HAL Id: hal-02555030
https://hal-enac.archives-ouvertes.fr/hal-02555030
Submitted on 27 Apr 2020
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Automatic Detection of Epileptic Spikes in Intracerebral
EEG with Convolutional Kernel Density Estimation
Ludovic Gardy, Emmanuel Barbeau, Christophe Hurter
To cite this version:
Ludovic Gardy, Emmanuel Barbeau, Christophe Hurter. Automatic Detection of Epileptic Spikes
in Intracerebral EEG with Convolutional Kernel Density Estimation. 4th International Conference
on Human Computer Interaction Theory and Applications, Feb 2020, Valletta, Malta. pp.101-109,
�10.5220/0008877601010109�. �hal-02555030�
Automatic Detection of Epileptic Spikes in Intracerebral EEG with
Convolutional Kernel Density Estimation
Ludovic Gardy
1,2,3 a, Emmanuel J. Barbeau
1,2 band Christophe Hurter
3 c1University of Toulouse, UPS, Centre de Recherche Cerveau et Cognition, Toulouse, France 2CNRS, CerCo, Purpan Hospital, Toulouse, France
3French Civil Aviation University, ENAC, Avenue Edouard Belin, Toulouse, France
{ludovic.gardy, emmanuel.barbeau}@cnrs.fr, christophe.hurter@enac.fr
Keywords: Electroencephalography, EEG, Time Series Visualization, Signal Processing, Kernel Density Estimation, Convolution, Noisy Signal, Event Detection, Epilepsy, Accessibility.
Abstract: Analyzing the electroencephalographic (EEG) signal of epileptic patients as part of their diagnosis is a very long and tedious operation. The most common technique used by medical teams is to visualize the raw signal in order to find pathological events such as interictal epileptic spikes (IESs) or abnormal oscillations. More and more efforts are being adopted to try to facilitate the work of doctors by automating this process. Our goal was to analyze signal density fields to improve the visualization and automatic detection of pathological events. We transformed the EEG signal into images on which we applied a convolution filter based on a Kernel Density Estimation (KDE). This method that we propose to call CKDE for Convolutional Kernel Density Estimation allowed the emergence of local density fields leading to a better visualization as well as automatic detection of IESs. Future work will be necessary to make this technique more efficient, but preliminary results are very encouraging and show a high performance compared to a visual inspection of the data or some other automatic detection techniques.
1
INTRODUCTION
Epilepsy is the name of a brain disorder character-ized predominantly by recurrent and unpredictable in-terruptions of normal brain function, called epileptic seizures (Fisher et al., 2005). Treating this disease sometimes requires the patient to undergo a record of his brain activity by Electroencephalography (EEG) in order to characterize the epileptogenic network, i.e., the brain area(s) involved in the seizures. EEG results from the electric signal generated by the co-operative action of brain cells (Blinowska and Durka, 2006). When epilepsy is drug-resistant, the solution to cure the patient is to surgically remove the area of his brain that causes seizures. To locate this area, it is often necessary to go through a stereoelectroen-cephalography (SEEG) consisting of the deep intrac-erebral implantation of electrodes. The depth EEG of the patient is recorded 24 hours a day for 6 to 15 days and then examined later by epileptologists. The electrophysiological markers and criteria for
de-a https://orcid.org/0000-0002-2977-8831 b https://orcid.org/0000-0003-0836-3538 c https://orcid.org/0000-0003-4318-6717
termining whether a cerebral area is impaired are still being defined (Valero et al., 2017; Roehri et al., 2017; Frauscher et al., 2017; Roehri et al., 2018). How-ever, some markers such as interictal epileptic spikes (IESs) are characteristic of the epileptogenic network (Talairach and Bancaud, 1966). They appear sponta-neously between periods of seizures and are of high amplitude for a duration ranging from 30 to 100 mil-liseconds (De Curtis and Avanzini, 2001) followed by a slow component for around 150 to 200 millisec-onds (Staley and Dudek, 2006). IESs are generated by synchronous discharges of a group of neurons in a region referred to as the irritative zone (Latka et al., 2003). While some would argue that the neural net-work that produces IESs is not always identical to the seizure onset zone, IESs are useful to support the di-agnosis of epilepsy (Staley and Dudek, 2006). Other markers, more recent and not used in clinical practice yet such as fast-ripples, are being studied by several teams around the world and could be essential mark-ers of the seizure onset zone (Staba et al., 2002; Ibarz et al., 2010). There is currently no official tool that can be used in clinical practice to detect these dis-crete and transient events, such as IES or fast-ripples.
Moreover, their characteristics can differ from one pa-tient to another and vary in shape, duration, or fre-quency, making them very difficult to detect by an au-tomatic tool. Many attempts have been made to create such devices, but they’ve had a relatively low impact and have not passed the gates of research laboratories. However, we address that question with a completely new approach that could provide an efficient solution. The amounts of data to be analyzed are so huge; we estimate it at around 317 billions of electrophysiolog-ical values for a 15-day, or 360 hours, recording in one patient. We can no longer let clinicians manu-ally look at the signal and search inside for patho-logical events without help. The technique we pro-pose to visualize and automatically tag pathological events in this massive amount of noisy EEG signal is based on image processing. Our tests show that IESs have a particular signature in the field of densi-ties. We transformed the signal into an image using Kernel Density Estimation (Silverman, 2018) and ap-plied a convolution onto this image to build density fields based on the spacing of pixels. The brief and rapid amplitude changes during spikes lead to a larger gap between activated pixels which is characterized by a low density after convolution. These low densi-ties are the ones that can be visualized and even au-tomatically recorded. We analyzed the intracerebral EEG recordings of a patient treated in the epilepsy department of CHU Purpan in Toulouse, France. Our method allowed us to obtain an improved visualiza-tion of the EEG signal enabling a fast localizavisualiza-tion of the IESs but also automatic detection of these patho-logical events. There is still much effort to be made to generalize this approach and make it functional for clinical implications, but the preliminary results we obtain using it are very encouraging. In this paper, we will define epilepsy and diagnostic problems before presenting the related works about EEG visualization and automatic pattern detection, then present the re-sults we have obtained and discuss them.
2
RELATED WORKS
This work builds upon recent studies covering time-series visualization (Wang et al., 2017) as well as the automatic search for pathological events such as inter-ictal epileptic spikes (IESs) in the EEG signal (Roehri et al., 2018). The question of IESs as markers of the epileptogenic network has been widely studied, but no study has yet made it easier to visualize them or de-tect them automatically. These techniques are either not reliable enough or too long or difficult to use.
2.1
EEG Visualization
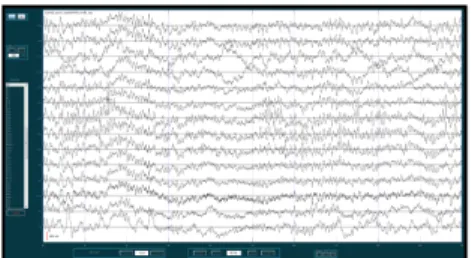
EEG recordings are most of the time represented as curve-shaped amplitudes varying over time (figure 1). These line graphs are a quick and easy way to describe and understand brain activity. Clinicians and neurol-ogists, in particular, are very attached to this visual-ization because they are used to it and understand it well.
Figure 1: Classical representation as a line graph of an EEG signal. Each line represents a channel from an intracerebral recording.
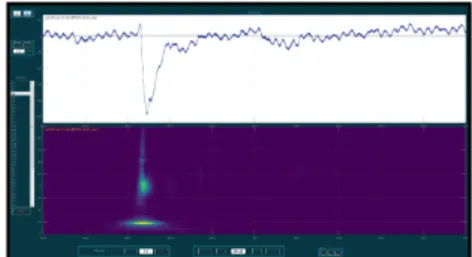
Time-frequency representation (figure 2) is an-other way to display the signal, seldom used in clini-cal practice but appreciated by neuroscientists. The frequency spectrum is a good indicator of ongoing cognitive processes and possibly epileptic events such as pathological oscillations (Bragin et al., 2002; Bra-gin et al., 1999a; H¨oller et al., 2015). The promising results of a time-frequency analysis of intracerebral EEG signals in epilepsy will probably make this a not to be missed step soon from a medical point of view as well.
Figure 2: Additional time-frequency representation for one channel. Top graph: raw data, bottom graph: the frequency values are represented along the y-axis and the power in these frequencies on a color scale.
2.2
Time Series Visualization
Many variants of time series representations have been proposed to synthesize or improve visualiza-tion (Saito et al., 2005; Javed and Elmqvist, 2010; Van Wijk and Van Selow, 1999; Weber et al., 2001; Lin et al., 2004; Kincaid and Lam, 2006; Hao et al., 2007; Zhao et al., 2011) but their effectiveness has been discussed, and the classical representation is still
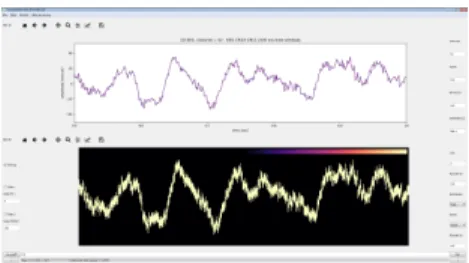
favored in practice. However, a recent study (Wang et al., 2017) proposed the application of Kernel Den-sity Estimation (KDE) to transform a signal into a density field (figure 3). They showed that it leads to a new type of signal, and in some cases, the visual-ization is improved compared to the original one. We think that the visual aspect is not the only one to be interesting, but that the whole notion of signal density is.
Figure 3: Density representation of a time series. Figure from (Wang et al., 2017).
2.3
Event Detection
Manually searching for pathological events such as interictal epileptic spikes (IESs) in the EEG signal is a very long and tedious operation for epileptolo-gists. During the past decades, many methods were proposed to detect theses events automatically (Birot
et al., 2013; Gotman, 1999). The noise makes it
difficult for an algorithm to be efficient in this task. Furthermore, nowadays technology and storage ca-pacities, allowing several days of 24/7 recordings on more than a hundred channels simultaneously, yield a considerable amount of data. Some algorithms work quite fast but are just as much sensitive to noise and no longer work when the pattern to research differs from the input signal. Some others are relatively effi-cient, but the computation time for large amounts of data is discouraging.
2.3.1 Dynamic Time Warping
In time series analysis, dynamic time warping (DTW) is one of the algorithms for measuring similarity be-tween two temporal sequences, for instance, two time series of the same length (Keogh et al., 2009; Ding et al., 2008). The difference with a pure Euclidean Distance is that the whole sequence is taken into ac-count to calculate the similarity between the pattern and the input signal, not just the sum of the distances between each point of the two sequences. DTW has been used for some biological applications (Rakthan-manon et al., 2012; Chadwick et al., 2011) or gesture recognition (Alon et al., 2008; Wobbrock et al., 2007) but the computation time on a massive amount of data
as well as the difficulty to detect an event when the in-put signal differs from the pattern can make it barely efficient. An improved version of the Trillion algo-rithm from the UCR (University of California, River-side) suite ((Rakthanmanon et al., 2012)) have been developped and tested (Jing et al., 2016) on a hun-dred patients, allowing a good recognition of IESs. Nevertheless, the necessity of having a pattern to re-search is still a problem. A user still needs to spec-ify a template, and there is a large variability of spike waveforms within and between patients among other factors.
2.3.2 Machine Learning
The past decade has seen the rise of machine learn-ing in EEG signal processlearn-ing, givlearn-ing birth to a rich literature. Work in this domain has focused as much on the automatic search for epileptic seizures (Song et al., 2012) as on that of epileptic markers such as pathological oscillations or IESs (Jrad et al., 2016; Birot et al., 2013). Many problems remain, start-ing with the fact that feature engineerstart-ing is a time-consuming and challenging task. Furthermore, pre-processing and cleaning the EEG signal requires to manipulate tools that are not easy to use, especially for clinicians. Deep learning methods have been more recently built with the promise to overcome these is-sues or at least lighten them as it can learn good fea-ture representations from raw data (Roy et al., 2019). It is essential to follow carefully this area of literature which is promising but not yet functional in practice.
2.3.3 Time-frequency
As shown in figure 4, sharp transients events present in depth-EEG signals (typically, the spike compo-nent of interictal epileptic spikes - IESs) are associ-ated with an abrupt increase of the signal energy in the lower and higher frequency bands (B´enar et al., 2010).
Figure 4: Example of an interictal epileptic spike (IES) in a one second time window. Top: raw signal, bottom: time-frequency representation.
An up-and-coming tool for the automatic detec-tion of IES and pathological oscilladetec-tions, based on
time-frequency analysis, is Delphos (Roehri et al., 2018; Roehri et al., 2016). However, we believe that a density analysis of the signal could lead to new pos-sibilities and compete or cooperate with the existing techniques in the challenge toward automatic detec-tion. Many false positives currently make the ex-ploitation of time-frequency data difficult, while this 2-dimensional signal may benefit from a density anal-ysis to sort the events detected by a threshold in the frequency-domain.
3
ESTIMATING DENSITIES
By converting a 1-dimensional EEG signal composed of a list of ordered values into 2-dimension, we ob-tain an image, which is an area of pixels. Thus, when the signal varies in amplitude or frequency, this area of pixels is affected. Two-dimensional convolution is a technique commonly used in image processing to apply effects such as blurring, sharpening, or edge detection. We used this technique, that affects pix-els whose value is different from 0, on our EEG sig-nal to emerge density fields that could witness the occurrence of pathological events. Sharp amplitude variations during IESs have the effect of spacing the points, and therefore the pixels representing the sig-nal on the image, from each other. It is this notion of spacing between points that we can exploit to iden-tify IESs in EEG. Typically the electrophysiological changes of the brain signal are gradual and homoge-neous, but during an IESs they become huge in a short time. When convolution is applied, non-pathological variations appear under the form of high densities and IESs under the form of low density because of the fast spacing between the points in the same amount of time.
3.1
Kernel Density Estimation
It was necessary to transform the 1-dimensional EEG signals into 2-dimensional images in order to apply a convolution. Using KDE allowed to convert each value (a float or an integer) of the 1-dimensional EEG signal into a vector representing a Gaussian-shaped density probability of finding this value within a range of values. The aggregation of these vertically trans-posed vectors along the time axis constitutes a 2-dimensional image of the original 1-2-dimensional EEG signal (figure 5).
Figure 5: Left: A randomly simulated 1-dimensional sig-nal where each timestamp is associated with a single value. Right: The same signal converted into a 2-dimensional im-age using KDE. Each timestamp is associated with a verti-cal vector representing a Gaussian-shaped density probabil-ity.
3.2
Convolution of the Imaged Signal
Convolution consists of applying a kernel on each of the pixels of the image. This technique has the effect of modifying the value of the current pixel by consid-ering the value of the neighboring pixels. Only pixels with a value different from 0 are affected. This pro-cess of adding each element of the image to its local neighbors, weighted by a kernel, leads to an improved visualization with additional density field. Let dij
de-note the pixel value of an image and V its value after processing, the convolution can be modeled by
V =|∑ q i=1(∑ q j=1fi jdi j) F | Where:
• fij= the coefficient of a convolution kernel at
po-sition i,j (in the kernel);
• dij = the data value of the pixel that corresponds
to fij;
• q = the dimension of the kernel, assuming a square kernel (if q = 3, the kernel is 3*3);
• F = either the sum of the coefficients of the kernel or 1 if the sum of coefficients is 0;
• V = the output pixel value;
The obtained density field, looking like an im-proved color-scaled line graph (figure 6), is affected both by variations in the amplitude and frequency of the original signal. If two pixels of value superior to 0 are side by side, then the two kernels overlap, and the original pixels are strengthened. If the pixels are distant, then they are not strengthened by their neigh-bors. The closer they are to each other, the higher the local density.
We called Convolutional Kernel Density Estima-tion (CKDE) the technique we used here, which aims to transform the time series into images and to apply a two-dimensional convolution on it.
Figure 6: After the application of a Gaussian kernel of size 11 and standard deviation 3 on the image, pixels that are close to each other are strengthened. On the contrary, pixels that are distant to each other are barely reinforced.
4
USER INTERFACE
4.1
Implementation
We have built a user interface employing PyQt5 in Python 3.6 to allow the user to visualize and interact-ing live with the data. The top panel of the main win-dow displays the 1-dimensional original signal and bottom panel the 2-dimensional imaged-signal (fig-ure 7). Graphical characteristics and window settings can be modified by the user using text boxes, combo boxes, toolbars, and menus on the top and sides of the window. Some possible actions are, for instance, pan-ning, zooming, changing colors, moving along the x and y axes, or saving the figures.
Figure 7: Main window of the GUI. The two panels at the center of the main window represent the 1-dimensional and the 2-dimensional signals. Live interactions are possible with both.
A user menu is also available for more sophisti-cated options such as creating a kernel for image con-volution. Opening the kernel window (figure 8) al-lows the user to generate a Gaussian kernel with or without customizing its parameters (size and standard
deviation). He can alternatively create a wholly cus-tomized kernel by filling a grid of the size he wants the values he wants.
Figure 8: The kernel window allows the setting and drawing of the kernel that will be employed for image convolution. Top: Gaussian kernel, bottom: custom kernel.
The user can travel forward or backward in the time dimension of the signals by using his keyboards’ left or right arrows or those located at the bottom of the window. Otherwise, it is possible to directly type a time value in seconds in the text box between the arrows.
4.2
User Feedback
The only user of the GUI described is the first author of this paper, accustomed to analyzing EEG signals on different clinical or research software. The GUI was designed to visualize the original signal and its associated image to identify possible errors in the al-gorithm, which is still under development. The main idea being a proof of concept that density analysis of EEG can be useful, only one recording channel at a time is displayed, to avoid missing any important de-tail (but all channels are processed). Navigation over time is instantaneous, as is the navigation from one channel to another or the enabling and disabling of the convolution, allowing a fast and efficient interac-tion. An upcoming more sophisticated interface will be developed soon with the ability to visualize multi-ple channels at once and a more straightforward de-sign so that the platform can be used instinctively by other users.
5
RESULTS
We have evaluated the efficiency of EEG signal 2-dimensional convolution to visualize and automati-cally detect interictal epileptic spikes (IESs). The data used are those of a patient who was hospitalized in 2015 and we know well. The EEG signal from the
same patient were used in a previous study for which the purpose was different (Despouy et al., 2019). For this last study, the EEG signal was carefully examined and tagged. We ran the current tests on a 10 minutes part of this tagged signal and compared previous man-ual detection of IESs to automatic detection using our new method. This data set was composed of a hun-dred intracerebral macroelectrodes recorded at 2048 Hz that we downsampled at 512 Hz.
5.1
Visualization of Events
The visual inspection of densities (figure 9) allowed an easy identification of IESs, which are character-ized by low densities due to the larger spacing be-tween pixels.
Figure 9: Convolution applied on one channel of the imaged EEG signal, which appears as a color-scaled line graph. Ex-ample of density decrease during an IES caused by the sig-nificant spacing between pixels during this sharp amplitude changing.
On the previous example, the size of the image was 512 (horizontal length) * 150 (vertical length), and the applied kernel was of size 7 * 7 with custom values, as follows: 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 10 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9 9
5.2
Automatic Detection of Events
A low pass filter was applied to the density field in order to remove everything from the image except the IESs (figure 10). This means that only the densities forming the IES are conserved since all the others are set to 0. The densities resulting from IESs thus be-come high densities. By summing the densities of each pixel of the resulting image, we discern if an IES is present or not. If no IES is present, the total density
Figure 10: Image after application of a low pass filter. All the high densities are removed to display only the IESs.
on the image will be around 0. If there is one, it will be higher, between 400 to more than 1000.
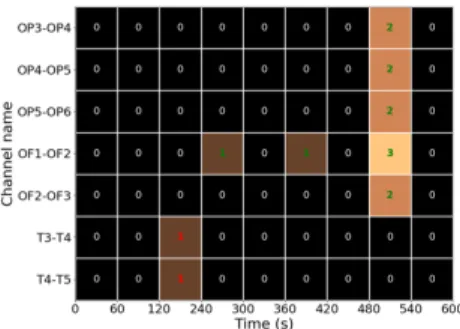
15 IESs were automatically identified in the por-tion of the analyzed signal. Among them, 13 were true IESs, while 2 were false positives (figure 11). 100% of the events that were tagged during previous visual inspection were automatically identified.
Figure 11: Map of detected events. The green text color indicates the correctly detected IESs, the red color the false positives. The names of the channels on which an event has been detected are represented along the y-axis. Time is represented in seconds along the x-axis. The more events detected in a given time period, the lighter the square corre-sponding to this time period for a given recording channel.
An IES is detected when the total density on the remaining image after removal of high densities by filtering exceeds a certain threshold. It works well to detect pathological events but also can lead to false positives, which are mainly due to two things (figure 12). Firstly, an accumulation of residual densities ac-companying rapid micro-changes in signal intensity. Secondly, an accumulation of low densities related to the borders of the density field not being eliminated during the filtering. We will expose in the Discussion a way to solve these problems.
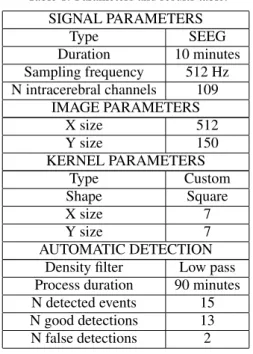
The main objective was to ensure the efficiency of detection. Thus, minimal effort was allocated to in-crease the speed of computation which currently lasts 90 minutes for a 10 minutes signal. Table 1 summa-rizes the parameters used for image generation from the original signal, kernel creation, and convolution, as well as the characteristics of the automatic detec-tion process.
Figure 12: Example of false detection. The total density on the image is high, as it is the case when an IES is detected, but this is only related to the accumulation of low scattered local densities. On the contrary, when it comes to a spike, the densities occur during a short duration.
Table 1: Parameters and results table.
SIGNAL PARAMETERS Type SEEG Duration 10 minutes Sampling frequency 512 Hz N intracerebral channels 109 IMAGE PARAMETERS X size 512 Y size 150 KERNEL PARAMETERS Type Custom Shape Square X size 7 Y size 7 AUTOMATIC DETECTION
Density filter Low pass
Process duration 90 minutes
N detected events 15
N good detections 13
N false detections 2
6
INTEREST OF THE CKDE
APPROACH
With this proof-of-concept, we have identified several interesting or novel aspects that are worth assessing in future work:
• Unlike other automatic detection methods, den-sity changes are neither affected by the signal ori-entation (positive or negative) nor by prominent but slow changes in amplitude, nor by unwanted oscillations.
• Using the parameters we proposed, only very abrupt variations are detected, like those caused by IESs, thus avoiding false detections driven by rise or fall of slow and non-pathological ampli-tudes.
• Furthermore, no training on the data is necessary
to the algorithm, and only a few or no preprocess-ing at all has to be done.
• From a visualization point of view, the represen-tation of densities in EEG is straightforward for the user to understand given that the data look a lot like a line graph.
• The preliminary results in automatic detection are encouraging with a 100% identification (13/13) of IESs and only two false positives inside real in-tracerebral recordings.
7
LIMITATIONS AND FUTURE
WORK
The main objectives of this study were to develop an efficient method to visualize SEEG data and au-tomatically detect pathological electrophysiological signals. We aim to compete with existing methods that still suffer from many gaps, but another pos-sibility could also be to use CKDE in cooperation with other routines to improve their efficiency. Fu-ture work will need to focus on a more precise and robust definition of the parameters such as the size and values of the kernel chosen for convolution as well as the low pass filter value. It is important to note that the parameters used here were hyper opti-mized after a preliminary visual analysis of the signal. In a future version, these parameters should be auto-matically determined so that the user does not have to spend time adjusting them. Furthermore, our tests have so far been carried out only on one patient im-planted with a hundred intracerebral recording chan-nels sampled at 512 Hz over 10 minutes. The first results lead us to be confident about the functional-ity of the technique and its potential performance, but we have to replicate them on a much more substantial amount of signal. This approach is currently under-way with data recorded from more than 60 epileptic patients implanted for SEEG examinations between 2013 and 2019. In addition, the detection of other types of pathological markers such as high-frequency oscillations (Bragin et al., 2002; Bragin et al., 1999b) and more specifically fast-ripples (Ibarz et al., 2010; Zelmann et al., 2009; Roehri et al., 2017) should be evaluated and compared with other detectors. The problem of false positives can be addressed by re-ducing the time window for automatic signal analy-sis. False positives are related to the accumulation of small local and scattered residual densities over a rela-tively long period, whereas the IESs are characterized by small densities as well, but stacked over a much shorter duration. By reducing the time window on
which the densities are summed from 1 second to 100 or 200 milliseconds, this problem should disappear. However, the calculation time should be increased.
REFERENCES
Alon, J., Athitsos, V., Yuan, Q., and Sclaroff, S. (2008). A unified framework for gesture recognition and spa-tiotemporal gesture segmentation. IEEE transac-tions on pattern analysis and machine intelligence, 31(9):1685–1699.
B´enar, C.-G., Chauvi`ere, L., Bartolomei, F., and Wendling, F. (2010). Pitfalls of high-pass filtering for detecting epileptic oscillations: a technical note on false ripples. Clinical Neurophysiology, 121(3):301–310.
Birot, G., Kachenoura, A., Albera, L., B´enar, C., and Wendling, F. (2013). Automatic detection of fast rip-ples. Journal of neuroscience methods, 213(2):236– 249.
Blinowska, K. and Durka, P. (2006). Electroencephalogra-phy (eeg). Wiley Encyclopedia of Biomedical Engi-neering.
Bragin, A., Engel Jr, J., Wilson, C. L., Fried, I., and Buzs´aki, G. (1999a). High-frequency oscillations in human brain. Hippocampus, 9(2):137–142.
Bragin, A., Engel Jr, J., Wilson, C. L., Fried, I., and Math-ern, G. W. (1999b). Hippocampal and entorhinal cor-tex high-frequency oscillations (100–500 hz) in hu-man epileptic brain and in kainic acid-treated rats with chronic seizures. Epilepsia, 40(2):127–137.
Bragin, A., Mody, I., Wilson, C. L., and Engel, J. (2002). Local generation of fast ripples in epileptic brain. Journal of Neuroscience, 22(5):2012–2021.
Chadwick, N. A., McMeekin, D. A., and Tan, T. (2011). Classifying eye and head movement artifacts in eeg signals. In 5th IEEE International Conference on Dig-ital Ecosystems and Technologies (IEEE DEST 2011), pages 285–291. IEEE.
De Curtis, M. and Avanzini, G. (2001). Interictal spikes in focal epileptogenesis. Progress in neurobiology, 63(5):541–567.
Despouy, E., Curot, J., Denuelle, M., Deudon, M., Sol, J.-C., Lotterie, J.-A., Reddy, L., Nowak, L. G., Pariente, J., Thorpe, S. J., et al. (2019). Neuronal spiking activ-ity highlights a gradient of epileptogenicactiv-ity in human tuberous sclerosis lesions. Clinical Neurophysiology, 130(4):537–547.
Ding, H., Trajcevski, G., Scheuermann, P., Wang, X., and Keogh, E. (2008). Querying and mining of time series data: experimental comparison of representations and distance measures. Proceedings of the VLDB Endow-ment, 1(2):1542–1552.
Fisher, R. S., Boas, W. V. E., Blume, W., Elger, C., Genton, P., Lee, P., and Engel Jr, J. (2005). Epileptic seizures and epilepsy: definitions proposed by the international league against epilepsy (ilae) and the international bu-reau for epilepsy (ibe). Epilepsia, 46(4):470–472.
Frauscher, B., Bartolomei, F., Kobayashi, K., Cimbalnik, J., van t Klooster, M. A., Rampp, S., Otsubo, H., H¨oller, Y., Wu, J. Y., Asano, E., et al. (2017). High-frequency oscillations: the state of clinical research. Epilepsia, 58(8):1316–1329.
Gotman, J. (1999). Automatic detection of seizures and spikes. Journal of Clinical Neurophysiology, 16(2):130–140. Gotman, J. and Gloor, P. (1976). Au-tomatic recognition and quantification of interictal epileptic activity in the human scalp eeg. Elec-troencephalography and clinical neurophysiology, 41(5):513–529.
Hao, M. C., Dayal, U., Keim, D. A., and Schreck, T. (2007). Multi-resolution techniques for visual exploration of large time-series data. In EUROVIS 2007, pages 27– 34.
H¨oller, Y., Kutil, R., Klaffenb¨ock, L., Thomschewski, A., H¨oller, P. M., Bathke, A. C., Jacobs, J., Taylor, A. C., Nardone, R., and Trinka, E. (2015). High-frequency oscillations in epilepsy and surgical outcome. a meta-analysis. Frontiers in human neuroscience, 9:574. Ibarz, J. M., Foffani, G., Cid, E., Inostroza, M., and de la
Prida, L. M. (2010). Emergent dynamics of fast rip-ples in the epileptic hippocampus. Journal of Neuro-science, 30(48):16249–16261.
Javed, W. and Elmqvist, N. (2010). Stack zooming for multi-focus interaction in time-series data visualiza-tion. In 2010 IEEE Pacific Visualization Symposium (PacificVis), pages 33–40. IEEE. Javed, W., McDon-nel, B., and Elmqvist, N. (2010). Graphical perception of multiple time series. IEEE transactions on visual-ization and computer graphics, 16(6):927–934. Jing, J., Dauwels, J., Rakthanmanon, T., Keogh, E., Cash,
S., and Westover, M. (2016). Rapid annotation of in-terictal epileptiform discharges via template matching under dynamic time warping. Journal of neuroscience methods, 274:179–190.
Jrad, N., Kachenoura, A., Merlet, I., Bartolomei, F., Nica, A., Biraben, A., and Wendling, F. (2016). Automatic detection and classification of high-frequency oscil-lations in depth-eeg signals. IEEE Transactions on Biomedical Engineering, 64(9):2230–2240.
Keogh, E., Wei, L., Xi, X., Vlachos, M., Lee, S.-H., and Protopapas, P. (2009). Supporting exact indexing of arbitrarily rotated shapes and periodic time series un-der euclidean and warping distance measures. The VLDB journal, 18(3):611–630.
Kincaid, R. and Lam, H. (2006). Line graph explorer: scal-able display of line graphs using focus+ context. In Proceedings of the working conference on Advanced visual interfaces, pages 404–411. ACM.
Latka, M., Was, Z., Kozik, A., and West, B. J. (2003). Wavelet analysis of epileptic spikes. Physical Review E, 67(5):052902.
Lin, J., Keogh, E., Lonardi, S., Lankford, J. P., and Nys-trom, D. M. (2004). Visually mining and monitoring massive time series. In Proceedings of the tenth ACM SIGKDD international conference on Knowledge dis-covery and data mining, pages 460–469. ACM. Rakthanmanon, T., Campana, B., Mueen, A., Batista, G.,
(2012). Searching and mining trillions of time se-ries subsequences under dynamic time warping. In Proceedings of the 18th ACM SIGKDD international conference on Knowledge discovery and data mining, pages 262–270. ACM.
Roehri, N., Lina, J.-M., Mosher, J. C., Bartolomei, F., and B´enar, C.-G. (2016). Time-frequency strategies for increasing high-frequency oscillation detectability in intracerebral eeg. IEEE Transactions on Biomedical Engineering, 63(12):2595–2606.
Roehri, N., Pizzo, F., Bartolomei, F., Wendling, F., and B´enar, C.-G. (2017). What are the assets and weaknesses of hfo detectors? a benchmark frame-work based on realistic simulations. PloS one, 12(4):e0174702.
Roehri, N., Pizzo, F., Lagarde, S., Lambert, I., Nica, A., McGonigal, A., Giusiano, B., Bartolomei, F., and B´enar, C.-G. (2018). High-frequency oscillations are not better biomarkers of epileptogenic tissues than spikes. Annals of neurology, 83(1):84–97.
Roy, Y., Banville, H., Albuquerque, I., Gramfort, A., Falk, T. H., and Faubert, J. (2019). Deep learning-based electroencephalography analysis: a systematic review. Journal of neural engineering.
Saito, T., Miyamura, H. N., Yamamoto, M., Saito, H., Hoshiya, Y., and Kaseda, T. (2005). Two-tone pseudo coloring: Compact visualization for one-dimensional data. In IEEE Symposium on Information Visualiza-tion, 2005. INFOVIS 2005., pages 173–180. IEEE. Silverman, B. W. (2018). Density estimation for statistics
and data analysis. Routledge.
Song, Y., Crowcroft, J., and Zhang, J. (2012). Automatic epileptic seizure detection in eegs based on optimized sample entropy and extreme learning machine. Jour-nal of neuroscience methods, 210(2):132–146. Song, Y. and Li`o, P. (2010). A new approach for epileptic seizure detection: sample entropy based feature ex-traction and extreme learning machine. Journal of Biomedical Science and Engineering, 3(06):556. Staba, R. J., Wilson, C. L., Bragin, A., Fried, I., and
Engel Jr, J. (2002). Quantitative analysis of high-frequency oscillations (80–500 hz) recorded in human epileptic hippocampus and entorhinal cortex. Journal of neurophysiology, 88(4):1743–1752.
Staley, K. J. and Dudek, F. E. (2006). Interictal spikes and epileptogenesis. Epilepsy Currents, 6(6):199–202. Talairach, J. and Bancaud, J. (1966). Lesion,” irritative”
zone and epileptogenic focus. Stereotactic and Func-tional Neurosurgery, 27(1-3):91–94.
Valero, M., Averkin, R. G., Fernandez-Lamo, I., Aguilar, J., Lopez-Pigozzi, D., Brotons-Mas, J. R., Cid, E., Tamas, G., and de la Prida, L. M. (2017). Mecha-nisms for selective single-cell reactivation during of-fline sharp-wave ripples and their distortion by fast ripples. Neuron, 94(6):1234–1247.
Van Wijk, J. J. and Van Selow, E. R. (1999). Cluster and calendar based visualization of time series data. In Proceedings 1999 IEEE Symposium on Information Visualization (InfoVis’ 99), pages 4–9. IEEE.
Wang, Y., Han, F., Zhu, L., Deussen, O., and Chen, B. (2017). Line graph or scatter plot? automatic selection of methods for visualizing trends in time series. IEEE transactions on visualization and computer graphics, 24(2):1141–1154.
Weber, M., Alexa, M., and M¨uller, W. (2001). Visualizing time-series on spirals. In Infovis, volume 1, pages 7– 14.
Wobbrock, J. O., Wilson, A. D., and Li, Y. (2007). Gestures without libraries, toolkits or training: a $1 recognizer for user interface prototypes. In Proceedings of the 20th annual ACM symposium on User interface soft-ware and technology, pages 159–168. ACM. Zelmann, R., Zijlmans, M., Jacobs, J., Chˆatillon, C.-E., and
Gotman, J. (2009). Improving the identification of high frequency oscillations. Clinical Neurophysiol-ogy, 120(8):1457–1464.
Zhao, J., Chevalier, F., Pietriga, E., and Balakrishnan, R. (2011). Exploratory analysis of time-series with chronolenses. IEEE Transactions on Visualization and Computer Graphics, 17(12):2422–2431.