was more mobile in the NE than in the ER (P < 0.05). This suggests that SUN3 may be involved in different protein complexes depending on its membrane localization and therefore may have different functions.
The SUN-domain protein family forms an interaction network at the NE
It has previously been shown that classical Cter-SUN-domain proteins can form homo- and hetero-oligomers (Zhou
et al., 2012a). These interactions have been well studied for human SUN2 by crystal structures (Zhou et al., 2012a). In
Arabidopsis, the Cter-SUN proteins AtSUN1 and AtSUN2
interact together with each other (Graumann et al., 2010). We investigated whether there is interaction within the five proteins of the AtSUN family or within the mid-SUN pro-teins using a split-ubiquitin based yeast two-hybrid (Y2H) system dedicated to membrane proteins, MYTH) (Snider
et al., 2010a,b). The MYTH system was previously used to
show a direct interaction between the SUN protein UNC-84 of Caenorhabditis elegans and the KASH proteins UNC-83 and KDP-1 (McGee et al., 2006, 2009).
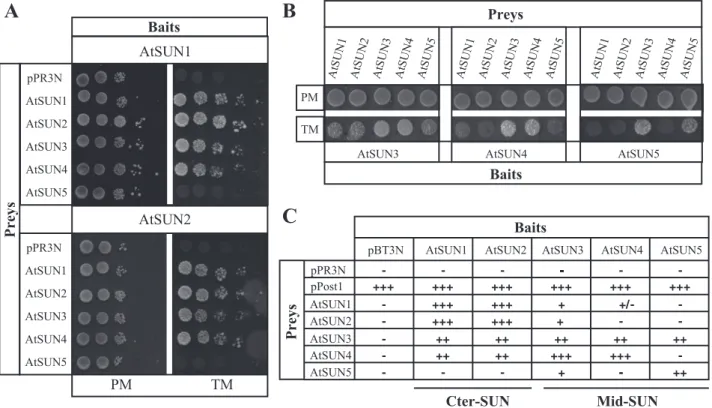
As a first step, AtSUN1 or AtSUN2 were used as baits. Although genetic interaction has been shown between Cter-SUN MPS3 and mid-Cter-SUN SLP1 proteins in yeast (Friederichs
et al., 2012), no physical interactions have so far been dem-onstrated between the two subfamilies of SUN proteins in higher organisms. The results obtained were very similar with both baits. As previously shown by acceptor photobleaching fluorescence resonance energy transfer (apFRET; Graumann
et al., 2010), homo and hetero Cter-SUN oligomers occur (Fig. 3A). Interestingly hetero-oligomers between the two Cter-SUN proteins and two of the mid-SUN proteins, AtSUN3 and AtSUN4, were also observed; however, they form with a slightly lower efficiency (Fig. 3A, 3C). AtSUN5 was not found to form hetero-oligomers with the two Cter-SUN proteins (Fig. 3A, 3C).
We then used mid-SUN AtSUN3 and AtSUN4 as baits. Again, interactions between AtSUN3 and AtSUN4 were stronger than between AtSUN3 and AtSUN1 or AtSUN2. Interactions with AtSUN5 could only be detected for AtSUN3 (Fig. 3B and 3C; Supplementary Figure S5).
Finally, AtSUN5, when used as bait, interacted strongly with itself and mid-SUN3 (Fig. 3B and 3C; Supplementary Figure S5). Altogether these results demonstrate the capacity of the mid-SUN proteins to form homo- and hetero-oligomers with other members of the SUN-domain protein family.
The protein interactions detected by MYTH were con-firmed to occur in planta using apFRET. For this, protein fusions of the mid-SUN and Cter-SUN proteins to YFP and CFP were co-expressed. In apFRET an increase in CFP fluo-rescence upon YFP bleaching indicates interaction between the YFP and CFP-fused proteins (Karpova et al., 2003). An increase of CFP fluorescence and therefore interactions were detected for the following combinations: CFP-AtSUN1 + YFP-AtSUN3, YFP-AtSUN1 + AtSUN4, CFP-AtSUN2 + YFP-AtSUN3, YFP-CFP-AtSUN2 + CFP-AtSUN4, and YFP-AtSUN3 + CFP-AtSUN4 (Table 1). Therefore, MYTH, confocal localization, and apFRET experiments confirm that Cter-SUN and mid-SUN proteins share simi-lar properties. They localize at the NE and are immobilized there. In addition, they are able to interact with each other. The interactions closely resemble the phylogenetic subdi-vision into Cter-SUN (AtSUN1 and AtSUN2) and mid-SUN (Atmid-SUN3 and Atmid-SUN4) subfamilies. Interactions are AtSUN1/AT5G04990 (Q9FF75) AtSUN2/AT3G10730 (Q9SG79) AtSUN3/AT1G22882 (F4I316) TM domain Coiled-coil domain SUN domain SUN SUN 471 aa 455 aa SUN 660 aa SUN 596 aa SUN 562 aa AtSUN4/AT1G71360 (F4I8I0) AtSUN5/AT4G23950 (F4JPE8) SUN A B Cter-SUN Mid-SUN ARATH_SUN1 ARATH_SUN2 HUMAN_SUN2 RAT_SUCO HUMAN_SUCO MOUSE_SUCO SCHPO_SLP1 YEAST_SLP1 DICDI_SUNB ARATH_SUN4 ARATH_SUN3 ARATH_SUN5 MAIZE_SUN5 MAIZE_SUN4 MAIZE_SUN3 0.98 0.9 0.98 0.84 0.88 0.98 0.97 0.88 0.5 Cter-SUN Mid-SUN
Fig. 1. Cter-SUN and mid-SUN protein lineages are distinct subfamilies. (A) Comparative protein organization of the Cter-SUN and mid-SUN protein subfamilies (lineages indicated at the right). UniProt protein IDs are indicated on the left between brackets. Protein lengths are indicated at the right of each protein. (B) Phylogenetic tree of selected plant and animal SUN proteins. SUN domains of Cter-SUN and mid-SUN proteins (lineages indicated at the right) were used to build up an ML phylogenetic tree. Cter-SUN proteins were used as outgroups. Bootstrap values are indicated on branches. This figure is available in colour at JXB online.
stronger within than between both subfamilies. Interestingly, AtSUN5 may be functionally distinct from the other mid-SUN proteins as suggested by the phylogenetic study and protein interactions (Supplementary Figures S1 and S2).
The CC domain of Cter-SUN is required to form hetero-oligomers with mid-SUN proteins
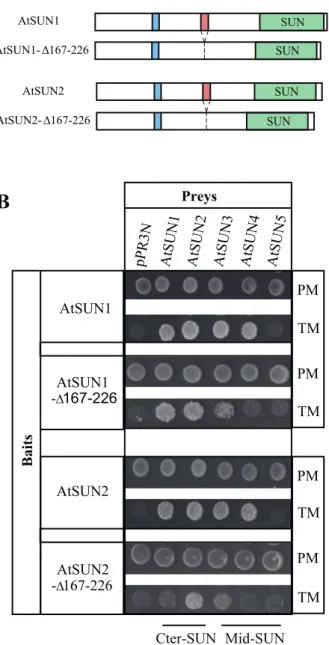
Preliminary investigations using the previously published CC- and SUN-domain deletions (Graumann et al., 2010) in a MYTH screen suggested that deletion of the CC domain significantly altered interactions between the AtSUN pro-teins while the SUN-domain deletion had no obvious effect (data not shown). The effect of the CC deletion construct was weak although reproducible. Therefore, we generated new CC deletions AtSUN1-∆167–226 and AtSUN2-∆167–226, respectively (Fig. 4A), after redefining the domain based on sequence alignment and the CC domain prediction software COILS (Lupas et al., 1991) (see Materials and Methods). These deletions were used as bait in subsequent interaction studies (Fig. 4B). Deletion of the CCs significantly reduced interactions between AtSUN1, AtSUN2, and AtSUN3, and fully abolished interactions with AtSUN4 (Fig. 4B). In addition, AtSUN1 and AtSUN2 deletion mutants, in which the N-terminus and SUN domain were deleted but the CC domain was still present, were able to interact with full-length AtSUN3 and AtSUN4 (Supplementary Figure S6). AtSUN5 was used as a negative control in these experiments as it does not interact with AtSUN1 and AtSUN2. A weak interac-tion between CFP-AtSUN2∆CC and YFP-AtSUN3 was also confirmed by apFRET (Table 1). These results confirm the involvement of the CC domain in Cter-SUN interactions (Graumann et al., 2010) and also demonstrate its importance for interactions between Cter-SUN and mid-SUN proteins.
Mid-SUN proteins play a role in nuclear morphology
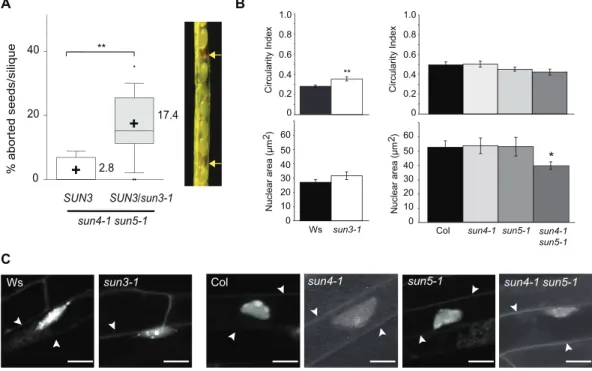
We selected single T-DNA insertion mutants and confirmed absence of full-length transcripts for AtSUN3, AtSUN4, and AtSUN5, respectively (Supplementary Figure S7). Mutant alleles are referred to as sun3-1, sun4-1, and sun5-1 hereaf-ter. The single mutants display no obvious growth or fertil-ity defects. While various combinations of double mutants are viable, the triple mutant sun3-1 sun4-1 sun5-1 was lethal under our growth conditions. Indeed, we screened large num-bers of seedlings for mid-SUN triple mutants in the progeny of sun4-1 sun5-1 SUN3/sun3-1 or sun3-1 sun4-1
SUN5/sun5-1 plants, but triple mutants were never recovered. Looking
carefully at siliques of SUN3/sun3-1 sun4-1 sun5-1 plants, aborted seeds were observed (Fig. 5A). Segregation ratios were not significantly different (P > 0.05) to 25% aborted seeds expected in plants heterozygous for a recessive embryo-lethal mutation. The results suggest that expression of mid-SUNs is needed during early embryo development. These observations further imply that the three mid-SUN proteins, which can be grouped into a monophyletic group (Fig. 1B), are essential for plant survival and that the loss of mid-SUN proteins cannot be functionally compensated by Cter-SUNs. R E E N ER NE N N YFP-AtSUN3 AtSUN4-YFP N AtSUN1-YFP GFP-Calnexin R E E N A B C D
GFP-Calnexin SUN1-YFP SUN2-YFP YFP-SUN3 SUN4-YFP 0.00 0.50 1.00 1.50 2.00 2.50 ER-NE localization ER/NE FI ratio Cter-SUN Mid-SUN ER enriched NE enriched *** ***
Construct Mobile fraction (%) Half time (sec)
YFP-AtSUN3 ER 23.20±1.94 4.32±0.51
YFP-AtSUN3 NE 29.22±1.70 4.21±0.45
AtSUN4-YFP ER 16.62±1.77 4.13±0.59
AtSUN4-YFP NE 13.17±3.44 4.30±0.68
Fig. 2. Localization of mid-SUN proteins. (A) Fluorescent protein fusions of NE marker AtSUN1-YFP and ER marker GFP-calnexin were transiently expressed in N. tabaccum leaf epidermal cells. (B) YFP-AtSUN3 and AtSUN4-YFP localize to both NE (arrow heads) and ER. N, nucleus; scale bar, 10µm. (C) ER/NE fluorescence intensity ratios as quantified for ER marker GFP-calnexin, NE markers AtSUN1-YFP and AtSUN2-YFP, as well as for YFP-AtSUN3 and AtSUN4-YFP when transiently expressed in N. tabaccum leaf epidermal cells. A ratio above 1 indicates ER accumulation (GFP-calnexin) and a ratio smaller than 1 indicates NE accumulation (AtSUN1 and AtSUN2). Values are means ± SEM (n = 25). AtSUN3 and AtSUN4 ER/NE fluorescence intensity ratios are significantly different from AtSUN1 and AtSUN2 (Student t-test; ***, P < 0.0001). (D) FRAP analysis of YFP-AtSUN3 and AtSUN4-YFP revealed varying proportions of mobile protein populations in the ER and NE (mobile fraction) while both fusion proteins move with a similar rate in these membranes (half time). Values are means ± SEM (n = 30). AtSUN3 is more mobile in ER and NE than AtSUN4 (Student’s t-test; P < 0.01).
Commonly, alteration of LINC complex components affect the nuclear shape of plant cells (Tatout et al., 2014). To dissect the impact on nuclear shape for the different mid-SUN proteins, we measured the Circularity Index (CI) and the nuclear area of the single mid-SUN mutants. Only the sun3-1 mutant showed more rounded nuclei by comparison to the wild type (P < 0.01; Fig. 5B). We then studied double mutant plants. However, whereas sun
3-1 is in the Wassilewskija (Ws) background, sun4-1 and sun5-1 were generated in the Columbia (Col) background.
As there are differences in nuclear morphology between Ws and Col (Fig. 5B), we only analysed the sun4-1
sun5-1 double mutant. sun4-sun5-1 sun5-sun5-1 shows weakly but
signifi-cantly smaller nuclei (P < 0.05; Fig. 5B) and altered nuclear morphology (Fig. 5C). The altered nuclear morphology observed for sun3-1 and sun4-1 sun5-1 mutants, which is AtSUN2 AtSUN1 AtSUN1 pPR3N AtSUN4 AtSUN3 AtSUN2 AtSUN5
AtSUN1 AtSUN2 AtSUN3 AtSUN4 AtSUN5
PM TM AtSUN4 AtSUN3 AtSUN1 AtSUN2 AtSUN3 AtSUN4 AtSUN5 pPR3N pPost1 pBT3N -+++ AtSUN2 -+++ +++ +++ ++ ++ AtSUN1 -+++ +++ +++ ++ ++ AtSUN3 ---+ + +++ +++ ++ + AtSUN4 - +/ -+++ +++ ++ AtSUN5 -+++ -++ ++ AtSUN5 AtSUN1 AtSUN2 AtSUN3 AtSUN4 AtSUN5 AtSUN1 AtSUN2 AtSUN3 AtSUN4 AtSUN5 PM TM Baits Preys
B
A
C
Baits Preys AtSUN1 pPR3N AtSUN4 AtSUN3 AtSUN2 AtSUN5 Baits Preys Cter-SUN Mid-SUNFig. 3. Interaction network within the SUN family. All constructs used pBT3N vector in order to produce a fusion protein between Cub and the N-terminal part of the SUN protein. Prey and bait are transformed in the NMY51 yeast strain and vectors are selected on PM (Permissive Medium: YNB without Leu and Trp). Interactions are assessed on TM (Test Medium: YNB without Leu, Trp, Ade, and His). The strength of the protein–protein interaction was evaluated by drop test (see Materials and Methods). (A) Interaction between AtSUN1 and AtSUN2 as baits and the whole members of the SUN-domain family as preys. (B) Interaction between AtSUN3, AtSUN4, and AtSUN5 (mid-SUNs) as baits and the five AtSUN proteins. Only the spots corresponding to 100 000 cells on Permissive (PM) and Test Media (TM) are shown. (C) Summary of protein interactions recorded by MYTH. Strength of interaction was established based on growth efficiency on solid test medium. The prey pOST1 and pPR3N were used as positive and negative controls, respectively.
Table 1. FRET efficiency as detected by apFRET illustrates interactions between Cter-SUN and mid-SUN proteins and the KASH
domain of AtTIK
Combinations EF control P-value t-test
Cter-SUNs and mid-SUNs
CFP-AtSUN1 + YFP-AtSUN3 2.33 ± 0.44 –2.05 ± 0.25 4.6E-10
YFP-AtSUN1 + CFP-AtSUN4 3.63 ± 0.56 –1.74 ± 0.41 5.7E-05
CFP-AtSUN2 + YFP-AtSUN3 2.29 ± 0.36 –0.99 ± 0.24 3.6E-10
YFP-AtSUN2 + CFP-AtSUN4 3.17 ± 0.75 –3.15 ± 0.51 3.1E-07
YFP-AtSUN3 + CFP-AtSUN4 2.41 ± 0.33 –2.12 ± 0.22 1.1E-14
CFP-SUN2ΔCC + YFP-AtSUN3 0.96 ± 0.56 –1.07 ± 0.30 0.00262
Cter-SUNs and AtTIK CFP-AtTIK-KASH + YFP-AtSUN1 3.73 ± 1.93 –2.36 ± 0.74 0.0123
CFP-AtTIK-KASH + YFP-AtSUN2 2.48 ± 1.76 –2.08 ± 0.75 0.03616
Mid-SUNs and AtTIK/AtWIP1
CFP-AtWIP1 + YFP-AtSUN3 2.05 ± 0.70 –1.18 ± 0.27 0.00013
CFP-AtTIK-KASH + YFP-AtSUN3 1.67 ± 0.72 –1.12 ± 0.26 0.00077
CFP-AtWIP1 + AtSUN4-YFP 0.00 ± 0.90 –0.57 ± 0.26 0.54822
CFP-TIK-KASH + AtSUN4-YFP 1.21 ± 0.56 –0.31 ± 0.22 0.01717
reminiscent of phenotypes in sun1 sun2 double mutants (Zhou et al., 2012b), suggests that Cter-SUNs and mid-SUNs have some overlapping function, perhaps as part of the same LINC complex.
Mid-SUN proteins interact with the KASH domain of AtWIP1
A key characteristic of Cter-SUN-domain proteins is their abil-ity to interact with KASH proteins to form a bridge, the LINC complex, that spans the nuclear envelope (Crisp et al., 2006;
Tapley et al., 2011). Recently, Zhou et al., (2012b) identified the first plant KASH-domain proteins, termed AtWIPs. Their study demonstrates, by co-immunoprecipitation, that both AtSUN1 and AtSUN2 Cter-SUN proteins interact with AtWIP1.
To assess whether mid-SUN proteins are also able to inter-act with members of the KASH-domain protein family to bridge the INM and the ONM, we took advantage of the MYTH system to investigate the interactions between the mid-SUN proteins and AtWIP1. First, AtSUN1 and AtSUN2 were used as bait and AtWIP1 as prey in order to confirm the SUN-KASH interactions described by Zhou et al., (2012b) in the MYTH system. Interactions could not be detected using the full length AtWIP1 (Fig. 6B). This could be explained by a localization of the full-length AtWIP1 incompatible with the bait to visualize the interaction in yeast (Snider et al., 2010a,b). However interactions were observed when the KASH domain alone (AtWIP1-KASH) was used as prey, although interactions were weaker with AtSUN2 compared to AtSUN1 (Fig. 6B). Interactions with the KASH domain of AtWIP1 were detected with the three mid-SUN proteins as bait (Fig. 6B). In planta interactions between full-length WIP1 (CFP-WIP1) and YFP-AtSUN3 were also observed (Table 1). However, in apFRET no CFP fluorescence increase was observed for CFP-WIP1 and AtSUN4-YFP. This may be due to protein topology causing the YFP and CFP moi-eties to be too far apart for fluorescence energy transfer to occur despite the interaction. In summary, both MYTH and apFRET demonstrate the capacity of the mid-SUN proteins to interact with a KASH-domain of plant KASH proteins.
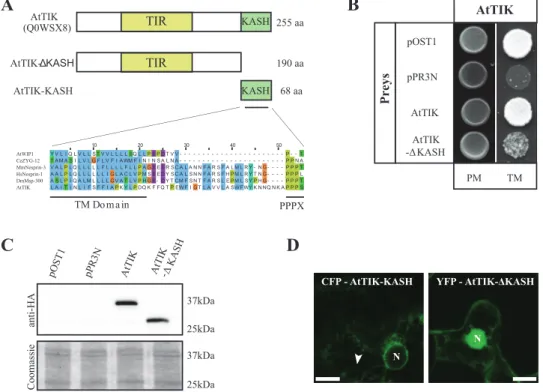
AtTIK, a novel KASH-domain protein localized to the NE
To identify novel SUN-domain protein interaction part-ners in Arabidopsis, a MYTH screen was performed with the two Cter-SUN proteins AtSUN1 and AtSUN2 as bait (see Materials and Methods). Among positives, several proteins containing TM domains were identified. One of them (accession number AT5G44920) is a protein of unknown function containing a C-terminal TM domain upstream of a PPPS motif, a characteristic signature of the KASH domain (Fig. 7A) (Starr and Fischer, 2005; Starr, 2011). In addition (and unlike AtWIP1) the protein con-tains a number of amino acids homologous to mammalian nesprins and Drosophila Msp-300 (Starr and Fischer, 2005) between (or within) the TM domain and the PPPS motif (Fig. 7A). It does not, however, show homology to any known KASH proteins in the remainder of the sequence as generally observed for KASH proteins (Starr and Fischer, 2005). However, the DORY programme (Zhou
et al., 2014), developed to detect KASH-domain proteins, was successfully used to identify this protein. AT5G44920 encodes two protein isoforms of 242 and 255 amino acids, respectively, both containing the putative KASH-domain and a putative Toll-Interleukin-Resistance (TIR) domain characteristic of a large family of proteins conserved in eukaryotes (Mitcham et al., 1996) (Fig. 7A). Although highly conserved, we found the presence of a KASH Fig. 4. The CC domain is essential for interaction between SUN-family
members. (A) Schematic representation of the deletion derivatives AtSUN1-∆167-226 and AtSUN2-∆167-226 in comparison to full length AtSUN1 and AtSUN2. (B) Interaction between AtSUN1 and AtSUN2 or their deletion derivatives AtSUN1-∆167-226 and AtSUN2-∆167-226 as bait and the five SUN-domain proteins as preys in the MYTH system. Only the spots corresponding to 100 000 cells on Permissive (PM) and Test Media (TM) are shown. This figure is available in colour at JXB online.
domain in a member of the TIR-domain family only in a few Brassicaceae species (Arabidopsis thaliana, Arabidopsis
lyrata and Thellungiella halophila). AT5G44920 has been
renamed AtTIK (for Arabidopsis thaliana TIR-KASH protein) and is referred to as such below. The longer iso-form of AtTIK was isolated in our two-hybrid screen, and was subsequently used for all the experiments described here. AtTIK is expressed highly in root, and at lower lev-els in other tissues (Genevestigator data; Supplementary Figure S8). Binding properties of AtTIK in the MYTH system reveals that it is able to form homomers (Fig. 7B). Furthermore, truncation of the KASH domain (deletion from amino acids 191 to 255, including the PPPS motif and the TM domain upstream; Fig. 7A), hereafter called AtTIK-∆KASH, significantly reduced these protein inter-actions, demonstrating the involvement of the KASH domain itself in this process (Fig. 7B). We confirmed that the loss of homomers was not a consequence of lower expression levels or instability of the deletion construct as both AtTIK and AtTIK-∆KASH were expressed at similar levels in the MYTH (Fig. 7C).
To investigate whether AtTIK is localized at the NE, fluo-rescent protein fusions of the full-length protein were gen-erated and transiently expressed in plant tissue, driven by a 35S promoter. Unfortunately the full-length protein did not express. Instead, two truncations of the protein were fused to fluorescent proteins and transiently expressed. The first was AtTIK-∆KASH and the second, hereafter referred to as AtTIK-KASH, consisted of the KASH domain (TM
domain and the PPPS motif) alone. YFP-AtTIK-∆KASH was soluble and predominately accumulated in the nucleus (Fig. 7D). On the other hand, CFP-AtTIK-KASH local-ized at the nuclear periphery and in the ER of some brightly fluorescent cells (Fig. 7D). The combination of those two truncations strongly indicates that AtTIK is NE-localized, which is typical for a member of the KASH protein fam-ily and consistent with the localization of its interacting partners.
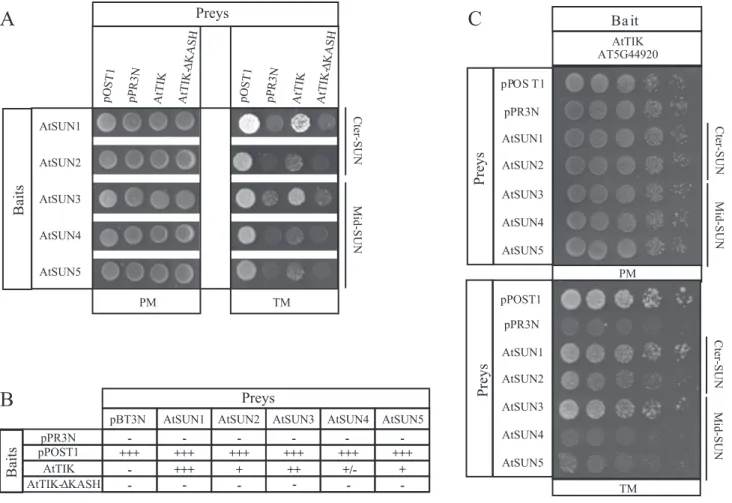
The KASH-domain of AtTIK is essential for the SUN-KASH interaction
We used the MYTH system to establish whether AtTIK inter-acts with mid-SUN proteins and whether its putative KASH domain is required for interaction (Fig. 8A). Used as bait, the two Cter-SUN proteins interact with AtTIK, although weakly in the case of AtSUN2. Interestingly the interaction is completely abolished when the putative KASH domain is deleted (AtTIK-∆KASH) (Fig. 8A). This result validates the putative KASH domain of AtTIK as functional. Secondly, we tested the three mid-SUN proteins as bait to determine if these proteins are also able to interact with AtTIK. As shown in Fig. 8A and 8B, the mid-SUN proteins indeed interact with the KASH-domain protein. Additionally these interactions are completely abolished when the KASH domain (AtTIK-∆KASH) is deleted. All these interactions were subsequently confirmed by using AtTIK as bait with the two SUN sub-families as a prey (Fig. 8C).
SUN3 sun4-1 sun5-1 0 20 40 % aborted seeds/silique SUN3/sun3-1 ** 17.4 2.8 + + A B C 0 0.2 0.4 0.6 0.8 1.0 0 0.2 0.4 0.6 0.8 1.0
Circularity Index Circularity Index
Ws sun3-1 Col sun4-1 sun5-1 sun4-1 sun5-1 ** Nuclear area (µm )2 0 20 10 30 40 50 60 0 20 10 30 40 50 60 Nuclear area (µm )2 *
Ws sun3-1 Col sun4-1 sun5-1 sun4-1 sun5-1
Fig. 5. Functional significance of the mid-SUN subfamily. (A) Percentage of aborted seeds per silique in different mid-SUN mutants. Range bars represent the 10–90% range of mean values, boxes represent interquartile distances, the horizontal line across range bars represents the median, and ‘+’ the mean values (also indicated at the right of each box). Kruskal Wallis non-parametric tests were applied to determine significant differences between mean values (**, P < 0.01). A representative silique from SUN3/sun3-1 sun4-1 sun5-1 mutant plants is shown at the right. Arrows indicate aborted seeds. (B) Circularity Index (CI) and nuclear area calculated from ~25 confocal images of the mutants indicated with their respective control ecotype. Error bars present SEM and the t-test was applied to determine the statistical significance (*, P < 0.05; **, P < 0.01). (C) Representative confocal images of ethidium bromide-stained root epidermal nuclei of mid-SUN mutants and their control ecotypes; arrow heads point to the outline of cells. Size bar, 10µm.
In addition to the MYTH, the functionality of the KASH domain in mediating SUN–KASH interactions in
planta was investigated. Using apFRET, interactions were
detected between CFP-AtTIK-KASH and AtSUN1-YFP, AtSUN2-YFP, YFP-AtSUN3, and AtSUN4-YFP (Table 1). Altogether, interactions between mid-SUN and KASH pro-teins (AtWIP1 and AtTIK), via the KASH domain, dem-onstrates a conserved functionality of the Cter-SUN and mid-SUN subfamilies in respect to NE bridging.
AtTIK is involved in root growth and nuclear morphology
The predominant expression of AtTIK in roots led us to investigate root growth in a T-DNA insertion line. For the homozygous Salk_205745 mutant line (tik-1) no full-length transcript for AtTIK was detected (Fig. 9A). Observation of seedling growth over a 14-day time period revealed that tik-1 had shorter roots (Fig. 9B). No other obvious growth pheno-types were observed. In metazoans, tissue-specific expression of KASH proteins such as KASH5 has been observed, and previously AtSINE1 was identified to be guard cell specific.
The expression pattern and mutant phenotype of AtTIK imply that it is also a tissue-specific KASH protein linked to processes in root development.
As mentioned previously, plant KASH and SUN proteins are commonly linked to nuclear morphology. While the CI of root nuclei in the tik-1 mutant was similar to the wild type, reduced width and length axes as well as a smaller nuclear area were observed (Fig. 9C). This indicates that AtTIK, like other plant LINC complex components, plays a role in nuclear morphology, specifically nuclear size.
Discussion
Eukaryotic LINC complexes are structurally and functionally diverse protein complexes, but all have at their core the SUN-KASH interaction that bridges the membranes of the NE and physically connects nuclear and cytoplasmic elements. While previous evidence has supported the existence of these SUN-KASH interactions in plants, this paper highlights the molecular diversity of LINC complexes at the plant NE. We show that both types of SUN-domain proteins, Cter-SUNs and mid-SUNs, as well as several types of KASH-domain proteins, are involved in the NE bridging.
Our characterization of the Arabidopsis mid-SUN pro-teins highlights similarities and differences to the Cter-SUN-domain proteins. While both contain the highly conserved SUN domain, are membrane intrinsic, localized in the NE, interact with each other, and have functions in nuclear mor-phology, their internal structures and binding affinities differ. The presence of the mid-SUNs in the ER may point towards a non-LINC-related function and evidence from yeast suggests that ER-localized mid-SUNs function in NE localization of Cter-SUNs (Friederichs et al., 2012). The three TM domains of the mid-SUNs allow for various membrane topologies and so far it is not clear how these proteins are inserted in the mem-brane. However, it can be speculated that the CC domain and SUN domain are localized in the periplasmic space, similar to the Cter-SUNs, to mediate SUN–KASH and SUN–SUN (via the CC) interactions. Interestingly, although members of the two subfamilies interact with themselves and with each other, differences in binding affinities indicate stronger inter-actions between either Cter-SUNs or mid-SUNs but weaker binding between Cter-SUNs and mid-SUNs. The greatest difference was found for AtSUN5, which only interacts with itself and AtSUN3. While AtSUN1–AtSUN4 are expressed more or less ubiquitously, AtSUN5 expression is tissue spe-cific – it is spespe-cifically found in anther, pollen, and endosperm tissue indicating that its interactions are related to specific functions. Murphy and Bass (2012) have previously suggested that ZmSUN3 plays a role in maize meiosis. As AtSUN5 is expressed in meiotically active tissue, it can be hypothesized that complexes containing AtSUN5 play a specific function during meiosis.
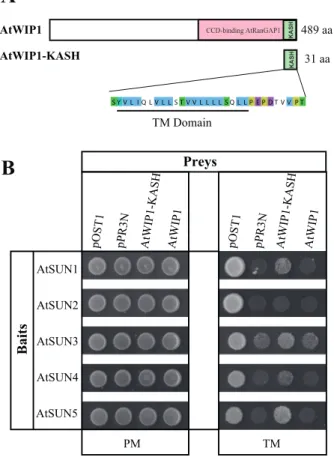
The SUN proteins may not be the only components that bring tissue-specificity to the plant LINC complex. AtTIK, here identified as a novel plant KASH protein, is mainly expressed in root tissue. Apart from an internal TIR domain, AtWIP1 CCD-binding AtRanGAP1
KAS H KAS H AtWIP1-KASH TM Domain 489 aa 31 aa
A
AtSUN4 AtSUN3 AtSUN1 AtSUN2 AtSUN5 pPR3N pOST 1 pPR3N AtWIP1-KASH pOST 1 PM TM Baits AtWIP1 AtWIP1 Preys AtWIP1 -KASHB
Fig. 6. Interaction between the SUN proteins and the KASH domain of AtWIP1. (A) Schematic representation of the full-length KASH-domain protein AtWIP1 and the deletion construct AtWIP1-KASH. (B) Interaction between AtWIP1 or the KASH domain alone (including the TM domain) as preys and the SUN-domain proteins in the membrane two-hybrid system. Only the spots corresponding to 100 000 cells on Permissive (PM) and Test Media (TM) are shown. The prey pOST1 and pPR3N were used as positive and negative controls, respectively. This figure is available in colour at JXB online.
it contains a KASH domain including the C-terminal TM domain, which anchors it to the NE. AtTIK specifically binds to all SUN proteins but interacts more strongly with AtSUN1 and AtSUN3. In animal and plant kingdoms, the TIR domain is present in proteins involved in the immune response. In plants a TIR domain associated with a nucleo-tide binding leucine-rich repeat (NB-LRR) is present in plant disease resistance proteins (R proteins) that have been shown to be involved in the defence mechanism against microbial infection (McHale et al., 2006; Głowacki et al., 2011). The
Arabidopsis genome comprises more than 135 proteins
con-taining TIR domains, of which 30 (including AtTIK) do not contain the NB-LRR domain characteristic of the R pro-teins (Jebanathirajah et al., 2002; Meyers et al., 2002). The role of this subfamily, called TIR-X (Meyers et al., 2002), is unknown, but they are likely to be involved in mediating protein–protein interactions and may function as adapter proteins interacting with the R proteins to transduce signals (Meyers et al., 2002; Jebanathirajah et al., 2002). This tempts speculation that AtTIK-containing LINC complexes may be involved in signalling events at the NE. A precedent for this comes from C. elegans, where matefin/Sun1 is involved in apoptosis signalling events (Tzur et al., 2006).
We found that both subfamilies of SUN-domain pro-teins can interact with the KASH domain of AtWIP1 and AtTIK. This has several implications. Firstly, this provides
evidence that mid-SUN proteins are not merely ‘accessory’ proteins of plant LINC complexes through their interac-tions with Cter-SUNs, but that they are active compo-nents of the SUN–KASH bridges. Secondly, the presence of three AtWIPs, two AtSINEs, five AtSUN proteins, and the AtTIK significantly increases the binding diversity of these bridges at the plant NE. As discussed, this is likely to facilitate tissue, developmental, and functional specific-ity and regulation of these complexes. Interestingly, while the KASH domains of AtTIK, AtWIP, and AtSINE can interact with the SUNs, they significantly differ in their last four amino acid motifs. The plant-specific WIP pro-teins and SINEs contain a conserved xVPT motif, while AtTIK’s PPPS motif more closely resembles the highly con-served PPPX motif of animal KASH domains (Starr and Fridolfsson, 2010). Why plants have two separate types of KASH domains, and perhaps even more, remains unclear but could be linked to tissue specificity and binding affin-ity. Some animal KASH proteins are known to be expressed in specific tissues (Razafsky et al., 2013). Previously, Sosa
et al., (2012, 2013) have suggested that despite the diver-gence of KASH domains, the penultimate proline is neces-sary for SUN–KASH interactions – as is S641 of the SUN domain (Zhou et al., 2012a); both critical amino acids are conserved in the AtTIK/AtWIP/AtSINE KASH domains and SUN domains, respectively.
Fig. 7. AtTIK is a new putative KASH-domain protein. (A) Schematic representation of AtTIK and the deletion constructs AtTIK-∆KASH and AtTIK-KASH. AtTIK contains a conserved interleukin-1 receptor/Toll-like receptor (TIR) domain represented in yellow and one KASH domain (including a TM domain) in green at the C-terminal part of the protein. Protein lengths are indicated at the right; UniProt protein ID is indicated on the left between brackets. The alignment of selected KASH domains from plants and animals is shown with the TM domain and the PPPX motif underlined. B) Interaction in the MYTH system between AtTIK as bait and AtTIK at full length or with its putative KASH domain deleted. The prey pOST1 and pPR3N were used as positive and negative controls, respectively. Only the spots corresponding to 100 000 cells on Permissive (PM) and Test Media (TM) are shown. (C) Protein levels of AtTIK and AtTIK-∆KASH preys were determined by western-blot analysis using anti-HA antibody. Coomassie staining shows equal loading of samples. (D) Subcellular localization of AtTIK truncations. Truncations of AtTIK (CFP-AtTIK-KASH and YFP-AtTIK-∆KASH) fused to fluorescent proteins and transiently expressed in tobacco leaf epidermal cells. Size bar, 10µm; N, nucleus; arrowhead, ER.
500 bp AT5G44920 Salk_205745 tik-1 Col tik-1 ACT TIK
C
B
A
Days post germination
Root length in cm Coltik-1
0 2 4 6 0 7 10 15 *** ** 0 2 4 6 Col tik-1 Nuclei width (µm) 0 10 20 30 40 50 60 70 Nuclei area (µm )2 Col tik-1 *** Col tik-1 0 4 8 12 16 * Nuclei length (µm)
Fig. 9. AtTIK plays a role in root development. (A) Schematic representation of AtTIK showing the position of the T-DNA insertion Salk_205745 (tik-1) used in this work (arrowhead). Transcript levels of AtTIK in wild type and homozygous tik-1 mutant plants were assessed using primers indicated as arrows. Primer sequences are available in Supplementary Table S1. Actin (ACT) was used as a reference. (B) Kinetics of root growth in Col (wild type) and tik-1 (mutant). Values are means ± SEM (n = 40). (C) Nuclear morphology parameters in root epidermal cells of Col and tik-1. Values are means ± SEM (n = 45). Statistical significance determined by Student’s t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Fig. 8. Interaction between AtTIK and the SUN-domain proteins. (A) MYTH assays between the SUN-domain proteins as bait and the full-length AtTIK or the KASH domain deletion as preys. pOST1 and pPR3N were used as positive and negative control prey, respectively. Spots correspond to 100 000 cells on Permissive and Test Media. (B) Summary of protein interactions using full length AtTIK and the KASH domain deletion as bait and pBT3N as negative control bait. Strength of interaction was established based on growth efficiency on solid test medium. (C) Interaction between AtTIK as bait and the five SUN proteins as prey. pOST1 and pPR3N were used as control preys. Spots from left to right correspond to 100 000, 10 000, 1000, 100 and 10 cells. Drops are spotted on Permissive (PM) and Test Media (TM).
The constituents and molecular functions of the NE in plants are still poorly understood. To date, LINC complex components such as the AtWIPs and Cter-SUNs remain one of the better-studied NE components. Adding to this, the mid-SUNs and AtTIK as KASH protein greatly enhance our knowledge of the plant NE proteome. It is becoming apparent that diverse protein complexes involving membrane intrinsic, nucleoplasmic, cytoplasmic, and nuclear pore components are active at the plant NE (Tatout et al., 2014). Dissecting their molecular functions and regulation, including those of the mid-SUNs and AtTIK, will be an exciting and important challenge for the future in this field.
Supplementary material
Supplementary data can be found at JXB online.
Supplementary Table S1. Oligonucleotides used in this study.
Supplementary Figure S1. Alignment of mid-SUN pro-teins from 13 selected dicot species.
Supplementary Figure S2. Phylogenetic tree of mid-SUN proteins from 13 selected dicot species.
Supplementary Figure S3. AtSUN expression patterns from Genevestigator.
Supplementary Figure S4. ER/NE fluorescence intensity analysis.
Supplementary Figure S5. Interaction network within the SUN family.
Supplementary Figure S6. The CC domain is sufficient for interaction between SUN family members.
Supplementary Figure S7. Mid-SUN T-DNA mutants. Supplementary Figure S8. AtTIK expression patterns.
Funding
Katja Graumann is funded by an Early Career Fellowship from the Leverhulme Trust. This work was supported by the CNRS, INSERM, and Blaise Pascal and Auvergne Universities.
Acknowledgements
The authors would like to thank Delphine Leroux, Julien Gerard, Karim Housseini B. Issa and Julien Vieu for their technical support as well as Iris Meier and Xiao Zhou (Ohio State University) for the use of DORY. We also thank the two anonymous reviewers for their fruitful comments.
References
Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17, 540–552.
Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. 2006. Coupling of the nucleus and cytoplasm role of the LINC complex. The Journal of Cell Biology 172, 41–53.
Field M, Horn D, Alsford S, Koreny L, Rout MP. 2012. Telomeres, tethers and trypanosomes. Nucleus 3, 0–8.
Friederichs JM, Gardner JM, Smoyer CJ, Whetstine CR, Gogol M, Slaughter BD, Jaspersen SL. 2012. Genetic analysis of Mps3
SUN domain mutants in Saccharomyces cerevisiae reveals an
interaction with the SUN-like protein Slp1. Genes Genomes Genetics 2, 1703–1718.
Głowacki S, Macioszek VK, Kononowicz AK. 2011. R proteins as fundamentals of plant innate immunity. Cellular and Molecular Biology Letters 16, 1–24.
Graumann K. 2014. Evidence for LINC1-SUN associations at the plant nuclear periphery. PLoS One 9, e93406.
Graumann K, Irons SL, Runions J, Evans DE. 2007. Retention and mobility of the mammalian lamin B receptor in the plant nuclear envelope. Biology of the Cell 99, 553–562.
Graumann K, Runions J, Evans DE. 2010. Characterization of SUN domain proteins at the higher plant nuclear envelope. The Plant Journal 61, 134–144.
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307–321.
Jebanathirajah JA, Peri S, Pandey A. 2002. Toll and interleukin-1 receptor (TIR) domain-containing proteins in plants: a genomic perspective. Trends in Plant Science 7, 388–391.
Karpova TS, Baumann CT, He L, Wu X, Grammer A, Lipsky P, Hager GL, McNally JG. 2003. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. Journal of Microscopy 209, 56–70.
Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. Journal of Molecular Biology 305, 567–580. Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252, 1162–1164.
McGee MD, Rillo R, Anderson AS, Starr DA. 2006. UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Molecular Biology of the Cell 17, 1790–1801.
McGee MD, Stagljar I, Starr DA. 2009. KDP-1 is a nuclear envelope KASH protein required for cell-cycle progression. Journal of Cell Science 122, 2895–2905.
McHale L, Tan X, Koehl P, Michelmore RW. 2006. Plant NBS-LRR proteins: adaptable guards. Genome Biology 7, 1–11.
Meyers BC, Morgante M, Michelmore RW. 2002. TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. The Plant Journal 32, 77–92.
Mitcham JL, Parnet P, Bonnert TP, Garka KE, Gerhart MJ, Slack JL, Gayle MA, Dower SK, Sims JE. 1996. T1/ST2 signaling establishes it as a member of an expanding interleukin-1 receptor family. Journal of Biological Chemistry 271, 5777–5783.
Murphy SP, Bass HW. 2012. The maize (Zea mays) desynaptic (dy) mutation defines a pathway for meiotic chromosome segregation, linking nuclear morphology, telomere distribution and synapsis. Journal of Cell Science 125, 3681–3690.
Murphy SP, Simmons CR, Bass HW. 2010. Structure and expression of the maize (Zea mays L.) SUN-domain protein gene family: evidence for the existence of two divergent classes of SUN proteins in plants. BMC Plant Biology 10, 269.
Oda Y, Fukuda H. 2011. Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. The Plant Journal 66, 629–641.
Oldenburg KR, Vo KT, Michaelis S, Paddon C. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Research 25, 451–452.
Punta M, Coggill PC, Eberhardt RY et al. 2011. The Pfam protein families database. Nucleic Acids Research 40, D290–D301. Razafsky DS, Ward CL, Kolb T, Hodzic D. 2013. Developmental regulation of linkers of the nucleoskeleton to the cytoskeleton during mouse postnatal retinogenesis. Nucleus 4, 399–409.
Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675.
Snider J, Kittanakom S, Curak J, Stagljar I. 2010a. Split-ubiquitin based membrane yeast two-hybrid (MYTH) system: a powerful tool for identifying protein-protein interactions. Journal of Visualized Experiments 36, 1698. Snider J, Kittanakom S, Damjanovic D, Curak J, Wong V, Stagljar I. 2010b. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nature Protocols 5, 1281–1293.
Sohaskey ML, Jiang Y, Zhao JJ, Mohr A, Roemer F, Harland RM. 2010. Osteopotentia regulates osteoblast maturation, bone formation, and skeletal integrity in mice. The Journal of Cell Biology 189, 511–525. Sosa BA, Kutay U, Schwartz TU. 2013. Structural insights into LINC complexes. Current Opinion in Structural Biology 23, 285–291.
Sosa BA, Rothballer A, Kutay U, Schwartz TU. 2012. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149, 1035–1047.
Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols 1, 2019–2025.
Starr DA. 2011. KASH and SUN proteins. Current Biology 21, R414–R415. Starr DA, Fischer JA. 2005. KASH ‘n Karry: The KASH domain family of cargo-specific cytoskeletal adaptor proteins. BioEssays 27, 1136–1146. Starr DA, Fridolfsson HN. 2010. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annual Review of Cell and Developmental Biology 26, 421–444.
Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I. 2013. Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Current Biology 23, 1776–1781.
Tapley EC, Ly N, Starr DA. 2011. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Molecular Biology of the Cell 22, 1739–1752.
Tatout C, Evans DE, Vanrobays E, Probst AV, Graumann K. 2014. The plant LINC complex at the nuclear envelope. Chromosome Research 22, 241–252.
Tzur YB, Margalit A, Melamed-Book N, Gruenbaum Y. 2006. Matefin/ SUN-1 is a nuclear envelope receptor for CED-4 during Caenorhabditis elegans apoptosis. Proceedings of the National Academy of Sciences,
USA 103, 13397–13402.
Zhou Z, Du X, Cai Z, et al. 2012a. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. Journal of Biological Chemistry 287, 5317–5326.
Zhou X, Graumann K, Evans DE, Meier I. 2012b. Novel plant SUN– KASH bridges are involved in RanGAP anchoring and nuclear shape determination. The Journal of Cell Biology 196, 203–211.
Zhou X, Graumann K, Wirthmueller L, Jonees J, Meier I. 2014. Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants. The Journal of Cell Biology 205, 677–692.