ORIGINAL ARTICLE
A novel clade of sporocarp-forming species
of glomeromycotan fungi in the
Diversisporales lineage
Dirk Redecker&Philipp Raab&Fritz Oehl&
Francisco J. Camacho&Regis Courtecuisse
Received: 13 October 2006 / Revised: 12 January 2007 / Accepted: 12 January 2007 / Published online: 14 February 2007 # German Mycological Society and Springer-Verlag 2007
Abstract In the early times of taxonomy of arbuscular mycorrhizal fungi (Glomeromycota), exclusively sporo-carpic species were described. Since then the focus has mainly shifted to species forming spores singly. For many of the sporocarpic species, no molecular data have been made available, and their phylogenetic position has remained unclear. We obtained small subunit ribosomal rDNA and internal transcribed spacer data from specimens of glomeromycotan sporocarps from tropical areas that were assigned to three morphospecies. The complete sequence of the 18S small rDNA subunit sequence, internal transcribed spacers (ITS) 1 and 2 and 5.8S rDNA subunit, was determined from a sporocarp of Glomus fulvum. Partial sequences of the small subunit and the other regions were obtained from Glomus pulvinatum and the newly described species Glomus megalocarpum. Molecular phylogenetic analyses placed all species analyzed as a monophyletic sister group to the Diversispora spurca/Glomus versiforme clade group (“Glomus group C”) within the
Diversis-porales. The phylogenetic divergence from other known species suggests that this clade may constitute a new genus. These findings will have important consequences for taxon definition in the Diversisporales. They will facilitate identification of these fungi using rDNA sequences within colonized roots or the environment.
Introduction
Arbuscular mycorrhiza is a symbiosis formed by the vast majority of land plants. In this interaction, the plant partner provides carbohydrates to the fungal symbiont, which improves the uptake of mineral nutrients using its fine hyphal network. Approximately 200 morphospecies of arbuscular mycorrhizal fungi in ten genera have been described, mostly based on spore morphology. These fungi were previously placed in the family Endogonaceae (Gerdemann and Trappe1974), later in the order Glomales of the Zygomycota (Morton and Benny1990). Recently, a separate phylum Glomeromycota was established for them (Schüßler et al. 2001). As the spore morphology is rather simple in many species, molecular phylogenetic methods have helped to clarify the evolutionary relationships in the Glomeromycota (Redecker and Raab 2006). One result of these analyses was that the largest genus Glomus, with about 70 species, is polyphyletic (Redecker et al. 2000a; Schwarzott et al. 2001). Fungi falling within the previous morphological definition of Glomus have been shown to be in at least four different monophyletic lineages. In fact, some dimorphic species even form glomoid spores as well as other types of spores (Redecker et al. 2000a). Some of these lineages were separated in the new genera (Morton
DOI 10.1007/s11557-007-0524-2
Taxonomic novelties: Glomus megalocarpum D. Redecker D. Redecker (*)
:
P. Raab:
F. OehlBotanical Institute, University of Basel, Hebelstrasse 1,
Basel 4056, Switzerland e-mail: dirk.redecker@unibas.ch F. J. Camacho
Department of Environmental Science, University of California, Riverside, CA 92521, USA
R. Courtecuisse
Faculté des sciences pharmaceutiques et biologiques, Département de Botanique,
B.P. 83,
and Redecker2001), partly based on molecular characters. One remaining clade of Glomus comprises Glomus group A (e.g. G. intraradices, G. mosseae) and group B (e.g. G. etunicatum, G. claroideum), which are rather distantly related but still form a monophyletic group together (Schwarzott et al.2001).
Another Glomus clade, referred to as Glomus group C, is more closely related to the Acaulosporaceae in rDNA phylogenies. It comprises the species G. versiforme, G. spurcum, and G. aurantium, as well as an isolate fitting the morphological definition of G. etunicatum, which is otherwise thought to belong to group B. Glomus group C was recently defined as a new family Diversisporaceae with Diversispora spurca as type species of the new genus (Walker and Schüßler2004). Other species have not been formally renamed. Because of the scarcity of morphological characteristics on the light microscopic level that could aid separation from other Glomus species, signature sequences in the 18S ribosomal subunit were used for the definition of Diversispora as well as the corresponding family and order. Numerous Glomus species form sporocarps, which mostly are relatively undifferentiated assemblages of spores. An exception are some of the species previously placed in the genus Sclerocystis. However, it was shown that Sclerocystis is an advanced clade within a group of well-defined nonsporocarpic Glomus group A species (Redecker et al. 2000b). Generally, the sporocarp-forming habit is not well correlated with phylogeny, and sporocarpic as well as nonsporocarpic representatives can be found in different phylogenetic groups. Very often, sporocarp for-mers are closely related to species forming spores singly in the soil, or a single species is known to form sporocarpic and nonsporocarpic spores (e.g. G. mosseae, G. corona-tum). Early taxonomy of the Glomales focused very much on sporocarpic species, a trend that has not continued recently. The phylogenetic relationships of many of the sporocarpic species listed in the groundbreaking study of Gerdemann and Trappe (1974) have not been elucidated. Therefore, it is impossible to recognize these taxa if they are detected by molecular methods within colonized roots. In this study, we sequenced regions of the ribosomal rDNA of some sporocarpic glomeromycotan species col-lected in tropical regions to elucidate their phylogenetic relationships and to allow the design of specific primers for molecular detection of these species.
Materials and methods
Specimen AC/Pohn99-001 was collected by Francisco J. Camacho in 1999 in a native forest at the ridge trail near the top of Sokehs Mountain on the pacific island of Pohnpei, Federated States of Micronesia (6.97°N 158.22°E). Other
sporocarps were collected on the Caribbean islands of Guadeloupe and Martinique in August/September 2005 by Christophe Lécuru. Details were as follows: specimen CL/ Mart05-35: Ste. Luce, Forêt départementalo-domaniale de Montravail, on forest floor; specimen CL/Mart05-049: Case pilote, Crête Jean Louis (Martinique), fruiting on dead wood; specimen CL/Guad05-51: Vieux-Fort (Guadeloupe), Ravine Blondeau, mesophile forest, on forest floor, close to Boletellus sp.; specimen CL/Mart05-111: Forêt départa-mentalo-domaniale de La Source Berry (Martinique), plantation of mahogany with indigenous species.
Slides to examine the spore morphology were obtained by scratching a few spores from the surface of cross-sections and embedding them into Polyvinyl alcohol lactoglycerol (PVLG, Koske and Tessier 1983). Pictures of the whole sporocarps or parts of them were taken using a Hewlett Packard ScanJet 5470 flatbed scanner at maximum resolution (2,400 dpi). Microscopical pictures of the spores were taken with an Olympus DP70-CU CCD camera attached to a Zeiss Axioplan compound microscope equipped with differential interference contrast optics. For size mea-surements, 20 spores of each specimen were used, unless indicated otherwise. Colors were described as a percentage of CMYK values using the INVAM color chart (http://invam.
caf.wvu.edu/otherinfo/articles/colorchart.htm).
DNA was extracted from 10 mg subsamples using a Qiagen DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Polymerase chain reaction (PCR) was conducted in two steps as a nested PCR using various primer combinations. For amplifying the complete 18S small subunit of specimen 1, universal primers were used (White et al. 1990). Preliminary experiments with universal primers had shown that the specimens belonged to the Diversisporales clade. Therefore, a primer was designed for specific amplification of the 3′ end of the small subunit and the ITS of this lineage (GLOC1355: TCTTAGAGGGACTATTGGCATTT). It was used in the second step of the nested PCR together with ITS4i (Redecker et al.2003), as second primer. In the first PCR step primers NS5/ITS4 (White et al.1990) were used. Cycling parameters were as described by Hijri et al. (2006). PCR products were ligated into pGEM-T (Promega, Wallisellen, Switzerland) and used to transform competent Escherichia coli DH5alpha. Clones were screened by blue/ white screening and colony PCR and sequenced in both directions using a Applied Biosystems Prism Dye Termi-nator 3.1 kit on an ABI 310 capillary sequencer (Applied Biosystems, Foster City, USA).
Sequence Navigator (Applied Biosystems) and Bio Edit (Hall 1999) were used to proofread and assemble the sequences. The near-complete sequence of the small subunit was assembled from sequenced fragments using variable regions for verifying that the fragments were from the same organism. To exclude chimaera formation, the
fragments were subjected separately to phylogenetic anal-ysis in PAUP* 4 (Swofford2001). Moreover, the sequences were tested using Chimaera Check (Cole et al. 2003) and Bellerophon (Huber et al.2004).
The small subunit sequence was included into a dataset comprising 39 selected taxa from the Glomeromycota with special focus on the Diversisporales. The alignment was done manually, and some regions were excluded from analysis because they could not be aligned unambiguously, resulting in 1,541 positions that were eventually analyzed. Phylogenetic trees were obtained under distance and maximum likelihood criteria using PAUP* 4. The neighbor joining and heuristic search algorithms were used, respec-tively. A gamma shape parameter of 0.5 was assumed for neighbor joining. Bayesian analyses were performed using MrBayes 3.1.1. (Ronquist and Huelsenbeck 2003) for Macintosh. Four chains were run over 9.2×106generations under a general time-reversible model. Every 500th tree was sampled, and the first 10% of trees were discarded as a “burnin” before constructing a 50% consensus tree.
Partial small subunit sequences from the 3′ end of the gene from more specimens were included into another dataset, comprising 31 taxa and 314 characters, containing sequences from spores of representative species of all glomeromycotan clades, as well as numerous partial 18S subunit sequences obtained from colonized roots. For Bayesian analysis, 5×106 generations were run, and the trees were processed as described above. For maximum likelihood analysis, the GTR+G+I model was used as determined using Modeltest 3.5 (Posada2004).
ITS1/5.8S subunit/ITS2 sequences were placed in a dataset containing all available sequences from the Diver-sispora/Glomus group C clade. Some sequences from the public databases labeled G. intraradices were excluded because of probable misidentification. The 5.8S and ITS2 regions were used for phylogenetic analysis. The dataset comprised 22 taxa, and a total of 352 characters were used for the analysis. For Bayesian analysis, 5×106generations were run, and the trees were processed as described above. The GTR+I model was used for maximum likelihood analyses.
DNA sequences were submitted to the European Molec-ular Biology Laboratory database under the accession numbers provided in Table1. The alignments were deposited in TreeBase with the accession numbers M3072 and M3073.
Results
Morphological analyses
Specimen AC/Pohn99-001 from Micronesia (Fig.1) had a size of 7×7×4 mm. The peridium was smooth and had a
tan to yellowish-brown color (20/60/100/10 on the INVAM color chart). One flat side of the sporocarp was not entirely covered by the peridium (“umbilicate sporocarp”). Spores were not embedded into hyphae. The spore mass was yellowish-brown (20/40/80/0).
Spores were oval to oblong to slightly claviform with a size of 80–138×63–85 μm (average 126×81 μm). The spore content was yellowish-brown and had a dense, fibrous appearance. The spore wall was 2.5 to 5 μm thick and layered, the sublayers were not separating under pressure. The hyphal attachment was 7.5 to 10 μm broad, as well as the hyphae themselves. Spore contents were occluded by a prominent, bulging septum, apparently originating from the innermost wall layer(s) (Fig. 2d). These morphological features place the sporocarp into the species Glomus fulvum (Berk. & Broome) Trappe and Gerd.
Two specimens from the Caribbean (CL/Mart05-049 and CL/Mart05-111; Fig. 2a and b) were morphologically similar and also fit the description of G. fulvum. The sporocarps had diameters of 7 and 10 mm, respectively. However, their spores were generally shorter than in the Micronesian specimen (dimensions, 65–95×75–95 μm and 80–95×65–75 μm, respectively). Two layers of the spore wall could be clearly distinguished in some spores. The spore wall was 2.5–4 μm thick, the layers were 1.2 to 2.4μm wide.
The remaining two Caribbean specimens (CL/Mart05 05–35 and CL/Guad05–51) were less strongly pig-mented. Sporocarp CL/Mart05 05–35 (Fig. 2c) had a diameter of 5 mm and fit the description of Glomus pulvinatum (P. Hennigs) Trappe and Gerd. The spore mass had a color of pale tan to ochre (0/20/40/0). Its spores had a diameter of 60–85 μm differing from G. fulvum by their globose to subglobose shape. The hyphal attachment was equally broad (8–10 μm) as in G. fulvum. The spore wall was 3 to 5 μm thick with no apparent subdivision into layers.
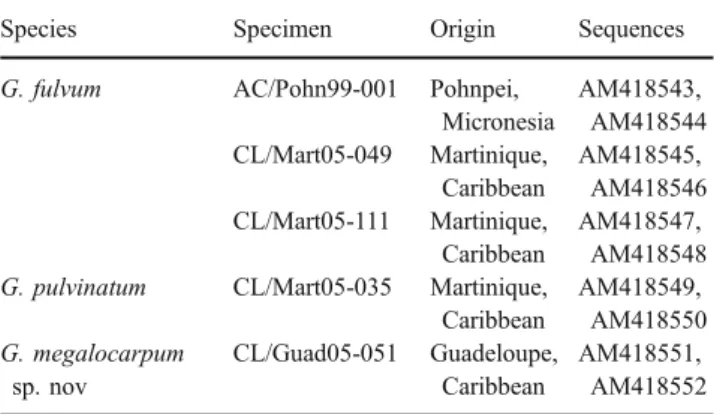
Table 1 Specimens studied and sequences obtained from them Species Specimen Origin Sequences G. fulvum AC/Pohn99-001 Pohnpei,
Micronesia AM418543, AM418544 CL/Mart05-049 Martinique, Caribbean AM418545, AM418546 CL/Mart05-111 Martinique, Caribbean AM418547, AM418548 G. pulvinatum CL/Mart05-035 Martinique,
Caribbean AM418549, AM418550 G. megalocarpum sp. nov CL/Guad05-051 Guadeloupe, Caribbean AM418551, AM418552
Specimen CL/Guad05-51 was a sporocarp considerably larger than the others (Fig.3). It could not be assigned to a known morphospecies and is therefore described as Glomus megalocarpum sp. nov. None of the specimens mentioned showed a reaction with Melzer’s reagent in the spores or attached hyphae.
Taxonomy
Glomus megalocarpum D. Redecker sp. nov.
Sporocarp firm, irregularly lobate, length approximately 3.8×2 cm, and with a brownish rind, whitish-velvety on the
Fig. 2 Spores of the sporocarp specimens from the Caribbean. a G. fulvum, specimen CL/ Mart05-049, b G. fulvum, spec-imen CL/Mart05-111, c G. pulvinatum, specimen CL/ Mart05-035, d G. megalocar-pum sp. nov. CL/Guad05-51 Fig. 1 Morphology of G. ful-vum AC/Pohn99-001 from Micronesia. a Two halves of the sporocarp, the top left part shows the cutting plane, the bottom right the exterior surface. b Separated spores scraped from the sporocarp, showing the var-iation in spore shape. Note the dense spore contents. c Spore showing the hyphal attachment with the septum. d Close-up of the septum showing the wall layers
outside. No smell perceptible. Composite spore mass in the interior pale tan to ochre (0/20/30/10 on the INVAM color chart). Spores obovate to pyriform or slightly irregular with dimensions of (56)65–95(115)×38–57(62) μm (arithmetic mean, 81 × 47 μm; n=40). Spore wall single-layered, hyaline, and 1.7–4 μm thick. Hyphal attachments usually not persistent. Spores occluded by a prominent septum, 7–8 μm broad, continuous with an inner lamina of the spore wall, separating a triangular cavity from the remain-der of the spore lumen. Sporogenic hyphae thin-walled, usually not remaining attached. Gleba indistinct, spores tightly packed in irregular arrangement. Peridial hyphae rust-colored at higher magnification. Distinct by character-istic ribosomal DNA sequences: e.g., TTGAAGTTTGG GAGCGG in the 18S small subunit.
Sporocarpium firmum, irregulare, lobatum, 38×20 mm, interiore pallide fulva vel ochracea, peridio brunneo, superficie tomentosa, inolens. Sporae obovatae, piriformes, (56)65–95(115)×38–57(62) μm, tunicis hyalinis, 1.7– 4 μm crassis, strato singulare. Hyphae sporophorae ephem-erae, tunicis tenuibus. Gleba indistincta, sporis ordinatione densa et irregulare. Sporae septo occlusae, cavitatem
triangularem 7–8 μm latam segregante. Hyphae peridii ferrugineae. Sequentia typica acidi desoxyribonucleici ribosomatum: TTGAAGTTTGGGAGCGG.
This species differs from other sporocarpic Glomus species by a number of characters, among them the large sporocarp size. G. fulvum and G. pulvinatum are distinct by the shape and color of their spores, as well as their rDNA sequences (see below). G. canadense (Thaxt.) Trappe and Gerd. was reported to have hyaline to pale yellowish spores with an oval to subglobose shape (Thaxter1922), whereas
Fig. 3 Morphology of G. megalocarpum sp. nov. a Sporocarp CL/Guad05-51 in the field: approximate size along its long axis, 38 mm. b Spores in PVLG, note the triangular cavity separated by the septum (asterisk). c Dried slices of the sporocarp. d Spores in PVLG, showing variation in shape. Scale bars 50μm in b, 1 cm in c, and 100μm in d
Distribution and habitat
Known only from type collection Mycorrhizal
associations
Unknown
Etymology Referring to the unusually large sporocarp Collection
examined
Guadeloupe, Vieux-Fort, Ravine Blondeau, mesophile forest, on forest floor, leg. C. Lécuru, 5.9. 2005. Holotype CL/Guad05-051 (LIP).
the spores of G. megalocarpum consistently have a higher length/width ratio. G. fragile (Berk. & Broome) Trappe and Gerd. also differs in spore shape and size and was reported to have a two-layered wall. G. flavisporum (M. Lange & Lund) Trappe and Gerd. has larger spores with thick, brown walls. G. boreale (Thaxt.) Trappe and Gerd. has red-brown spores (Gerdemann and Trappe 1974). Sporocarps of G. cuneatum McGee & A. Cooper contain dark-colored spores and differ in their unique sporocarp anatomy (McGee and Trappe2002).
Molecular phylogeny
A sequence comprising the complete 18S small subunit, ITS1, 5.8S subunit, and ITS2 regions was obtained from specimen AC/Pohn99-001 (G. fulvum, Micronesia). From the other specimens, a region of 315 base pairs at the 3′ end of the small subunit, as well as ITS1, 5.8S subunit, and ITS2 were sequenced. Two clones spanning these regions were analyzed from each specimen to estimate the amount of intraspecific diversity.
AJ276088 Glomus versiforme X86687 Glomus versiforme
Y17650 Diversispora spurca Z14009 Gigaspora albida Z14010 Gigaspora gigantea
AJ242729 Scutellospora projecturata AJ276092 Scutellospora aurigloba U31997 Scutellospora castanea
AJ306444 Scutellospora weresubiae AB041344 Scutellospora cerradensis Z14012 Scutellospora pellucida
AJ276094 Scutellospora gilmorei Z14005 Acaulospora rugosa
AJ306439 Acaulospora longula -/60
Y17633 Acaulospora laevis
Z14006 Entrophospora colombiana Z14004 Acaulospora spinosa
AJ619955 Pacispora scintillans
X58725 Glomus intraradices Z14007 Glomus mosseae
AJ006800 Archaeospora trappei
AJ006766 Paraglomus occultum 0.1 72/81 57/75 81/76 67/97 88/75 74/100 99/100 98/96 73/88 86/100 84/90 -/52 91/96 -/72 -/53 99/100 -/96 -/71 61/83 80/100 94/99
AM418547 Glomus fulvum CL/Mart05-111
AM418548 Glomus fulvum CL/Mart05-111 AM418545 Glomus fulvum CL/Mart05-049
AM418546 Glomus fulvum CL/Mart05-049
AM418544 Glomus fulvum AC/Pohn99-001 AM418549 Glomus pulvinatum CL/Mart05-035
AM418550 Glomus pulvinatum CL/Mart05-035 AM418551 Glomus megalocarpum CL/Guad05-051
AM418552 Glomus megalocarpum CL/Guad05-051 Fig. 4 Phylogenetic tree of the
Diversisporales and outgroups based on Bayesian analysis of partial sequences (314 base pairs) of 18S small ribosomal subunit sequences. The tree was rooted with Archaeospora/ Paraglomus. The first/second numbers on the nodes denote neighbor-joining bootstrap val-ues from 1,000 replications or posterior probabilities from Bayesian analysis, respectively
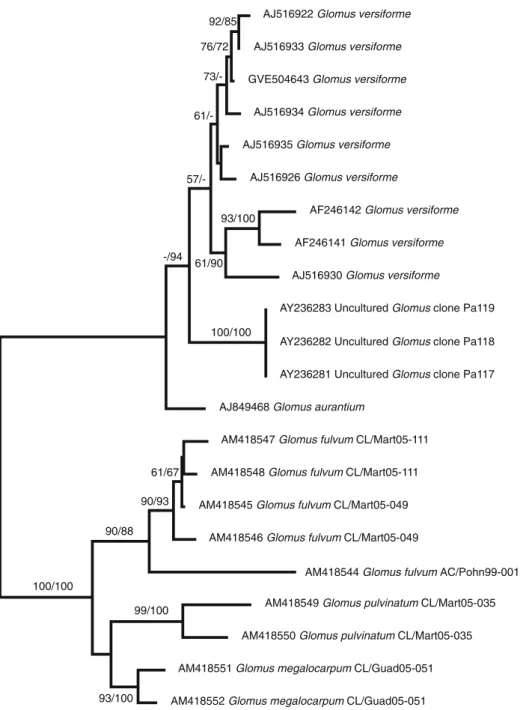
Figure4shows a phylogenetic tree obtained by Bayesian analysis of partial 18S subunit sequences. The sequences from all sporocarp specimens form a monophyletic sister group to D. spurca/G. versiforme within the Diversis-porales. The common clade as well as the sister group relationship received medium to high bootstrap support. The topology of the tree concerning these clades was verified using maximum likelihood analyses. In the analyses using the full-length small subunit sequence of G. fulvum AC/Pohn99-001 (results not shown), higher support (97% bootstrap support in distance analysis and 100% posterior probability in Bayesian analysis) could be
obtained for the clade uniting G. fulvum and D. spurca/ G. versiforme.
In the ITS tree (Fig. 5), the sporocarp specimens are clearly separated from the Diversispora clade, which is represented here by G. versiforme and G. aurantium. ITS sequences of D. spurca are not available in the databases. In some regions, the ITS are difficult to align between the two major clades in the tree. The branching order among the sporocarp specimens is almost identical to the small subunit tree. The different clones obtained from specimens G. fulvum CL/Mart05-049 and CL/Mart05-111 are extremely similar (1 to 6 positions different out of 314 in the 18S subunit
90/88 90/93 99/100 93/100 57/- 61/-100/100 73/-76/72 92/85 61/90 93/100
AJ516922 Glomus versiforme
AJ516933 Glomus versiforme
GVE504643 Glomus versiforme
AJ516934 Glomus versiforme
AJ516935 Glomus versiforme
AJ516926 Glomus versiforme
AF246142 Glomus versiforme
AF246141 Glomus versiforme
AJ516930 Glomus versiforme
AY236283 Uncultured Glomus clone Pa119
AY236282 Uncultured Glomus clone Pa118
AY236281 Uncultured Glomus clone Pa117
AJ849468 Glomus aurantium
AM418547 Glomus fulvum CL/Mart05-111
AM418548 Glomus fulvum CL/Mart05-111
AM418545 Glomus fulvum CL/Mart05-049
AM418546 Glomus fulvum CL/Mart05-049
AM418544 Glomus fulvum AC/Pohn99-001
AM418549 Glomus pulvinatum CL/Mart05-035
AM418550 Glomus pulvinatum CL/Mart05-035
AM418551 Glomus megalocarpum CL/Guad05-051
AM418552 Glomus megalocarpum CL/Guad05-051 0.01 substitutions/site 61/67 100/100 -/94
Diversispora/ Glomus
group C
Glomus fulvum
group
Fig. 5 Phylogenetic tree of the Glomeromycota based on dis-tance analysis of 5.8S rDNA and ITS2 sequences (351 base pairs) by neighbor joining. The tree was rooted by midpoint rooting. The first/second num-bers on the nodes denote neigh-bor-joining bootstrap values from 1,000 replications or pos-terior probabilities from Bayes-ian analysis, respectively
and 1 to 9 positions out of 351 in 5.8S subunit and ITS2) and cannot be separated conclusively. Their intermingled grouping in the tree proves that they belong to the same species. The sequence from G. fulvum AC/Pohn99-001 is closely associated with the latter two sequences, but shows some sequence differences. G. megalocarpum CL/Guad05-51 and G. pulvinatum CL/Mart05-35 are clearly separated from the G. fulvum sequences and from each other.
No root colonization could be detected by Trypan Blue staining 6 months after the inoculation of Plantago media with G. fulvum. Therefore, its mycorrhizal status remains to be elucidated.
Discussion
The morphological differences among the sporocarps are in concordance with the rDNA phylogenetic trees. The G. fulvum isolates have spores similar in shape, size, and color, whereas the other two can be easily distinguished.
G. fulvum belongs to a group of sporocarpic species of Glomus that were described at a time when taxonomy of that genus was very much focused on sporocarp formers (Thaxter 1922; Gerdemann and Trappe 1974). G. fulvum was originally described as Paurocotylis fulva by Berkeley and Broome (1873), and transferred to Glomus by Gerdemann and Trappe (1974). Thaxter (1922) and Gerdemann and Trappe (1974) also listed several similar, putatively related species (G. canadense, G. pulvinatum). In particular, Thaxter (1922) highlighted the septum as a distinctive characteristic of G. fulvum and relatives.
Interestingly, G. fulvum was reported to be common in tropical areas and to have a wide distribution (Sri Lanka,
Caribbean, Brazil, South Pacific; Gerdemann and Trappe
1974). The present specimens from Micronesia and the Caribbean fit well into that context. G. pulvinatum was reported from tropical America, Tasmania, and New Zealand (Gerdemann and Trappe 1974). Some other morphologically related species mentioned by these authors (G. boreale, G. canadense, G, flavisporum) were reported from temperate/boreal habitats.
In rDNA-based molecular phylogenies, Diversispora/ Glomus group C forms a clade well separated from other Glomus groups (Schwarzott et al.2001). However, it is not united by any obvious morphological characteristics on the light microscopic level that could aid distinguishing it from other Glomus species. Walker and Schüßler (2004) reported that D. spurca can be differentiated from Glomus species by a flexible inner wall. However, this is not undisputed (Kennedy et al. 1999), and there is no clear evidence that the other species in the Diversisporaceae share this putative morphological feature. No clear evidence for such an inner flexible wall could be found in the sporocarpic species analyzed here, even in spores with cracked walls.
All species in the G. fulvum group share the ability to form relatively large, compact sporocarps. The sporocarp of G. megalocarpum may be one of the largest glomeromy-cotan sporocarps reported so far. The spores of fungi of the G. fulvum clade show a prominent septum that closes a broad hyphal attachment. A collar-like remnant of the spore wall often protrudes beyond the septum. The spore content is often extremely dense, sometimes appearing stringy.
The prominent septum that occludes the mature spore appears as a potential uniting morphological feature between the Diversispora and G. fulvum clades. G. versiforme also has a rather prominent septa that look
Table 2 Signature sequences defined by Walker and Schüßler (2004), and their occurrence in the partial 18S subunit sequences presented in this study
similar to the ones in G. fulvum. In G. spurcum, it is apparently more difficult to see, in contrast to a putative relative G. eburneum (Kennedy et al. 1999). However, many other Glomus groups also feature a septum between hypha and spore. In many cases, it is not as obvious as in the G. fulvum group but the broad hyphal attachment of the latter may contribute to the visibility of the septum.
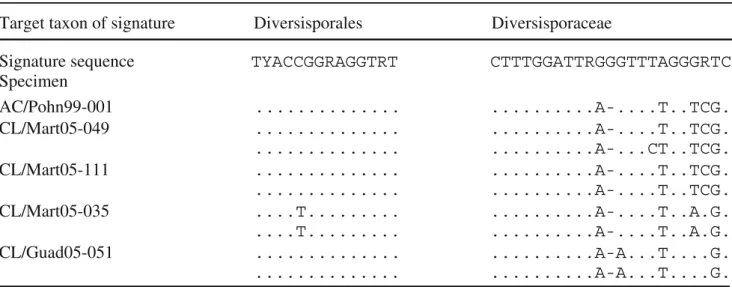
The sequence signatures used by Walker and Schüßler (2004) to define Diversispora and the corresponding higher taxa only partly match the new sequences presented here. In the 18S subunit sequence of AC/Pohn99-001, the first signature sequence GGTTTH characteristic for the order Diversisporales is present, as well as the second (TYACCG GRAGGTRT). However, the sequence signatures for the family Diversisporaceae show numerous changes (three changes out of 16 positions in the first signature, 5/20 in the second, and 1/14 in the third). The same is true for the genus-specific positions (4/23, 1/16, 6/23). From the two signatures located in the gene region sequenced from all specimens in this study, the order-specific signature is relatively well conserved in the taxa presented in this study, in contrast to the family-specific signature (Table2).
These findings indicate that either these taxa have to be revised to comprise the sporocarpic species analyzed here or a new taxa have to be established. The G. fulvum group is phylogenetically clearly distinct from Diversispora/ Glomus group C, and it shows some uniting morphological features. Therefore, it may be preferable to erect a separate genus rather than include it into Diversispora, which has been highly problematic to define morphologically. How-ever, we will await the analysis of more potential members of the clade before taking formal action and, if possible, additional gene sequences.
Mycorrhizal associations of G. fulvum are unknown, and it was reported to fruit on leaves and rotten wood (Gerdemann and Trappe 1974). We did not succeed in establishing it in mycorrhizal symbiosis but of course this does not prove that the fungus is not able to form mycorrhizas. It should be noted that ectomycorrhizal fungi fruiting on pieces of wood were mistaken for saprophytes for a long time until they were detected in the symbiosis by molecular methods (Gardes and Bruns1996).
In the past few years, a large database of environmental glomeromycotan rDNA sequences that originate from molecular field studies of colonized roots has accumulated. Although the G. fulvum group should be detected by the primer pair NS31/AM1, which was used in a number of studies (Helgason et al. 1998), no close matches to G. fulvum etc. could be detected among those sequences by Basic Local Alignment Search Tool searches or phyloge-netic analyses. Because of the sampling locations of the respective studies, most database sequences were found in temperate regions, but it should be noted that Glo36, the
closest match to the G. fulvum group detected in an extensive study of a tropical forest in Costa Rica (Husband et al.2002), is much closer to G. versiforme/spurcum than to G. fulvum. Therefore, the nutritional mode of G. fulvum and relatives remains to be demonstrated. We used the new sequence data to design the primer GLOC1355, which will be refined to obtain a primer that can be applied to identify members of this clade within colonized roots. More environmental studies, in particular from the tropical areas where the sporocarps were found, are necessary to under-stand the ecological role of this glomeromycotan clade.
We demonstrated that a fascinating and spectacular group of species of glomeromycotan fungi that have been known for a long time constitutes a major, previously unknown monophyletic clade. These findings reiterate that glomeromycotan diversity is still far from being exhaus-tively characterized on the molecular level.
Acknowledgments The authors would like to thank Christophe Lécuru and Pierre-Arthur Moreau for collecting and providing sporocarps. The Swiss National Science Foundation (SNF) is gratefully acknowledged for funding through grant no. 3100-066688 to Redecker. The specimens from the Caribbean were collected during the course of the research program “Inventaire mycologique des Petites Antilles: biodiversité, écologie et protection” granted to Courtecuisse. Oehl would like to thank Jim Trappe for providing specimens of sporocarps from his collection. The continuing support by Andres Wiemken and Thomas Boller at the Botanical Institute, University of Basel is gratefully acknowledged.
References
Berkeley M, Broome C (1873) Enumeration of the fungi of Ceylon. Part II. J Linn Soc London Bot 14:137
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM (2003) The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442–443
Gardes M, Bruns TD (1996) Community structure of ectomycorrhizal fungi in a Pinus muricata forest: above- and below-ground views. Can J Bot 74:1572–1583
Gerdemann JW, Trappe JM (1974) Endogonaceae in the Pacific Northwest. Mycol Mem 5:1–76
Hall TA (1999) Bioedit: user-friendly biological sequence alignment and analysis program for Windows 95/98/NT. North Carolina State University, USA
Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW (1998) Ploughing up the wood-wide web? Nature 394:431
Hijri I, Sykorova Z, Oehl F, Ineichen K, Mäder P, Wiemken A, Redecker D (2006) Communities of arbuscular mycorrhizal fungi in arable soils are not necessarily low in diversity. Mol Ecol 15:2277–2289 Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to
detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Husband R, Herre EA, Turner SL, Gallery R, Young JPW (2002) Molecular diversity of arbuscular mycorrhizal fungi and patterns of host association over time and space in a tropical forest. Mol Ecol 11:2669–2678
Kennedy LJ, Stutz JC, Morton JB (1999) Glomus eburneum and G. luteum, two new species of arbuscular mycorrhizal fungi, with emendation of G. spurcum. Mycologia 91:1083–1093
Koske RE, Tessier B (1983) A convenient, permanent slide mounting medium. Newslett Mycol Soc Am 34:59
McGee P, Trappe JM (2002) The Australian zygomycetous mycorrhizal fungi. II. Further Australian sporocarpic Glomaceae. Aust Syst Bot 15:115–124
Morton JB, Benny GL (1990) Revised classification of arbuscular mycorrhizal fungi (Zygomycetes): a new order Glomales and Gigasporineae and two new families Acaulosporaceae and Gigasporaceae with an emendation of Glomaceae. Mycotaxon 37:471–491
Morton JB, Redecker D (2001) Two new families of Glomales, Archaeosporaceae and Paraglomaceae, with two new genera Archaeospora and Paraglomus, based on concordant molecular and morphological characters. Mycologia 93:181–195
Posada D (2004) Modeltest 3.5. Facultad de Biologia, Universidad de Vigo, Vigo, Spain
Redecker D, Morton JB, Bruns TD (2000a) Ancestral lineages of arbuscular mycorrhizal fungi (Glomales). Mol Phylogenet Evol 14:276–284
Redecker D, Morton JB, Bruns TD (2000b) Molecular phylogeny of the arbuscular mycorrhizal fungi Glomus sinuosum and Sclerocystis coremioides. Mycologia 92:282–285
Redecker D, Hijri I, Wiemken A (2003) Molecular identification of arbuscular mycorrhizal fungi in roots: perspectives and problems. Folia Geobot 38:113–124
Redecker D, Raab P (2006) Phylogeny of the Glomeromycota (arbuscular mycorrhizal fungi): recent developments and new gene markers. Mycologia 98:885–895
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 Schüßler A, Schwarzott D, Walker C (2001) A new fungal phylum,
the Glomeromycota: phylogeny and evolution. Mycol Res 105:1413–1421
Schwarzott D, Walker C, Schüßler A (2001) Glomus, the largest genus of the arbuscular mycorrhizal fungi (Glomales), is non-monophyletic. Mol Phylogenet Evol 21:190–197
Swofford DL (2001) PAUP*. Phylogenetic analysis using Parsimony (*and other methods). Sinauer Associates, Sunderland, MA Thaxter R (1922) A revision of the Endogoneae. Proc Am Acad
Arts Sci 57:291–351
Walker C, Schüßler A (2004) Nomenclatural clarifications and new taxa in the Glomeromycota. Mycol Res 108:981–982
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic, New York, pp 315–322