HAL Id: hal-03096510
https://hal.archives-ouvertes.fr/hal-03096510
Submitted on 5 Jan 2021HAL is a multi-disciplinary open access
archive for the deposit and dissemination of sci-entific research documents, whether they are pub-lished or not. The documents may come from teaching and research institutions in France or abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est destinée au dépôt et à la diffusion de documents scientifiques de niveau recherche, publiés ou non, émanant des établissements d’enseignement et de recherche français ou étrangers, des laboratoires publics ou privés.
Interleukin 7 regulates switch transcription in
developing B cells
Audrey Dauba, Fatima-Zohra Braikia, Chloé Oudinet, Ahmed Amine
Khamlichi
To cite this version:
Audrey Dauba, Fatima-Zohra Braikia, Chloé Oudinet, Ahmed Amine Khamlichi. Interleukin 7 regu-lates switch transcription in developing B cells. Cellular and molecular immunology, Nature Publishing Group/Chinese Society of Immunology, 2020, �10.1038/s41423-020-0430-y�. �hal-03096510�
Cellular and Molecular Immunology
1
Correspondence
2
Interleukin 7 regulates switch transcription in developing B cells
3
Audrey Dauba, Fatima-Zohra Braikia, Chloé Oudinet, and Ahmed Amine Khamlichi* 4
Institut de Pharmacologie et de Biologie Structurale, IPBS, Université de Toulouse, CNRS, Université 5
Paul Sabatier, 31077 Toulouse, France. 6
7
* Correspondence: Ahmed Amine Khamlichi (ahmed.khamlichi@ipbs.fr). Tel: +33 5 61 17 55 22 8
9
2
Class switch recombination (CSR) enables activated mature B cells to change immunoglobulin 11
(Ig) isotype expression from IgM to IgG, IgE or IgA. The process requires activation-induced cytidine 12
deaminase (AID) and transcription of the target sequences at the Ig heavy chain (IgH) constant locus, 13
composed of multiple constant (CH) genes each specifying an Ig isotype.1 The CH genes are organized
14
in transcription units made up of I promoters/exons upstream of highly repetitive sequences called 15
switch (S) sequences and the CH exons. Signal-dependent induction of switch transcription at
16
downstream S sequences is required for recombination with the constitutively transcribed Sµ sequence, 17
the universal switch donor site.1 CSR can also occur in developing B cells, albeit at lower frequency.
18
The signaling pathways and the transcriptional regulatory elements that control CSR in developing B 19
cells are still ill-known. There is evidence that signaling through Toll-like receptors induces AID 20
expression and CSR at early developmental stages,2-4 and some cis-acting elements are involved in
21
preventing switch transcription in developing cells.5
22
Interleukin 7 (IL7) is a non-redundant cytokine that plays an important role in early B cell 23
development.6,7 IL7, through its receptor (IL7R) signaling, regulates multiple fundamental and
24
pathological processes including proliferation, survival, V(D)J recombination, gene expression, auto-25
immunity and leukemia.e.g.8-12 But its role in the regulation of switch transcription is unknown.
26
To investigate the role of IL7 in switch transcription, we set up a stromal cell-free culture 27
system that contains IL7 alone (see additional information). IL7R is expressed in pro-B cells and is 28
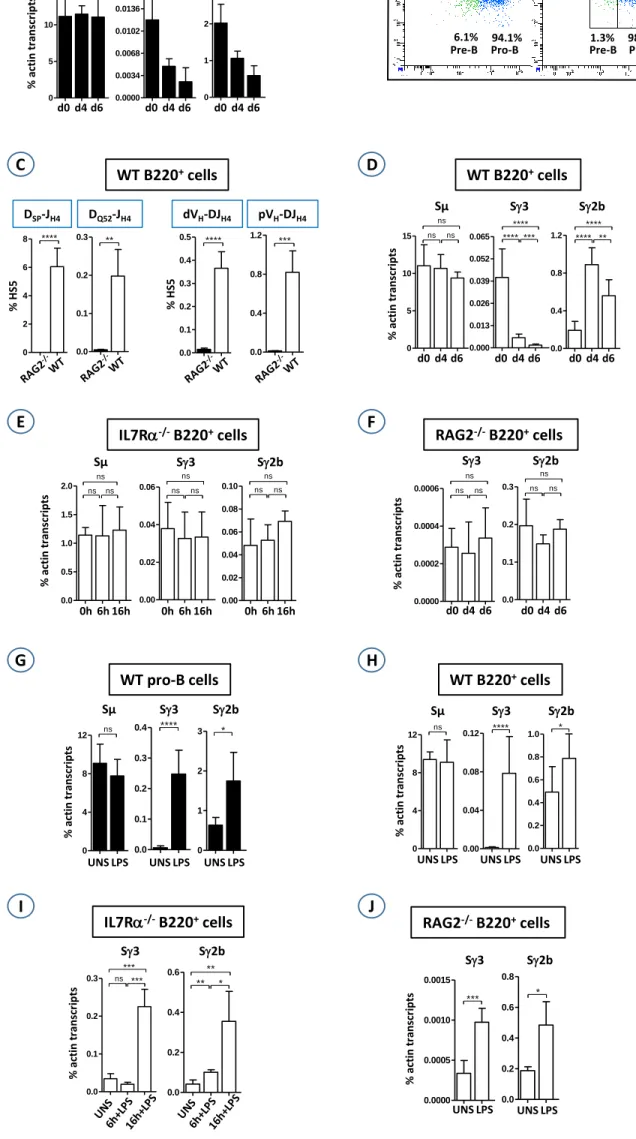
down-regulated in pre-B cells. 6,7 Consequently, at day 2, and in stark contrast to sorted pro-B cells
29
which proliferated in the IL7 medium, all sorted pre-B cells were dead (not shown). In pro-B cells, 30
transcript levels of S1, S2a, S and S were extremely low to undetectable (not shown). We therefore 31
focused on S3 and S2b transcripts. Unexpectedly, S3 and S2b transcripts levels steadily decreased 32
with time while Sµ transcripts levels did not vary (Fig. 1A). We also adapted the IL7 culture system to 33
the whole medullar B220+ population, a mixture of B cell precursors, immature and circulating cells.
We reasoned that IL7R-expressing B cells (pre-pro-B and pro-B cells), though representing only a 35
small fraction (8-15% of B220+ cells) will proliferate whereas IL7R-negative cells will die. Indeed,
36
pro-B cells proliferated vigorously (Fig. S1A) and represented almost the whole population at day 6 (> 37
98 %) (Fig. 1B and Fig. S1B). The propagated cells have undergone V(D)J recombination on the IgH 38
locus (Fig. 1C and Fig. S1C). Again, Sµ transcript levels did not vary, but S3 levels steadily dropped 39
until they reached background levels at day 6. The levels of S2b transcripts increased until day 4 then 40
significantly decreased (Fig. 1D). Our interpretation is that, when starting with B220+ population, the
41
actual effect of IL7 on pro-B cells becomes obvious only after 4 days of culture (see supplementary 42
discussion). Altogether, the above data reveal that IL7 inhibits S3 and S2b transcription in ex vivo 43
propagated pro-B cells. 44
Since IL7 conveys its effects through IL7R, switch transcripts levels would be expected to not 45
vary in the absence of the receptor. The IL7R is composed of the common chain and the chain. 6,7
46
Mice devoid of IL7R chain display a severe block at early pro-B cell stage.13 B220+ cells, containing
47
mainly pre-pro-B cells and potentially some early pro-B cells, were sorted from IL7R-/- BMs.
48
Preliminary experiments revealed that cultured IL7R-deficient B220+ cells died after 24 hours (not
49
shown). Therefore, we were compelled to quantify S3 and S2b transcripts at earlier time points. 50
Interestingly, S3 and S2b transcript levels did not significantly vary after 6h of culture (Fig 1E), 51
suggesting that IL7R was required to inhibit S3 and S2b transcription. 52
IL7-mediated inhibition of S3 and S2b transcription was seen in pro-B cells that had 53
rearranged their IgH loci (Fig. 1C). To explore if IL7 effect on switch transcription required V(D)J 54
recombination, we quantified S3 and S2b transcripts levels in cultured RAG2-deficient pro-B cells. 55
We found no obvious effect on S3 and S2b transcription (Fig. 1F). We conclude that IL7 inhibits 56
switch transcription in cultured pro-B cells that underwent V(D)J recombination. 57
4
S3 and S2b transcription is induced by LPS. To explore the interplay between LPS and IL7 58
on switch transcription, sorted pro-B cells or BM B220+ cells were cultured in the presence of IL7 in
59
the presence or not of LPS. LPS readily induced switch transcription, though the effect on S3 transcript 60
levels was stronger (Fig. 1G, 1H). Induction of S3 and S2b transcription led to CSR to S3 and S2b 61
respectively (Fig. S2A). Aicda gene (encoding AID) transcription was also induced by LPS (Fig. S2B). 62
Induction of switch transcription was not restricted to S3 and S2b as S1 and S transcripts, which 63
were undetectable in unstimulated pro-B cells after 6 days of IL7 culture, were readily induced by 64
LPS+IL4 (Fig. S2C). 65
S3 transcript levels increased in LPS-activated IL7R-deficient B220+ cells, though only after
66
16 h, whereas the increase of S2b transcript levels was already significant after 6 h (Fig. 1I). In 67
activated RAG2-deficient pro-B cells, S3 and S2b transcription was moderately induced (Fig. 1J), 68
while the induction was stronger for S1 and S transcription (Fig. S2D). 69
Overall, activation of cultured pro-B cells induced switch transcription, strongly suggesting that 70
the inhibitory effect conveyed by IL7R signaling is overridden by the pathways that induce CSR. 71 72 73 74 75 ADDITIONAL INFORMATION 76
Supplementary information is available at CMI’s website. 77
ACKNOWLEDGEMENTS 79
We thank Amy L. Kenter and Cornelis Murre for help with RAG-deficient pro-B cell cultures, and the 80
IPBS animal facility staff and the Imaging Core Facility TRI-IPBS, in particular Emmanuelle Näser, 81
for their excellent work. This work was supported by the Agence Nationale de la Recherche [ANR-16-82
CE12-0017], the Institut National du Cancer [INCA_9363, PLBIO15-134], the Fondation ARC pour 83
la Recherche sur le Cancer [PJA 20191209515], and the Ligue Contre le Cancer (Ligue Régionale : 84
comités de l’Ex Région Midi-Pyrénées). FZB was supported by a fellowship from the INCa. CO is a 85
fellow of the Ministry of Higher education & Research and is recipient of a fellowship from the 86
“Fondation pour la Recherche Médicale”. Tri-IPBS has the financial support of ITMO Cancer Aviesan 87
(National Alliance for Life Science and Health) within the framework of Cancer Plan. 88
89
AUTHOR CONTRIBUTIONS 90
Conceptualization, AAK. Methodology, AD, FZB, CO and AAK. Investigation, AD, FZB, and CO. 91
Handling mouse lines, AD. Writing—original draft, AAK. Writing-review and editing, AD, FZB, 92
CO and AAK. Supervision, AAK. Funding acquisition, AAK. 93
94
COMPETING INTERESTS 95
The authors declare no competing interests. 96
6
REFERENCES 98
1. Yu, K. & Lieber, M. R. Current insights into the mechanism of mammalian immunoglobulin class
99
switch recombination. Crit. Rev. Biochem. Mol. Biol. 54, 333-351 (2019). 100
2. Han, J. H. et al. Class switch recombination and somatic hypermutation in early mouse B cells are
101
mediated by B cell and Toll-like receptors. Immunity 27, 64-75 (2007). 102
3. Edry, E., Azulay-Debby, H. & Melamed, D. TOLL-like receptor ligands stimulate aberrant class
103
switch recombination in early B cell precursors. Int. Immunol. 20, 1575-1585 (2008). 104
4. Swaminathan, S. et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia.
105
Nat. Immunol. 16, 766-774 (2015).
106
5. Braikia, F. Z. et al. Inducible CTCF insulator delays the IgH 3' regulatory region-mediated activation
107
of germline promoters and alters class switching. Proc. Natl. Acad. Sci. U S A. 114, 6092-6097 (2017). 108
6. Ceredig, R. & Rolink, A. G. The key role of IL-7 in lymphopoiesis. Semin. Immunol. 24, 159-164
109
(2012).
110
7. Clark, M. R., Mandal, M., Ochiai, K. & Singh, H. Orchestrating B cell lymphopoiesis through
111
interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 14, 69-80 (2014). 112
8. Corcoran, A. E. et al. The interleukin-7 receptor alpha chain transmits distinct signals for
113
proliferation and differentiation during B lymphopoiesis. EMBO J. 15, 1924-1932 (1996). 114
9. Chowdhury, D. & Sen, R. Transient IL-7/IL-7R signaling provides a mechanism for feedback
115
inhibition of immunoglobulin heavy chain gene rearrangements. Immunity 18, 229-241 (2003). 116
10. Malin, S. et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination
117
during pro-B cell development. Nat. Immunol. 11, 171-179 (2010). 118
11. Heltemes-Harris, L. M. & Farrar, M. A. The role of STAT5 in lymphocyte development and
119
transformation. Curr. Opin. Immunol. 24, 146-152 (2012). 120
12. Dooms, H. Interleukin-7: Fuel for the autoimmune attack. J. Autoimmun. 45, 40-48 (2013).
121
13. Peschon, J. J. et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-122
deficient mice. J. Exp. Med. 180, 1955-1960 (1994). 123 124 125 126 127 ADDITIONAL INFORMATION 128
Supplementary information is available at CMI’s website. 129
Figure legend 131
Fig. 1. (A). Sorted WT pro-B cells were cultured for 6 days in the IL7 medium. At days 0, 4 and 6, 132
total RNAs were extracted, reverse transcribed and subjected to qPCR for the indicated switch 133
transcripts. The histograms show the standard deviation. (B). WT B220+ cells were sorted and cultured
134
for 6 days in the IL7 medium. At days 4 and 6, cells were stained with CD19, CD43, and anti-135
IgM, and gated on IgM- population. (C). Genomic DNAs were prepared at day 6 and were assayed for
136
D-JH4 rearrangements, and for VH-DJH4 rearrangements. (D-F). WT (D), IL7R-/- (E), and RAG2-/- (F)
137
B220+ cells were sorted and cultured in the IL7 medium. At the indicated time points, total RNAs were
138
extracted and assayed for switch transcripts as in (A). (G-J). Sorted WT pro-B (G) or sorted B220+
139
cells from WT (H), IL7R-/- (I), and RAG2-/- (J) BMs were cultured for 4 days in the IL7 medium,
140
then in the presence (LPS) or not (UNS) of LPS for additional 2 days, except for IL7R-/- B220+ cells
141
(6h and 16 hours in IL7+LPS medium). Switch transcripts were quantified as in (A). (n≥4 for A and 142
G, n≥8 for B-F, n>8 for H, n=4 for I, and n>4 for J)(****, p<0.0001, ***, p<0.001; **, p<0.01; *,
143
p<0.05; ns, not significant).
Fig. 1 Sg3 IL7 0-6-16h mo FIG3 IL7R _J0 IL7R -6h NS IL7R -16h NS x x x x x x x 0.00 0.02 0.04 0.06 ns ns ns % act in t ranscr ipt s Sg2b Sg3 Sg2b IL7 0-6-16h mo FIG3 IL7R _J0 IL7R -6h NS IL7R -16h NS x x x x x x x 0.00 0.02 0.04 0.06 0.08 0.10 ns ns ns 0h 6h 16h Sµ IL7R 0 à 16h NS Re la ti v e l e v e l to a ct in IL7R _J0 IL7R -6h NS IL7R -16h NS x x x x x x x 0.0 0.5 1.0 1.5 2.0 ns ns ns Sµ 0h 6h 16h 0h 6h 16h
IL7Ra-/-B220+cells RAG2-/-B220+cells
% act in t ranscr ipt s Sg3 RAG d0-d4-d6 mo FIG3 RA G J0 RA G J4 +IL7 RA G J6 +IL7 x x x x x x x 0.0000 0.0002 0.0004 0.0006 ns ns ns Sg2b RAG d0-d4-d6 mo FIG3 J0 J4 J6N S x x x x x x x 0.0 0.1 0.2 0.3 ns ns ns Sg2b Sg3 d0 d4 d6 d0 d4 d6
WT pro-B cellsSg3 WT triés d6NS-LPS+IL7 FIG4
WT J6 NS T ri WT J6 LPS +IL7 Tri x x x x x x x x 0.0 0.1 0.2 0.3 0.4 ****
Sg2b WT triés d6NS-LPS+IL7 FIG4
WT J6 NS Tri WT J6 LPS +IL7 Tri x x x x x x x x 0 1 2 3 * Sg2b Sµ Sg3 % act in t ranscr ipt s
Sµ WT triés d6NS-LPS+IL7 FIG4A
WT J6 NS T ri WT J6 LPS +IL7 Tri x x x x x x x x 0 4 8 12 ns UNS LPS UNS LPS UNS LPS WT B220+cells % act in t ranscr ipt s Sg2b Sµ Sg3 Sµ WT mo d6NS-LPS+IL7 FIG4 WT J6 NS mo WT J6 LPS +IL7 mo x x x x x x x x 0 4 8 12 ns Sg3 WT mo d6NS-LPS+IL7 FIG4 WT J6 NS mo WT J6 LPS +IL7 mo x x x x x x x x 0.00 0.04 0.08 0.12 **** Sg2b WT mo d6NS-LPS+IL7 FIG4 WT J6 NS mo WT J6 LPS +IL7 mo x x x x x x x x 0.0 0.2 0.4 0.6 0.8 1.0 * UNS LPS UNS LPS UNS LPS RAG2-/-B220+cells
Sg3 RAG mo d6NS-LPS+IL7 FIG5
RA G J6 +IL7 RA G J6 LP S+I L7 x x x x x x x x 0.0000 0.0005 0.0010 0.0015 ***
Sg2b RAG mo d6NS-LPS+IL7 FIG5
RAG J6+I L7 RAG J6 L PS+I L7 x x x x x x x x 0.0 0.2 0.4 0.6 0.8 * Sg3 Sg2b % act in t ranscr ipt s UNS LPS UNS LPS IL7Ra-/-B220+cells % act in t ranscr ipt s
Sg3 IL7R mo 0-6-16hNS-LPS+IL7 FIG5
IL7R _J0 IL7R 6h LPS +IL7 IL7R -16h LP S+I L7 x x x x x x x 0.0 0.1 0.2 0.3 ns *** *** Sg3 Sg2b
Sg2b IL7R mo 0-6-16hNS-LPS+IL7 FIG5
IL7R _J0 IL7R -6h LPS +IL7 IL7R -16h LP S+I L7 x x x x x x x 0.0 0.2 0.4 0.6 ** * ** WT B220+cells Sµ d0-d4-d6 WT mo FIG2 mo d0 mo d4 mo d6 x x x x x x x 0 5 10 15 ns ns ns Sµ Sg3 Sg2b d0-d4-d6 WT mo FIG2 mo d0 mo d4 mo d6 x x x x x x x 0.0 0.4 0.8 1.2 **** ** **** Sg3 d0-d4-d6 WT mo FIG2 mo d0 mo d4 mo d6 x x x x x x x 0.000 0.013 0.026 0.039 0.052 0.065 **** *** **** Sg2b d0 d4 d6 % act in t ranscr ipt s d0 d4 d6 d0 d4 d6 B 6.1% 94.1% 1.3% 98.8% C D 19 CD43 d4 d6 Pro-B Pro-B Pre-B Pre-B WT B220+cells WT pro-B cells % act in t ranscr ipt s Sµ Sg3 Sg2b Sµ d0-d4-d6 WT triés FIG2C trié s d0 trié s d4 trié s d6 x x x x x x x 0 5 10 15 ns ns ns Sg3 d0-d4-d6 WT triés FIG2C trié s d0 trié s d4 trié s d6 x x x x x x x 0.0000 0.0034 0.0068 0.0102 0.0136 0.0170 ** * **** Sg2b d0-d4-d6 WT triés FIG2C trié s d0 trié s d4 trié s d6 x x x x x x x 0 1 2 3 ** ** **** d0 d4 d6 d0 d4 d6 d0 d4 d6 A DQ52-JH4 WT-RAG DQ52-JH4 FIG1 RA G WT x x x x x x x x 0.0 0.1 0.2 0.3 ** WT-RAG DSP-JH4 RA G WT x x x x x x x x 0 2 4 6 8 **** % H S5 DSP-JH4 WT-RAG 558-JH4 RA G WT x x x x x x x x 0.0 0.1 0.2 0.3 0.4 0.5 **** dVH-DJH4 % H S5 WT-RAG 7183-JH4 RA G WT x x x x x x x x 0.0 0.4 0.8 1.2 *** pVH-DJH4 WT B220+cells D C F E H G J I