THÈSE EN CO-TUTELLE
pour obtenir le grade de
DOCTEUR DE UNIVERSITÉ MONTPELLIER II ET
DOCTEUR DE UNIVERSIDADE DE BRASÍLIA
Formation doctorale: EERPG - Evolution, écologie, ressources génétiques, paléontologie
ECOLE DOCTORALE SIBAGHE
Systèmes intégrés en Biologie, Agronomie, Géosciences, Hydrosciences et Environnement
Formation doctorale: Biologia Animal
PROGRAMA DE PÓS-GRADUAÇÃO EM BIOLOGIA
présentée et soutenue publiquement par
Renata SANTOS DE MENDONÇA
le 23 novembre 2010
Estudos taxonômicos de ácaros Tetranychidae no Brasil e filogenia e estrutura genética do ácaro rajado, TETRANYCHUS URTICAE Kock, inferidas a
apartir de sequencias do DNA ribosômico e mitocondrial
Études taxonomiques des acariens Tetranychidae au Brésil, en particulier sur la phylogenie et la structure genetique des populations de l´acarien jaune, TETRANYCHUS URTICAE koch, inferées à partir des sequences d´ADN
ribosomique et mitochondrial.
JURY:
Mme Maria NAVAJAS, Directeur de Recherche INRA/CBGP, Montpellier, France Mme Ivone R. DINIZ , Professeur Université de Brasília, Brésil
M Gilberto J. de MORAES, Professeur Universidade de São Paulo, ESALQ/USP, Brésil M Francisco J. FERRAGUT Professeur Université de Politecnica de Valencia, Spain Mme Marie-Stéphane TIXIER, Professeur Montpellier SupAgro / UMR CBGP, France Mme Denise NAVIA Directeur de recherche Embrapa Cenargen, Brésil
Directrice de Thèse Co-Directrice de thèse Rapporteur Rapporteur Examinateur Examinateur
UNIVERSIDADE DE BRASÍLIA - BRÉSIL
INSTITUTO DE CIÊNCIAS BIOLÓGICAS UNIVERSITÉ DE MONTPELLIER II - FRANCE SCIENCES ET TECHNIQUES DU LANGUEDOCii
Thesis partially supported by:
CAPES, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil and
CNPq, Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil
Performed on:
Center for Biology and Management of Populations
Centre de Biologie pour la Gestion des Populations – CBGP CBGP, Campus International de Baillarguet CS 30016 34988 Montferrier-sur-Lez cedex, France
http://www.ensam.inra.fr/cbgp
FRANCE
Embrapa Resources Genetics and Biotechnology Embrapa Recursos Genéticos e Biotecnologia
Parque Estação Biológica - PqEB - Av. W5 Norte (final) Caixa Postal 02372 - Brasília, DF - Brasil - 70770-917
http://www.cenargen.embrapa.br
BRAZIL
November 2010 Montpellier, France
iii
Université de Montpellier II – Sciences et Techniques du Languedoc and Universidade de Brasília allowing the unparalleled oportunity to replayed training simultaneously at both universities due to a a join doctorate supervision agreement
signed between France and Brazil
November 2010 Montpellier, France
iv
TABLE OF CONTENTS
TABLE OF CONTENTS ……….………..……….. ACKNOWLEDGEMENTS ……….. PREFACE ……….. ABSTRACT ………... RESUMO ………... RESUMÉ ……… Introduction et contexte de la thèse ……....………...……...…….…………....….. Objectifs et approches ………...……...…….………….…..PART - I: Introduction and context of the thesis ………..
INTRODUCTION ……….. General objectives ………. Strategies ………....…. Specific objectives ………. THE CONTEXT OF THE THESIS ………. Pest status of the two-spotted spider mite, Tetranychus urticae Koch 1836 ………
Outbreaks and economical impact in worldwide agriculture …………...……… Pest control ………...……….. Biological traits of T urticae favouring its pest status ………...………
The presence of biotypes ………. The use of molecular markers in Acarology ………... Phytophagous mites - nuclear ribosomal DNA and mitochondrial DNA ……… Genetic data to study ecological differenciation in Tetranychidae ……….. Methods in molecular phylogenetic analysis ………. REFERENCES ………...
PART - II: Chapters 1, 2, 3 and 4 ………..………
CHAPTER 1
Two new spider mites (Acari: Tetranychidae) from Brazil: a Monoceronychus McGregor (Bryobiinae) from fingergrass and an Oligonychus Berlese (Tetranychinae) from grape.
International Journal of Acarology. Article accepted in International Journal of Acarology
………..…………...
CHAPTER 2
Notes on Brazilian spider mites (Acari: Tetranychidae) – new hosts and localities. Article submitted to the Journal of Insect Science ………..…
iv vii viii ix xi xiii xv xvi 1 2 3 4 5 5 6 6 8 8 8 10 11 13 15 16 31 32 55
v CHAPTER 3
A critical review on some closely related species of Tetranychus sensu stricto Tuttle & Baker (1968) (Prostigmata: Tetranychidae) in the public DNA sequences database. Article submitted to the Experimental and Applied Acarology ………...…….………..
CHAPTER 4
Phylogeography, and genetic structure in Tetranychus urticae Koch (Acari:
Tetranychidae) populations inferred from ITS2 and COI DNA sequences. Article in progress ……….………… ABSTRACT ………... INTRODUCTION ……….. SPECIFIC OBJECTIVES ……… MATERIAL AND METHODS ………. Field samples ……….. Morphological mite species identification ……….. DNA extraction, PCR amplification and sequencing ……… Sequence retrieval and dataset ……….. Sequence and phylogenetic analyses ……….. Analysis of Molecular Variance (AMOVA) and genetic structure accessed by COI
sequences of Tetranychus urticae ……… RESULTS AND DISCUSSION ……….. The ITS region of ribosomal DNA ……….. The COI region of mitochondrial DNA ……….. AMOVA results - Genetic structure of Tetranychus urticae populations related to
geographic origin and host plants based on COI mitochondrial sequences ……… Distribution and frequency of ITS2 sequences ...………..……. Distribution and frequency of COI haplotypes ………..……… REFERENCES ………. MAIN RESULTS ……….……….………. CONCLUSION ……….………. 91 139 139 140 142 143 143 148 148 149 151 151 152 153 155 159 162 163 167 177 180
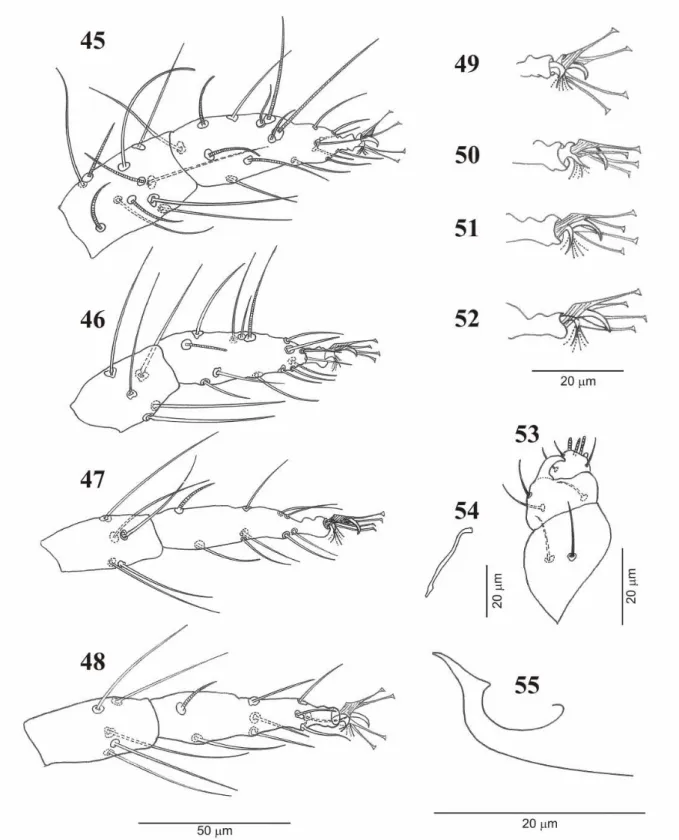
vi LIST OF FIGURE- CHAPTER 4
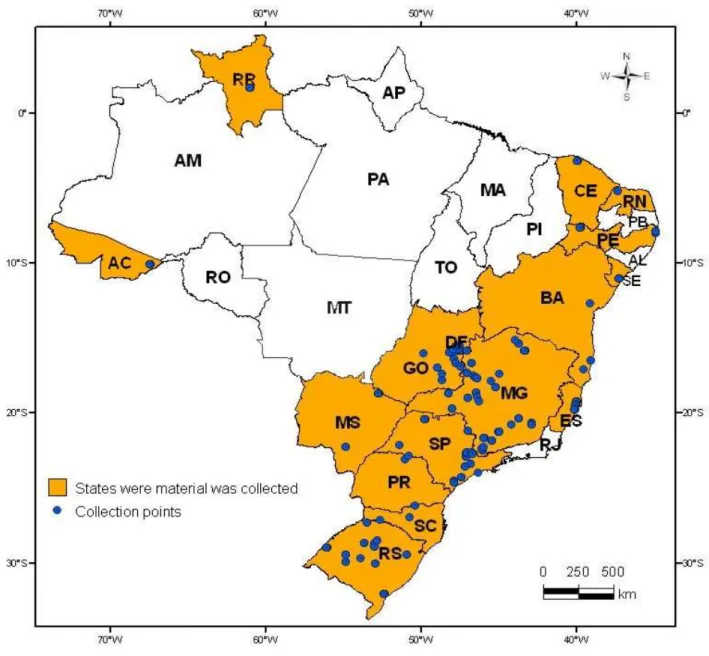
Figure 1. States and their respective sampling points for collecting Tetranychus urticae in Brazil between October 2004 and July 2008 ……….………... Figure 2. A. World map of Tetranychus urticae survey between 10/2004 and 07/2008. B. Detail of the studied sites in France and Spain ……….... Figure 3. Phylogeny tree inferred from mtDNA COI sequences of Tetranychus urticae retrieved from Genbank and obtained in this study from T. urticae field samples. The colors highlight the haplotypes 1, 2 and 10.The haplotype number and Genbank accessions are given (see Table 6). The Neighbor-Joining (NJ) method was used based on distances calculated using Kimura’s two-parameter (K2P) correction. Bootstrap values (higher than 50%) are near to the branches. The scale bar indicates the number of changes per site in NJ(K2P) inference ………. Figure 4. Distribution and frequency of Tetranychus urticae ITS2 haplotypes (ribosomal DNA) in the world. ITS sequences retrieved from Genbank and those obtained in this study were
included ………..………... Figure 5. Distribution and frequency of Tetranychus urticae COI haplotypes (mitochondrial DNA) in the world. COI sequences retrieved from Genbank and those obtained in this study were included ……….………... 144 146 157 165 166 LIST OF TABLES - CHAPTER 4
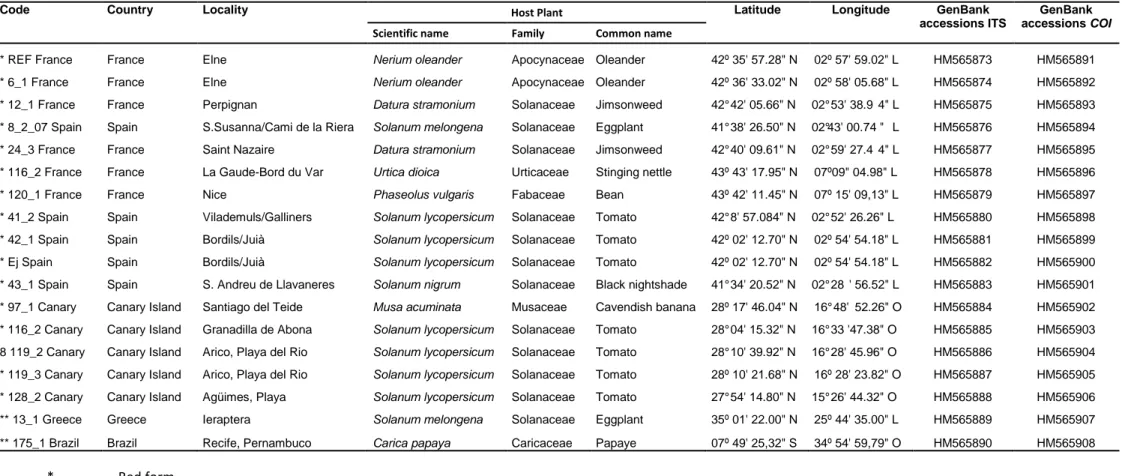
Table 1. Collection records of Tetranychus urticae mites from Brazil sequenced in this study sequenced and respective information on amplified region……….….. Table 2. Collection records of Tetranychus urticae mites from Paleartic region and Canada sequenced and respective information on amplified region……….……….. Table 3. PCR and sequencing primers used to obtain nuclear ribosomal ITS and mitochondrial
COI sequences of Tetranychus urticae………..
Table 4. Descriptive analyses and molecular diversity indices. Intraspecific information based on ITS1 and ITS2 regions of rDNA for Tetranychus urticae sequences……… Table 5. Descriptive analyses and molecular diversity indices. Intraspecific information based on sequences of the COI region of Tetranychus urticae specimens……….. Table 6. Accession numbers of COI sequences retrieved from Genbank for a set of Tetranychus
urticae. The haplotype number attributed in this study and the respective country and host plant
in Genbank are indicated. The sequences obtained in this study were included………... Table 7. Analysis of molecular variance (AMOVA) of Tetranychus urticae populations from several host plants from two distinct geographical regions, Mediterranean and Asian regions, based on COI sequences of the mtDNA……… Table 8. Analysis of molecular variance (AMOVA) of sets from Tetranychus urticae populations collected on several host plants in Asian region……….. Table 9. Exact test of Sample Differentiation based COI haplotypes frequencies of Tetranycus
urticae from Asian and Mediterranean regions, the Netherlands and Brazil………....
145 147 150 154 155 158 160 161 162
LIST – SUPPLEMENTAL MATERIAL – CHAPTER 4
Supplemental Material. Appendix 1. List of Tetranchus urticae ITS sequences retrieved from Genbank. Accession numbers were copied from Genbank. Information on host plants (common and family names), sample localities, the size of the sequence (bp) and original bibliographic references are given ……… Supplemental Material. Appendix 2. List of Tetranchus urticae COI sequences retrieved from Genbank. Accession numbers were copied from Genbank. Information on host plants (common and family names), sample localities, the size of the sequence (bp) and original bibliographic references are given ………...….
174
vii
Acknowledgements
I must start by acknowledging my supervisors Maria Navajas and Ivone Rezende Diniz. With their enthusiasm and sound advice they helped to accomplish my PhD. Their comments and suggestions on this thesis were most valued.
I am grateful to Denise Navia. Her work drove most of my initial interest and curiosity in mites which proved strong enough to have lead to my PhD. studies, and continues to do so. She provideds encouragement, good company, and lots of good ideas. I am grateful to the University of Montpellier II, University of Brasília, CBGP and Embrapa Cenargen staff for assisting me in many different ways.
I thank Alain Migeon, for help in mite collections and mite identification and Sandrine Cros-Arteil, for technical assistance in molecular techniques. I am grateful to Philippe Auger for his support and valuable comments on the thesis.
I wish to thank my colleagues Nathalie, Jean-Francois, Anne-Laure, Philippe Audio, Cecile, Frank, Gwenaelle, Silvy Hart, Christine Silvy, Roula, Jean-Philippe and Marie-Stephanie and Mirreille that were part of the CBGP team that helped me. We had fun together working in Montferrier-sur-Lez, France.
I would like to thanks the members of the PhD committee, Denis Bourguet, Elisabeth Fournier, George Roderick, Isabelle Olivieri and Benoît Facon for valuable suggestions on the research.
I am indebted to many of my friends who supported me to go through the difficult times overseas, Alain, Angham, Mirreille, Astrid, Sandrine, Vieira, Karen, Julia, Rosemaire and Francisco.
Deserve special mention all my best friends in the Laboratory for Plant Quarantine for all the emotional support, comraderie, entertainment, and caring they provided.
I am gratful to researchers, professors and PhD students from Brazil and Europe for helping with mite collections and providing samples.
I am thankful to Carlos H. W. Flechtmann, CNPq, ESALQ, USP, Samuel Paival and Patrícia Ianella, Embrapa Cenargen. With their great efforts to explain things clearly and simply they helped me in my research.
Last, but definitely not least, I owe my husband Dener, my boys Felipe e Diego and my family the utmost gratitude for all the love and understanding during the long years of my Ph.D. Thank you for enduring my absence. To them I dedicate this thesis.
viii
PREFACE
This thesis has been written in chapters, each one structured as a scientific article. It comprises two parts: the first includes an introduction and presents the context of the thesis as well as the literature referred to in the research. The second part encompasses four chapters: one scientific article that was submitted and accepted for publication in the International Journal of Acarology1; two articles that were submitted to the Journal of Insect Science2 and Experimental and Applied
Acarology3; and one article that is currently in progress4.
The data from the experiments was obtained by collecting mites from the Tetranychidae family in Brazil, in countries of the Palearctic region, and in Canada. The intention was to obtain samples of the Tetranychus urticae Koch.
The main tasks undertaken whilst carrying out research for the thesis were: processing and inspecting samples from the Tetranychidae mites that were collected in different regions and in host plants; preserving Tetranychidae mites on microscope slides; morphological identification of the samples collected; perform molecular analyses (extraction, PCR amplification and sequencing of DNA) of specimens identified as T. urticae; conduct phylogenetic analysis of sequence data. In order to obtain comprehensive information on the genetic variability, phylogeny and structure of the population of T. urticae in Brazil and elsewhere, the sequences obtained for this study were analyzed together with sequences of closely related species of
Tetranychus sensu stricto Tuttle and Baker (1968), available at Genbank, including those of T. cinnabarinus Boisduval, a supposed synonym.
1
Chapter 1. Renata S. Mendonça, Denise Navia, Carlos H. W. Flechtmann (2009). Two new spider mites (Acari: Tetranychidae) from Brazil - a Monoceronychus McGregor (Bryobinae) from fingergrass and an Oligonychus Berlese (Tetranychinae) from grape. International Journal of Acarology, 36(6) 2
Chapter 2 Renata S. Mendonça, Denise Navia, Ivone R. Diniz, Carlos H. W. Flechtmann (2010). Notes on Brazilian spider mites (Acari: Tetranychidae) – new hosts and localities. Journal of Insect
Science.
3
Chapter 3. Renata S. de Mendonça, Denise Navia, Ivone R. Diniz, Philippe Auger, Maria Navajas (2010). A critical review of some closely related species of Tetranychus sensu stricto Tuttle & Baker (1968) (Acari: Tetranychidae) in the public DNA sequences databases. . Experimental and Applied Acarology.
4
Chapter 4. Phylogeny, phylogeography and genetic structure in Tetranychus urticae Koch (Acari: Tetranychidae) populations inferred from ITS2 and COI DNA sequences. In progress.
ix
ABSTRACT
MENDONÇA, R. S. de. TAXONOMIC STUDIES OF TETRANYCHIDAE MITES IN BRAZIL WITH EMPHASIS IN THE PHYLOGENY AND POPULATION GENETIC STRUCTURE OF THE TWO-SPOTTED SPIDER MITE, TETRANYCHUS URTICAE KOCH, INFERRED FROM RIBOSOMAL AND MITOCHONDRIAL DNA SEQUENCES. 2010. 198 p. Thesis (Doctorate). SIBAGHE Systèmes Intégrés en Biologie, Agronomie, Géosciences, Hydrosciences, Environnement. Université Montpellier II Sciences et techniques du Languedoc, France.
The Tetranychidae family or spider mites comprises a large group of phytophagous mites with many of its species considered as important pests of agricultural crops worldwide. The two-spotted spider mite, Tetranychus urticae Koch (Prostigmata: Tetranychidae), an ubiquitous species causing outbreaks in many crops, is certainly the most studied species. Many aspects of its systematic, biology, feeding habits, and control have been studied. In Brazil, T. urticae is considered as one of the three main mite pests. Despite the unquestionable progress on tetranychid studies in the country, it is essential to gain in knowledge of these mites from unexplored regions or crops. The use of molecular tools to study phylogentic relationship among species and also population genetic structure of key pest species, such as T. urticae would enhance the understanding of the family. In this study we performed a survey of Tetranychidae mites from Brazil, including 15 States and the Federal District. A total of 550 samples of 120 different plant species were collected. Tetranychid mite infestations were confirmed in 207 samples, and 22 species belonging to seven genera of the Bryobiinae and Tetranychinae subfamilies were identified on 58 different host plants. Thirty-six new hosts for the Tetranychidae were recorded in Brazil, South America and worldwide for eleven species. New localities were registered for four tetranychid genera and a new record to South America was confirmed. Four species were identified as new for science: two belonging to the
Oligonychus Berlese genus, on grape (Vitis vinifera L.) and rose (Rosa sp.) from
Minas Gerais; and two belonging to the Monoceronychus McGregor and
Schizotetranychus Tragardh genera, both from weeping fingergrass (Eustachys distichophylla Lag. Nees) from Rio Grande do Sul. We also analyzed sequences of
the genus Tetranychus deposited in databases. We analyzed and evaluated the identity of 105 Genbank accessions of ITS2 rDNA and 138 COI mtDNA sequences which were deposited as T. urticae and as fourteen other taxa morphologically closely related to Tetranychus sensu stricto. In addition, ITS2 and COI sequences of 18 T. urticae samples unambiguously identified by morphological criteria were generated in this study and included in the analyzed dataset. Among the deposited sequences in the Genbank, numerous cases of apparently mistaken identities were identified in the group Tetranychus s. str., especially between T. urticae, T.
cinnabarinus, T. kanzawai and T. truncatus. The results suggest that nearly 30% of
the studied sequences may be unreliable (misidentified or dubious). In particularly this study sheds new light on the controversial taxonomic status of T. cinnabarinus and highlights the need of using combined morphological and molecular approaches to guaranty reliability of accessions in public databases. Information on genetic variability and structure of T. urticae populations from Brazil and worldwide was also performed. New sequences of ITS and COI were obtained from individuals collected in Brazil and some localities of the Palearctic Region (France, continental Spain, Canary Islands, Greece, Syria, Tunisia, Poland and Norway – plus one from
x
Canada). While significant differences were detected on population genetic structure of the analyzed samples according to the geographic region, any effect of the host plant was observed. Haplotype diversity inferred from both ITS and COI sequences was higher in samples from the Mediterranean basin. ITS sequences obtained from Brazil samples were homogenous, and two COI haplotypes were found, one of them also present in France, Spain and the Canary Islands and the other in Japan.
Key words: Acari, Tetranychinae, Bryobiinae, genetic variability, molecular markers, Neotropical region, misidentification.
xi
RESUMO
Mendonça, R. S. de. ESTUDOS TAXONÔMICOS DE ÁCAROS TETRANYCHIDAE NO BRASIL E FILOGENIA E ESTRUTURA GENÉTICA DO ÁCARO RAJADO,
TETRANYCHUS URTICAE KOCH, INFERIDAS A PARTIR DE SEQUÊNCIAS DO DNA RIBOSSÔMICO E MITOCONDRIAL. 2009 198 p. Tese (Doutorado), Instituto de Ciências Biológicas, Universidade de Brasília, DF.
A família Tetranychidae compreende um grande número de espécies estritamente fitófagas e inclui pragas importantes para a agricultura mundial. O ácaro rajado,
Tetranychus urticae Koch (Prostigmata: Tetranychidae) é uma espécie cosmopolita
que se destaca pelos elevados prejuízos ocasionados às lavouras. No Brasil, T.
urticae está entre os três principais ácaros pragas. Muitos aspectos sobre
sistemática, biologia, hábitos alimentares e controle têm sido investigados para a espécie. Apesar dos incontestáveis avanços procedentes de estudos com tetraniquídeos no Brasil, é importante expandir as pesquisas para novas regiões do país em plantas ainda não avaliadas. A exploração de novas áreas de conhecimento como os estudos moleculares que dão acesso a informações de variabilidade genética, filogenia e estrutura de populações podem também contribuir para o conhecimento dessa família de ácaros no país. Nesse estudo, realizou-se um levantamento de Tetranychidae no Brasil em 15 Estados e no Distrito Federal. Foram coletadas 550 amostras de 120 espécies vegetais. Infestações por ácaros tetraniquídeos foram confirmadas em 207 amostras e 22 espécies distribuídas em sete gêneros das subfamílias Tetranychinae e Bryobiinae foram identificadas em 58 hospedeiros diferentes. Foram registrados novos hospedeiros para tetraniquídeos no Brasil, na América do Sul e no mundo para 11 espécies. Quatro novas localidades foram registradas no Brasil para espécies presentes e uma nova ocorrência foi relatada no país. Quatro novas espécies foram encontradas: duas do gênero
Oligonychus Berlese, em uva (Vitis vinifera L.) e rosa (Rosa sp.) em Minas Gerais; e
duas dos gêneros Monoceronychus McGregor e Schizotetranychus Tragardh, ambas em capim coqueirinho (Eustachys distichophylla Lag. Nees) no Rio Grande do Sul. Visando a obtenção de informações sobre a variabilidade genética, filogenia e estrutura de populações de T. urticae no Brasil e no mundo foram realizadas análises filogenéticas e de variância molecular (AMOVA) baseadas em sequências do DNA ribossômico (ITS) e mitocondrial (COI) de 22 populações coletadas no Brasil e 33 na Região Paleártica (França, Espanha Peninsular, Ilhas Canárias, Grécia, Síria, Tunísia, Noruega e Polônia – e uma amostra extra do Canadá). Primeiramente, as sequências de T. urticae obtidas foram analisadas conjuntamente com aquelas de espécies próximas pertencentes ao grupo Tetranychus sensu stricto disponíveis no Genbank (105 ITS e 138 COI), incluindo o seu possível sinônimo, T.
cinnabarinus Boisduval. Nessa investigação foram detectadas incoerências que
levaram ao questionamento da confiabilidade de dados moleculares disponibilizados no Genbank. Entre as sequências analisadas, cerca de 30% de erro aparente de identificação taxonômica foi detectado, especialmente para T. urticae, T.
cinnabarinus, T. kanzawai Kishida e T. truncatus Ehara. As possíveis causas foram
discutidas e sugestões relacionadas a situação controversa do taxon T. cinnabarinus são apontadas. Em seguida, considerando um conjunto de dados constituído por sequências obtidas de exemplares de T. urticae inequivocamente identificados por critérios morfológicos, foram conduzidas as analises moleculares que indicaram a ocorrência de estruturação genética nas populações de T. urticae em função da
xii
localidade geográfica. O efeito da planta hospedeira não foi observado. A diversidade haplotípica, inferida pelas duas regiões do genoma (ITS e COI) analisadas foi maior entre os países banhados pelo mar Mediterrâneo. Não se observou variabilidade genética entre as populações de T. urticae coletadas nas cinco Regiões do Brasil quando se avaliou a região ITS. A presença de dois haplótipos mitocondriais (COI) foi constatada no Brasil, um compartilhado com a França, Espanha e Ilhas Canárias e outro com o Japão.
Palavras chaves: Acari, Bryobiinae, erros de identificação taxonômica, marcadores moleculares, região Neotropical, Tetranychinae, variabilidade genética.
xiii
RÉSUMÉ
MENDONÇA, R. S. de. ÉTUDES TAXONOMIQUES DES ACARIENS TETRANYCHIDAE AU BRÉSIL, EN PARTICULIER SUR LA PHYLOGENIE ET LA STRUCTURE GENETIQUE DES POPULATIONS DE L´ACARIEN JAUNE,
TETRANYCHUS URTICAE KOCH, INFERÉES À PARTIR DES SEQUENCES D´ADN RIBOSOMIQUE ET MITOCHONDRIAL. 2009, 198 p. Thèse (Doctorat), Institut des Sciences Biologiques, Université de Brasília, DF.
La famille des Tetranychidae est constituée d’environ 1250 espèces d’acariens phytophages. Parmi elles, une centaine est des ravageurs plus ou moins importants pour l’agriculture mondiale. L´acarien jaune, Tetranychus urticae Koch (Prostigmata: Tetranychidae) est une espèce ubiquiste qui est responsable de dommages importants qu´elle cause aux cultures. Divers aspects de la systématique, de la biologie et du contrôle des populations de cette espèce font actuellement l´objet de nombreuses recherches. Au Brésil, T. urticae fait partie des trois principaux acariens ravageurs des cultures. Malgré les avancées incontestables des études sur les tétranychidés, il est important d´étendre les recherches à de nouvelles régions du Brésil et pour des plantes hôtes n´ayant pas encore été évaluées. Les aspects moléculaires pour accedér des informations sur la relation phylogénétique entre d’espéces et sur la structure génétique des populations de ravageur, tel que T.
urticae, peut également améliorer la compréhension de la famille. Dans cette étude,
un inventaire des Tetranychidae du Brésil a été réalisé dans 15 États ainsi que dans le District Fédéral, avec 550 échantillons de 120 espèces végétales collectés. Des infestations par des acariens tétranychidés ont été confirmées dans 207 de ces échantillons. Vingt-deux espèces de tétranyques appartenant à sept genres de la sous-familles des Bryobiinae et Tetranychinae ont été identifiées chez 58 hôtes différents. Trente-six nouvelles plantes hôtes pour 11 espèces de tétranychidés ont été répertoriées au Brésil, en Amérique du sud ou dans le monde. De nouvelles localités ont été enregistrées au Brésil pour quatre tétranychides et une espèce a également été signalée pour la première fois en Amérique du sud. Quatre nouvelles espèces ont été découverte: deux appartenant au genre Oligonychus Berlese, présentes sur la vigne (Vitis vinifera L.) et le rosier dans le Minas Gerais; deux appartenant aux genres Monoceronychus McGregor et Schizotetranychus Tragardh, toutes deux présentes sur Eustachys distichophylla Lag. Nees, dans Rio Grande do Sul. La contribution à la systématique du genre Tetranychus a consisté à analyser conjointement des séquences d´ADN ribosomique (ITS) et mitochondrial (COI) de femelles de T.urticae, T. cinnabarinus (synonyme potentiel de T. urticae) et d’espèces très proches appartenant également au groupe Tetranychus sensu stricto. Les séquences on été obtenues à partir d’individus de T. urticae collectés dans cette étude et identifiés sans ambiguïté par des critères morphologiques et dans Genbank pour les autres espèces (105 ITS et 96 COI). Cette étude a mis en évidence des incohérences ce qui nous a amené à remettre en question la fiabilité des données moléculaires concernant le groupe Tetranychus s. str. disponibles dans Genbank. Nos résultats suggèrent que près de 30% des identifications taxonomiques parmi les séquences analysées sont fausses ou douteuses, surtout pour T. urticae, T.
cinnabarinus, T. kanzawai Kishida et T. truncatus Ehara. Cette étude apporte
également un éclairage nouveau sur le taxon controversé qu’est T. cinnabarinus et souligne la nécessité de l'utilisation combinée des approches morphologiques et moléculaires afin d’augmenter la fiabilité des séquences dans les banques de
xiv
données moléculaires publiques. Des données sur la variabilité génétique, la phylogénie et la structure de populations de T. urticae au Brésil et dans le monde sont ensuite présentées. De nouvelles séquences ITS et COI de 22 populations collectées au Brésil et de 37 prélevés dans la zone Paléarctique (France, Espagne, Iles Canaries, Grèce, Syrie, Tunisie, Pologne et Norvège – plus une du Canada) ont été analysés conjointement avec les séquences T. urticae déposées dans Genbank. Les résultats indiquent la présence significative d’une structuration génétique des populations de T. urticae par rapport à la localisation géographique. Cependant, l´effet de la plante hôte n´est pas observé. La diversité haplotypique, inférée à partir des ces deux régions du génome (ITS et COI) est plus élevée dans les pays du pourtour méditerranéen. La région ITS n´a pas permis d´observer de variabilité génétique entre les populations analysées. On a constaté la présence de deux haplotypes mitochondriaux (COI) au Brésil, l´un partagé avec la France, l´Espagne et les îles Canaries, et l´autre avec le Japon. Une discussion porte sur la nécessité de mener de nouvelles recherches en incluant d´autres régions du monde n´ayant pas encore été échantillonnées, ainsi que sur l’intérêt d´augmenter le nombre de populations dans certains pays déjà étudiés.
Mots-clés: Acari, Bryobiinae, Tetranychinae, erreurs d´identification, marqueurs moléculaires, région Néotropicale, variabilité génétique
xv
Introduction et contexte de la thèse
Tetranychus urticae Koch 1836 est une espèce qui présente une grande
importance agronomique au même titre que les autres de la famille Tetranychidae (Helle & Sabelis 1985). Cet acarien, qui est probablement originaire d´Eurasie, se serait disséminé par des moyens naturels dans les régions tempérées de l´hémisphère nord par le biais des activités humaines (Navajas et al. 1998). Cette espèce très polyphage a une distribuition cosmopolite (Bolland et al. 1998; Migeon & Dorkeld 2006). En conséquence il cause de sérieux dommages à de nombreuses cultures dans le monde entier et il est considéré comme l´un des acariens phytophages les plus importants (Helle & Sabelis 1985, Moraes & Flechtmann 2008). Au Brésil, en raison du nombre élevé d´espèces végétales à valeur économique qu’il attaque et en raison des dommages qu’il occasionne, T. urticae fait partie des trois principaux acariens ravageurs (Moraes & Flechtmann 2008).
De nombreux aspects portant sur la biologie et les habitudes alimentaires de
T. urticae ont fait l´objet de recherches. Cette espèce suscite un intérêt tout
particulier en ce qui concerne la sélection d’hôtes et les processus évolutifs puisque des différences sont rapportées entre des lignées (considérées comme des races) associées à des hôtes distincts (Gotoh et al. 1993). L´existence de différences génétiques parmi des populations en fonction de l’éloignement géographique, indépendamment de la plante hôte, est également mentionnée (Tsagkarakou et al. 1997). En plus de la différentiation génétique associée à la sélection par la plante hôte et par l’éloignement géographique, la présence de biotypes résistants à des pesticides est rapportée dans les pays où T. urticae a subi une intense pression de sélection par la lutte chimique (Miller et al. 1985; Edge & James 1986; Dennehy et al. 1987; Grafton-Cardwell et al. 1987; Flexner et al. 1988; Tian et al. 1992; Landeros et
al. 2006; Lee et al. 2006). Les problèmes de résistance de T. urticae aux acaricides
au Brésil ont été étudiés par Sato et al. (2000, 2002, 2004, 2007) et par Flores (2001). D’autre part, dans quelques ouvrages et publications, deux formes de l’espèce distinguées par la couleur du corps sont reconnues: la forme jaune correspondant à T. urticae et la forme rouge que certains acarologues considèrent comme une espèce à part entière appelée T. cinnabarinus (Boisduval 1867) (Boudreaux 1956; Boudreaux et Dosse 1963a, b, Meyer, 1974). Toutefois, les
xvi
différences morphologiques entre les deux formes étant très ténues et la coloration n'étant pas un critère taxonomique fiable, la validité du taxon T. cinnabarinus est rejetée par de nombreux auteurs (Pritchard et Baker 1955; Dupont 1979; Meyer, 1987; Baker & Tuttle 1994, Bolland et al., 1998). Actuellement, le problème de la synonymie entre ces deux espèces demeure un sujet de controverses.
Malgré l´importance économique de l´acarien jaune, il n´existe pas d´informations sur sa dissémination, sa sélection par les hôtes et sur la variabilité génétique dans différentes cultures et diverses régions géographiques au Brésil. Ces informations sont importantes car elles peuvent être à la base de l’élaboration de techniques spécifiques de contrôle de l´espèce notamment par le biais de la lutte intégrée contre les ravageurs. Elles sont également essentielles pour guider les mesures de mise en quarantaine visant à éviter l´introduction de nouveaux biotypes particulièrement agressifs, ou résistants à des pesticides.
Les populations de T. urticae et d’autres Tetranychidae sur les plantes cultivées sont devenues récurrentes que récemment notamment pour le soja cultivé dans le Rio Grande do Sul. Ces infestations semblent se produire plus fréquemment en fonction de l´avancée des zones de plantation mais aussi avec la mise en culture de nouvelles variétés ou en raison de modifications dans le système de conduite des cultures (Guedes et al. 2007).
Parmi les 1265 espèces de Tetranychidae répertoriées dans le Monde près de 100 sont signalées au Brésil (Bolland et al. 1998; Migeon & Dorkeld 2008). Comme la majeure partie des connaissances sur les tétranychidés s’applique à la région du Sud-est brésilien ou a des plantes cultivées, il est important d´étendre les connaissances faunistiques sur les acariens de cette famille aux autres régions du pays.
Objectifs et approches d’étude
Ce projet de recherche s’est fixé plusieurs objectifs: 1) appréhender la diversité des acariens Tetranychidae et leur spectre d’hôtes au Brésil par des inventaires réalisés dans différentes régions; 2) contribuer à la systématique moléculaire du genre Tetranychus; 3) participer à l’évaluation de la variabilité
xvii
génétique des populations de T. urticae à l´échelle mondiale en y incluant des populations du Brésil et 4) rechercher les effets liés à la géographie et à la plante hôte dans la structure génétique des populations de T. urticae.
Deux marqueurs moléculaires indépendants ont été utilisés pour les études de variabilité, de caractérisation génétique et de structuration des populations de T.
urticae: (1) un fragment de l´ADN mitochondrial, la région codant pour la Cytochrome oxydase I (COI); (2) la région du génome nucléaire “Internal Transcribed Spacer”
(ITS) de l´ADN ribosomique (Navajas 1998; Navajas et al. 1998; 2003). Les marqueurs génétiques sélectionnés dans ce travail sont les plus utilisés dans les recherches portant sur la systématique, la phylogénie des Tétranychidae. Lors de l’étude phylogénétique portant sur T. urticae, des séquences de cette espèce obtenues durant l´étude ont été analysées et comparées à celles d´espèces proches appartenant au groupe Tetranychus sensu stricto Tuttle & Baker (1968) (y compris T.
cinnabarinus son synonyme potentiel), disponibles dans la base de données
Genbank. Des incohérences au niveau de l’assignation de certaines séquences à certains taxa ont été révélées. Ces analyses sont présentées, discutées et des procédés visant à vérifier une plus grande fiabilité des bases de nucléotides à usage public sont suggérés.
Ce document de thèse est rédigé en anglais et il comporte deux parties: la première comprend l’introduction et la présentation du contexte de la thèse, la deuxième rassemble quatre chapitres correspondant à des articles publiés, soumis ou en préparation (un publié dans International Journal of Acarology1, un soumis à
Journal of Insect Science2, un à Experimental and Applied Acarology3 et un article qui est actuellement en cours de rédaction4).
1
Chapter 1. Renata S. Mendonça, Denise Navia, Carlos H. W. Flechtmann (2009). Two new spider mites (Acari: Tetranychidae) from Brazil - a Monoceronychus McGregor (Bryobinae) from fingergrass and an Oligonychus Berlese (Tetranychinae) from grape. International Journal of Acarology. 36(6). 2
Chapter 2. Renata S. Mendonça, Denise Navia, Ivone R. Diniz, Carlos H. W. Flechtmann (2010). Notes on Brazilian spider mites (Acari: Tetranychidae) – new hosts and localities. Journal of Insect
Science.
3
Chapter 3. Renata S. de Mendonça, Denise Navia, Ivone R. Diniz, Philippe Auger, Maria Navajas (2010). A critical review of some closely related species of Tetranychus sensu stricto Tuttle & Baker (1968) (Acari: Tetranychidae) in the public DNA sequences databases. . Experimental and Applied Acarology.
4
Chapter 4. Phylogeny, phylogeography and genetic structure in Tetranychus urticae Koch (Acari: Tetranychidae) populations inferred from ITS2 and COI DNA sequences. In progress.
1
2
INTRODUCTION
The two-spotted spider mite Tetranychus urticae Koch, 1836, is a very important species to agriculture, as others species from the Tetranychidae family (Jeppson et al. 1975; Helle & Sabelis 1985). This mite probably originated in Eurasia in light of the high frequency of samples from this region and from the variety of host plants on which it has been collected. It has been spread throughout the temperate regions of the northern hemisphere by wind and throughout the world via the transport of plants by man (Navajas 1998). Now it is a species of cosmopolitan distribution and highly polyphagous, infesting both cultivated and non-cultivated plants (Bolland et al. 1998; Migeon & Dorkeld 2006). For this reason and owing to the serious damage it causes to various agricultural crops throughout the world, it is regarded as one of the main phytophagous mites (Helle & Sabelis 1985). In Brazil, because of the high number of economically valuable vegetable species that are attacked and damaged, T. urticae is considered one of the three main pest mites in the country (Moraes & Flechtmann 2008) and its infestations has been recorded in many crops (Flechtmann 1985; Souza Filho et al. 1994; Gallo et al. 2002).
Many aspects of the biology and feeding habits of T. urticae have been the focal point of research. Results regarding host selection and evolutionary processes have revealed differences between strains of T. urticae, which in some cases were regarded as races associated with distinct hosts (Gotoh et al. 1993). However research also mentioned genetic differences among populations due to the distance between the geographical regions analyzed, regardless of the host plant (Tsagkarakou et al. 1997). Besides genetic differences due to host plant selection and geographic regions, the presence of biotypes resistant to pesticides has been reported in countries where T. urticae experienced intense selection pressure (Miller
et al. 1985; Edge & James 1986; Dennehy et al. 1987; Grafton-Cardwell et al. 1987;
Flexner et al. 1988; Tian et al. 1992; Landeros et al. 2006; Lee et al. 2006). Problems related to T. urticae resistance to insecticides/miticides in Brazil have been documented by Sato et al. (2000, 2002, 2004 and 2007) and Flores (2001). Despite some local differences among T. urticae populations, DNA sequence analysis of mites collected at a continental scale, tent to show high homogeneity of the species resulting from intense movements often human mediated (Navajse et al , 1988). Two forms of the species can be recognized by their green or red body colour. According
3 to De Boer (1985), the body colours observed in mites result from the pigments found in the hemolymph. However, the red type of T. urticae has been often called T.
cinnabarinus (Boisduval 1867) (Boudreaux 1956; Boudreaux & Dosse 1963a, b,
Meyer 1974). The accuracy of the taxon T. cinnabarinus has been questioned by many authors (Pritchard and Baker 1955; Dupont 1979; Meyer 1987; Baker & Tuttle 1994) and it remains to this day a point of discussion.
Despite the economic importance of the two-spotted spider mite, there is no information regarding its genetic variability in different crops and geographic regions of Brazil. This kind of information is important and would be helpful in adopting phytosanitary measures designed to manage the mite. It is also essential in justifying quarantine methods to prevent introducing new T. urticae biotypes that are adapted to and aggressive towards different hosts, or resistant to different pesticides, as reported in other countries.
Infestations by the two-spotted spider mite and other Tetranychidae have posed economic problems in crops where damage was previously either not observed or only limited to specific areas, as in soybeans cultivated in Rio Grande do Sul. These infestations probably take place because of expanding cultivated areas, adapting new varieties, and/or changes crops management (Guedes et al. 2007).
Approximately 100 described species of Tetranychidae have already been reported in Brazil (Bolland et al. 1998; Migeon & Dorkeld 2008). However, the most intensive search for Tetranychidae mites in the country has been concentrated mostly on the southeast region and usually pertains to economically important crops. Agriculture has expanded towards new regions in Brazil and is encroaching biological reserves, it is important to continue studying tetranychids in unexplored areas and to gather new information that would enhance our understanding of this family.
GENERAL OBJECTIVES
The general objectives of this research were: cooperate to the understanding of mites from the Tetranychidae family in Brazil; contribute to the molecular systematics of the genus Tetranychus; obtain information on the genetic variability of T. urticae populations in a global context, including new populations from Brazil and the Palearctic region, and investigate the geographic and the host plant effects upon the genetic structure of T. urticae populations.
4
STRATEGIES
To conduct studies on Tetranychidae mites in Brazil, samples of mites were collected from several hosts ranging from ornamental, fruit-bearing, vegetable, grass, oilseed, and large crop species plants found in five regions in Brazil that included 15 states and the Federal District. With the support of the Embrapa Team of Genetic Resources and Biotechnology, and collaborators from universities and research institutes, a wide range of sampling was carried out in the country. Plants were collected when they presented symptoms of tetranychid attacks, according to Moraes & Flechtmann (2008), and the target organism was the two-spotted spider mite, T.
urticae.
For studies on variability, genetic characterization, and population structure of
T. urticae, two target DNA fragments were PCR-amplified and sequenced: (1) the
intergenic “Internal Transcribed Spacer” (ITS) of the nuclear ribosomal DNA; (2) a fragment of mitochondrial DNA, the Cytocrome Oxidase I (COI) gene; (Navajas 1998; Navajas et al. 1998; Navajas & Boursot 2003). Genomic DNA was extracted from single adult females from each population. The genetic markers chosen in this study are the most commonly used for research on systematics, phylogeny and evolution, and are very useful for studying populations of Tetranychidae mites. Tetranychus
urticae populations used in the molecular studies were those collected in Brazil, plus
populations collected in seven Palearctic countries with the support of the acarology staff of the Center for Biology and Management of Populations (CBGP), Monferrier-sur-Lez, France, oher collaborators from Europe, and including one sample from Canada (specimens from the mite rearing reference of the Genoma T. urticae Project). The obtained DNA sequences were used to investigate genetic variability, phylogeny, phylogeographic patterns, and the association to host plants in T. urticae populations collected in several regions of the world.
During the course of the phylogenetic studies of the two-spotted spider mite, the sequences uncovered in this study and those available at GenBank were analyzed together with closely related species belonging to the group Tetranychus
sensu stricto Tuttle & Baker 1968 (including its possible synonym T. cinnabarinus),
obtained from Genbank. Inconsistencies were detected, leading to doubts on the reliability of data provided by GenBank.
5 This study reported new taxonomic information on tetranychid mites in Brazil. It has uncovered that several DNA sequences that were publically available and attributed to economically relevant taxa, are incorrectly named. It also sheds new light on the controversial taxonomic status of T. cinnabarinus and highlights the need to use combined morphological and molecular approaches to ensure the reliability of access to public databases. The genetic variability, phylogeographic patterns, and the association to host plants in T. urticae populations collected in several regions of the world were investigated.
SPECIFIC OBJECTIVES:
1) Conduct a survey of Tetranychidae mites in Brazil on different host plants and in different regions of the country
2) Describe new taxa for Tetranychidae family
3) Contribute to the molecular systematics of the genus Tetranychus
4) Investigate the factors affecting the genetic variability of T. urticae populations in a global context through phylogenetic analyses based on sequences of the nuclear and mitochondrial genome, including new sequences obtained from samples collected in Brazil and the Palearctic region, and inspect the geographic and host plants effects upon the genetic structure of T. urticae populations.
THE CONTEXT OF THE THESIS
The Tetranychidae Donnadieu family includes a large group of phytophagous mites (spider mites) (Jeppson et al. 1975; Helle & Sabelis 1985). Thus far, this family includes about 1,265 species, distributed in 76 genera. South America has recorded 186 tetranychid species, 105 of them being in Brazil (Bolland et al. 1998; Migeon & Dorkeld 2006).
Among the phytophagous mites of the Tetranychidae family, the genus
Tetranychus Dufour 1832 includes 148 valid species (Migeon & Dorkeld 2006) and
comprises several species of high economic and quarantine importance in agriculture. This genus provides a good example of organisms where species
6
identification might be misleading. Systematics has traditionally relied on the diversity of a few minute morphological characters, mainly based on microscopic examination. Not only males might be rare and difficult to find in field populations, but also the shape of the aedeagus (part of the male genitalia) must be known in order to discriminate closely related species. Thus, both sexes are often needed to confirm specie identification. As a result, morphologically based identification of Tetranychus species requires advanced taxonomic expertise, especially in morphologically similar taxa, as it is the case with species belonging to the group Tetranychus (Tetranychus) (Tuttle & Baker 1968), or Tetranychus sensu stricto, from the ninth species-group as defined by Flechtmann & Knihinicki (2002). In this group, the two-spotted spider mite,
Tetranychus urticae, a highly polyphagous species causing outbreaks in many crops
worldwide, is certainly the most studied species.
The wide range of geographical and ecological diversity of the two-spotted spider mite has prompted many studies on its morphological traits (Boudreaux 1956; Bund & Helle 1960; Boudreaux and Dosse 1963a, b, Pritchard & Baker 1955; Meyer 1974, 1987; Baker & Tuttle 1994; Ehara 1999; Carbonelle et al. 2007) and on its molecular systematics (Gotoh & Tokioka 1996; Navajas et al. 1996; Navajas et al. 1998, Ehara 1999; Hinomoto et al. 2001; Xie et al. 2008; Li et al. 2009).
Pest status of the two-spotted spider mite, Tetranychus urticae Koch 1836
Outbreaks and economical impact in worldwide agriculture
The two-spotted spider mite, Tetranychus urticae (Prostigmata: Tetranychidae) infests around 1,094 vegetable species in 109 countries around the world.. Among the 19 most important agricultural crops in world agriculture, six (citrus, cotton, grapes, apples, beans, and papaya) were reported as being hosts of the two-spotted spider mite (Yaninek & Moraes 1991). Damage caused by T. urticae is usually described in many other crops; however damage resulting by its infestations is not always quantified.
In cotton, T. urticae infestations have cause reductions of 14% to 44% in seed production, 4% in fat content, 17% in fiber weight (Cannerday & Arant 1964) as well
7 as 4% in fiber resistance (Oliveira 1972), and 25% in weight loss (Oliveira & Calcanhoto 1975).
In ornamental plants, damages brought upon cloves, for example, reached between 20% and 30% (Rajashekharappa & Mallik 2008).
Damage by T. urticae to citrus plants has been noted by McMurtry (1985) and infestations have been reported in Spain, Greece, Cuba, Morocco, and in Florida in the United States (Ramos et al. 1988; Cáceres & Childers 1991; Tsagkarakou et al. 1997; Allam et al. 1999; Miranda et al. 2003). In the Mediterranean region, damages have decreased the value of commercial fruits (Jacas & Garcia 2001; Aucejo et al. 2003; Urbaneja et al. 2008).
In Europe, T. urticae is regarded as one of the main pests attacking grapevines (Kassemeyer 1996) and its presence has been reported in France (Rambier 1958), Spain (Arias Giralda & Nieto Calderón 1982), Switzerland (Baillod et
al. 1982), and the former Czechoslovakia (Hluchy & Pospisil 1991).
Okra cultivation in India presented losses between 12% and 36% in fruit production (Prasad & Singh 2007) and, in Iran, caused losses of 13% to 70% in the weight of fruits (Anaraki et al. 2007). Damage to tomatoes has been outlined by various authors (Barbosa & França 1980; Minami 1980; Flechtmann 1985; Gonçalves et al. 1998; Aragão et al. 2002).
In the United States, T. urticae populations are responsible for reducing the amount of fruit in strawberry fields (Sances et al. 1982). In Africa, production loss can reach 50% (Coates 19741, cited by Wysoki 1985). In Brazil, the two-spotted spider mite is the main pest in strawberry fields in the states of São Paulo and Rio Grande do Sul, and is found especially during fruit-bearing periods and during harvests, rendering most of the harvest unviable, given the damage and suspected contamination from pesticides (Tanaka et al. 2000; Moraes & Braun 2002).
1
COATES, T.J.D. (1994). The influence of some natural enemies and pesticides on various populations of Tetranychus cinnabarinus (Boisduval), T. lombardinii Baker & Pritchard and T. ludeni Zacher (Acari: Tetranychidae) with aspects of their biologies. Entomology Memoir, 42. Department of Agricultural Technical Services, Republic of South Africa.
8
Pest control
Chemical methods are most common for controlling T. urticae and have been studied in detail (for example: Watanabe et al. 1994; Heungens & Tirry 2001; Botton
et al. 2002; Kimura et al. 2005; Barros et al. 2007). Alternatively, conventional
biological control using predator mites (Acari: Phytodeiidae) has been carried out in different countries (Khalequzzaman et al. 2007; Ferguson 2008; Moraes & Flechtmann 2008). In Brazil, predator mites Neoseiulus californicus (McGregor, 1954) and P. macropilis (Banks, 1905) are being used to control T. urticae on fruit-bearing plants (apple and strawberry), ornamental plants, and in greenhouse crops (Monteiro et al. 2002; Marchetti et al. 2003; Ferla 2004; Marchetti & Ferla 2004; Ferla
et al. 2007; Oliveira et al. 2007). Biological control along with chemical methods
where biotypes of predator mites resistant to certain pesticides are used has provided a promising alternative for T. urticae control in the United States, Ukraine, South Africa and Australia (Pringle 2001; Waite 2001). In Brazil, similar studies have been conducted on the predators N. californicus and P. macropilis (Banks) (Sato et
al. 2006, 2007; Poletti et al. 2008).
Biological traits of T urticae favouring its pest status
The reproduction of T. urticae occurs via facultative parthenogenesis. In the absence of sexual reproduction, females have the ability to reproduce asexually. The ovaries are undergo complete development producing only haploid males. Following the first copulations, females produce predominately diploid females, which lead to haplodiploidy, whereby males are produced by arrhenotokous parthenogenesis and females by sexual reproduction. Therefore, a single female can generate offspring (Helle & Pijnacker 1985). In addition, the species has great plasticity to change and feed on different host plants, which has probably contributed to the success of the worldwide colonization.
The presence of biotypes
Biotypes mean populations of a given species that have particular biological characteristics. The term “host race” represents a differentiated form, which may or
9 may not crossbreed, but that feeds on specific host plants. This term is often used in entomological research and the term biotype must take precedence over “host race” (Futuyma 1995). Differences in plant damaging observed among populations of phytophagous organisms can be linked to geographic distribution or host plant selection. For example, the red form of T. urticae infests and causes damage to citrus orchards in the Mediterranean region, particularly in Spain (Aucejo et al. 2003; Ansaloni et al. 2006; Abad et al. 2006, Urbaneja et al. 2008), that commercializing in
natura fruit, especially oranges, tangerines, and lemons (Neves & Marino 2002). However, T. urticae has not been reported damaging Brazilian citrus orchards. The reason why this species, which harms various hosts in Brazil, has not been found damaging citrus in the country remains unknown. This is an relevant example on the importante of studies on population genetic structure and population differenciation to address agricultural issues.
Another important factor in investigating genetic differences among T.
urticae populations is the development of resistance to insecticides and miticides that
results from excessive use of these products in attempts to control mites on crops. The development of resistance to pesticides in T. urticae populations can provide the emergence of new biotypes that embarrass control and frustrate attempts to adopt Integrated Pest Management practices.
The resistance of T. urticae to pesticides has been documented in different crops in several countries (Cranham & Helle 1985; Miller et al. 1985; Edge & James 1986; Dennehy et al. 1987; Grafton-Cardwell et al. 1987; Flexner et al. 1988; Tian et
al. 1992; Landeros et al. 2006; Lee et al. 2006). Croft & Hoyt (1983)2, cited by Prokopy & Croft (1994), stated that T. urticae are the most difficult and burdensome pest to control in apple orchards in North America, due to their capacity to develop resistance to insecticides. In Brazilian apple orchards, T. urticae is not one of the main pests. In Brazil, problems with resistant T. urticae population to pesticides have been reported in cotton plants, rose bushes, strawberry fields, and peach trees (Chiavegato et al. 1983; Suplicy Filho et al. 1994; Sato et al. 1994, 2000, 2002, 2004, 2007; Flores 2001). Nevertheless, the introduction of resistant populations to
2
CROFT, B.A.; HOYT, S. C. (1983) Integrated management of insect pests of pome and stone fruits. New York: Wiley Interscience. (454p.).
10
insecticides or aggressive T. urticae populations would compromise integrated pest management programs currently used for orchards in the south of the country and would cause negative impacts on the production chain of Brazilian apples.
The risk of introducing and establishing new T. urticae biotypes that can possibly worsen economic problems in the country by importing plant products must be taken into account (Ferreira 1997). Studies on genetic variability in Brazilian T.
urticae populations can help distinguish between populations already present in the
country and introduced biotypes.
The use of molecular markers in Acarology
Choosing genetic markers with the adequate variability to the taxonomical level (e.g. genus, species, populations) to be examined is the first step to any study based on genetic differenciation. For istance, when comparing populations, genetic markers must be highly polymorphic to allow the detection of individual movements, which might be minimal. It is necessary to be familiar with the general properties, the fields of application, and the limits of each marker in order to ensure a choice that is compatible with the research objectives.
Studies with molecular markers can aid in understanding systematics, phylogenetics, and population structures in Acarology, and molecular techniques are becoming more and more used tools for taxonomists and acarologists in general. They help not only in separating related species but also differentiating between subspecific categories and characterizing biotypes, thereby benefitting researchers involved in phytosanitary defence, biological control, parasitology, population biology, molecular ecology, control, and various other areas.
The first studies using molecular markers in acarology began in the 1990s (Kaliszewski et al. 1992; Navajas et al, 1994; Mozes-Koch & Gerson 1998). Since then, reviews on the use of molecular markers in systematic and phylogenetic studies of mites comprising information on the methods, the main results, and perspectives have been published by Navajas & Fenton (2000); Navajas (2001), Cruickshank (2002) and Navajas & Roderick (2008).
Mites species of agricultural importance and displaying great diversity, have been the focus of studies using molecular techniques. These mainly belong to the Tetranychidae and Eriophyidae phytophagous families, and to the Phytoseiidae
11 predator mites (Navajas et al. 1992, 1998, 1999, 2000; Tsagkarakou et al. 1999; Navajas & Fenton 2000; Noronha et al. 2003; Tixier et al. 2006, Navajas & Navia 2010).
As for many other arthropods, the most commonly used markers in molecular systematics of mites are mitochondrial DNA (mtDNA) and nuclear ribosomal DNA (rDNA). They differ considerably in terms of genomic organization (in that one is of mitochondrial and the other of nuclear), evolution mode (they present differential rates of evolution) and transmission (maternal and biparental transmission, respectively). Given the importance and utility of mtDNA and rDNA markers in Acarological research, their general characteristics, use, as well as the most important subjects related to them, have been revised.
Phytophagous mites - nuclear ribosomal DNA and mitochondrial DNA
Phylogenetic studies seek to reconstruct the evolutionary history of groups of organisms (Amorim 1994). Recent advances in population genetic theories along with rapid advances in molecular techniques have allowed access to detailed phylogenetic information on populations, in which different haplotypes represent operational taxonomic units (OTU) (Crandall & Templeton 1996; Harvey & Nee 1996). DNA sequences can be important tools in reconstructing evolutionary relationships among organisms (Weekers et al. 2001).
However, it is widely admitted that the phylogenetic content of a single genetic marker gives only a partial view of the phylogeny and the haplotype ancestry reconstruction of the species.. It is then important to consider the way in which the contrasting properties of markers can affect their capacities to construct phylogenies (Navajas & Boursot 2003), and that phylogenetic studies based on the results of a single molecular marker are unreliable and can lead to unfounded conclusions (Brower et al. 1996; Ross et al. 1999; Navajas & Boursot 2003). For example, phylogenetic analyses using sequences of cytochrome oxidase 1 regions (COI) of mtDNA and the internal transcribed spacer (ITS2) of rDNA of two closely related species of the genus Tetranychus, T. urticae and T. turkestani, showed that these two species are polyphyletic for mtDNA while monophyletic for rDNA. The nucleotide diversity was extensive for the mtDNA COI gene, 3-4%, and extremely low for the ITS2 region at less than 0.5%. The results indicated that, despite biparental
12
transmission and multiplicity of copies in the genome, nuclear rDNA had a smaller effective population size than mtDNA in these species (Navajas & Boursot 2003). The authors argue that the combination of efficient concerted evolution and/or gene conversion in the rDNA cluster, the haplodiploidy of these species and the female-biased sex ratio could account for this apparent contradiction in relation to the expected pattern. Likewise, the the phylogenetic contect of these to markers was contrasted when comparinig the the phylogeny of Japanese species in the
Panonychus Yokoyama genus, based on sequences of the COI region (mtDNA), and
those obtained using regions of the rDNA (Toda et al. 2000).
In general, the mutation rate of mtDNA sequences tend to be higher that those of nuclear genes and are appropriate for population studies. Because of the haploid nature of mitochondrial genes, the frequency of a single mtDNA haplotype within a population should fluctuate more rapidly than the frequencies of nuclear DNA. Thus, mtDNA should be more sensitive to founder events and small population sizes than nuclear DNA. Consequently, the probable loss or gaining of a mtDNA haplotype will be significant for small populations and only a small migration among populations would be sufficient to fix a mtDNA lineage in all populations (Roderick 1996).
Navajas et al. (1998) noted in T. urticae populations with different origins sampled in a wide geographic area, approximately 5% divergence of nucleotides in the COI region of the mtDNA, while no variation was found in the ITS2 region. The authors also compared the evolution rates of these two markers in one group of species of the same genus and did not obtain the same results, realizing that in other
Tetranychus species, the ITS2 region also varied. The results indicated that the ITS
region evolved 2.5 times faster than the COI region in Tetranychus species. The authors suggested that the homogeneity of ITS2 sequences in T. urticae was due to its capacity to colonize and consequently, to its relatively high gene flow.
The utility of these markers in tracing invasive species was shown in a study on Mononychellus progressivus Doreste (Navajas et al. 1994). The genetic structure of this invasive mite native to the Neotropical region and accidentally introduced to East Africa in the 1970s, was studied using COI and ITS2 sequences. The results suggested that M. progressivus was introduced from populations originating in Colombia (Navajas et al. 1994). This is relevant information to guide the search in the native are of the pest of adecuate biological control agents to be introduced in Africa.
13 Likewise, based on the analyses of ribosomal DNA sequences (ITS) and mitochondrial DNA sequences (16S and COI), Navia et al. (2005) studied the phylogeography of Aceria guerreronis Keifer (Eriophyidae) populations worldwide, and suggested that the species originated in America and determined the introduction routes in Africa/Asia.
Another important use of molecular data in Acarogly is to address taxonomic issues. The genus Amphytetranychus Oudemans was restored based on the convergence of molecular (ITS2 and COI) and morphological data (Navajas et al. 1997). The molecular approach was also useful to propose the species status of T.
pueraricola (Ehara & Gotoh) by discriminating it from T. urticae Koch (Navajas et al.
1998). Navajas et al. (2001) established the synonymy between T. kanzawai Kishida and T. hydrangea Pritchard & Baker, based on analyses of the ITS2 sequences and on crossbreeding experiments. Intraspescific variation was reported in T. kanzawai in studies with ITS and COI sequences (Hinomoto & Takafuji 2001).
Genetic data to study ecological differenciation in Tetranychidae
Relationships between phylogeny and geographic distribution represent the phylogeographic patterns for a given species. In addition, environemental conditions might select specific genotypes adapted to specific a habitat which drives population differenciation.
The genetic structure of a species can be inferred indirectly through genetic data. This data can be analyzed in two distinct ways: 1) populations are treated as genetic units and measurements of similarity among populations are estimated, and 2) phylogeographic analysis, in which relationships between genotypes (phylogenies of alleles or haplotypes) of one or more populations can be investigated according to its geographic location. Variations in DNA sequences are analyzed to establish relations between haplotypes of the same loco (Roderick 1996).
In Europe and Asia, genetic differentiation among T. urticae populations has been studied using genetic markers, together with the evaluation of biological, behavioural and morphological parameters (Gotoh et al. 1993; Hence et al. 1998; Navajas 1998; Tsagkarakou et al. 1999; Navajas et al. 2000). The results indicate that the variability of the species can be linked to geographic distribution but also to