OmpC and the
s
E
regulatory pathway are involved
in adhesion and invasion of the Crohn’s
disease-associated Escherichia coli strain LF82
Nathalie Rolhion, Frédéric Antonio Carvalho and Arlette Darfeuille-Michaud*
Univ Clermont 1, Pathogénie Bactérienne Intestinale, USC INRA 2018, Clermont-Ferrand F-63000, France.
Summary
Ileal lesions of 36.4% of patients with Crohn’s disease (CD), an inflammatory bowel disease in humans, are colonized by pathogenic adherent-invasive Escheri-chia coli (AIEC), and high levels of antibodies directed against E. coli OmpC are present in 37–55% of CD patients. We therefore investigated the expression of OmpC and its role in the interaction of CD-associated adherent-invasive E. coli strain LF82 with intestinal epithelial cells. High osmolarity induced a significant increase in the ability of LF82 bacteria to interact with Intestine-407 cells, which correlates with increased OmpC expression. Deletion of ompC gene markedly decreased the adhesion and invasion levels of the corresponding mutant. A LF82-DompR mutant impaired in OmpC and OmpF expression, showed decreased adhesion and invasion, and unlike a K-12-negative OmpR mutant did not express flagella and type 1 pili. Interestingly, the wild-type phenotype was restored when OmpC or OmpF expression was induced in the LF82-DompR mutant. Overexpression of RpoE in the LF82-DompR isogenic mutant restored a full wild-type phenotype without restoring OmpC expression. Increased expression of RpoE was observed in wild-type strain LF82 at high osmolarity. Hence, the role of OmpC in the AIEC LF82 adhesion and invasion is indirect and involves thesEregulatory
pathway.
Introduction
Crohn’s disease (CD) is an inflammatory bowel disease of unknown aetiology in humans (Duchmann et al., 1999). CD has features that might be the result of a microbial process in the gut (Sartor et al., 1996; Elson, 2000;
Podolsky, 2002; Shanahan, 2002). Various studies have addressed the hypothesis that pathogenic bacteria con-tribute to the pathogenesis of inflammatory bowel disease (Burke and Axon, 1988; Liu et al., 1995; Sartor et al., 1996; Schultsz et al., 1997; Lamps et al., 2003). Of the bacteria that may play a role in pathogenesis of CD, Escherichia coli strains have been assigned a putative role. E. coli are abnormally predominant (between 50% and 100% of the total number of aerobes and anaerobes) in early and chronic ileal lesions of CD and most E. coli strains isolated from the ileal mucosa of CD patients adhere to intestinal epithelial cells (Darfeuille-Michaud et al., 1998). In addition to their ability to adhere, E. coli strains isolated from 36.4% of CD patients are able to invade intestinal epithelial cells (Darfeuille-Michaud et al., 2004) and belong to a new pathogenic group of E. coli, designated AIEC for adherent-invasive E. coli (Boudeau et al., 1999). Intramucosal E. coli were found in 29% of CD patients versus 9% of controls and these E. coli strains possess invasive ability (Martin et al., 2004).
Electron microscopic examination of AIEC reference strain LF82-infected intestinal epithelial cells revealed a macropinocytosis-like process of entry dependent on actin microfilaments and microtubule recruitment, and characterized by the elongation of membrane extensions, which surround the bacteria at the site of contact between entering bacteria and epithelial cells (Boudeau et al., 1999). Type 1 pili-mediated adherence plays an essential role in the invasive ability of strain LF82 by inducing membrane extensions (Boudeau et al., 2001) and nucle-otide sequences of fim genes revealed that strain LF82 produces variant of type 1 pili compared with those of E. coli K-12. We also reported that these type 1 pili vari-ants have to be expressed in the genetic background of strain LF82 in order to promote bacterial uptake because their expression in E. coli strain K-12 is not sufficient to confer invasiveness. Flagella also play a direct role in the adhesion-invasion process of AIEC strain LF82 via motil-ity, and an indirect role in the interaction between bacteria and epithelial cells by downregulating the expression of type 1 pili and of unknown factor(s) involved in invasive-ness that remain(s) to be investigated (Barnich et al., 2003). In addition, the lipoprotein NlpI, which is probably located in the inner-membrane, is thought to operate in a Accepted 15 January, 2007. *For correspondence. E-mail
arlette.darfeuille-michaud@u-clermont1.fr; Tel. (+33) 4 73 17 79 97; Fax (+33) 4 73 17 83 71.
regulatory pathway involved in the synthesis of flagella, type 1 pili, and other virulence factors yet to be identified (Barnich et al., 2004). We also found that the YfgL lipo-protein is required to confer invasive ability on AIEC strain LF82 independently of type 1 pilus and flagellum expres-sion, but in correlation with release of outer membrane vesicles in which bacterial effectors are entrapped and contribute to the invasion process (Rolhion et al., 2005).
Recent studies have reported an abnormal reactivity to microbial components, including E. coli outer membrane protein OmpC, in patients with CD. High levels of antibod-ies directed against E. coli OmpC are present in 37–55% of patients with CD, whereas no more than 5% of non-inflammatory bowel disease individuals express anti-OmpC (Arnott et al., 2004; Beaven and Abreu, 2004; Mow et al., 2004). Reactivity to E. coli OmpC is associated with severe CD characterized by small bowel involvement, frequent disease progression, longer disease duration, and greater need for intestinal surgery (Landers et al., 2002; Mow et al., 2004). Thus, the aim of the present study was to investigate the expression of OmpC and its role in the interaction of CD-associated adherent-invasive E. coli strain LF82 with human intestinal epithelial cells.
Results
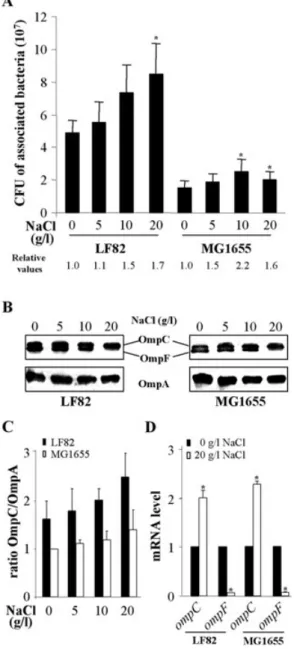
Increased adhesion and expression of OmpC in AIEC strain LF82 after bacterial growth at high osmolarity A significant increase in the number of AIEC LF82 bacteria associated with intestinal epithelial cells was observed when bacteria were cultured in the presence of 20 g l-1
NaCl compared with those grown without NaCl (Fig. 1A). The K-12 strain MG1655 showed lower adhesion levels than those of strain LF82. However, relative values of the number of cell-associated bacteria after growth at increased osmolarities compared with growth in the absence of NaCl indicated similar increases in the adhe-sion levels for K-12 and AIEC LF82 bacteria (Fig. 1A). Analysis of the expression of OmpA, OmpC and OmpF for AIEC LF82 and MG1655 bacteria grown at various osmo-larities was performed by Western immunoblotting using anti-OmpA and anti-OmpC/F antibodies. After growth of LF82 or MG1655 bacteria in the presence of increased concentrations of NaCl from 5 to 20 g l-1, OmpA expression
was not affected, that of OmpF decreased, and that of OmpC increased (Fig. 1B). As shown by the determination of the ratios OmpC/OmpA (Fig. 1C), a 1.5-fold increases in OmpC expression was observed for the AIEC strain LF82 when bacteria were grown at high osmolarity compared with bacteria grown in the absence of NaCl. For K-12 strain MG1655, a 1.4-fold increase in OmpC expression was observed under such conditions. Comparison of OmpC expression in AIEC strain LF82 and K-12 strain MG1655
Fig. 1. Effects of high osmolarity in AIEC strain LF82 and K-12
strain MG1655.
A. Effects of bacterial growth in LB with increased concentrations of NaCl (0–20 g l-1) on adhesion to Intestine-407 cells. Cell-associated
bacteria were quantified after a 3 h infection period. Each value is the mean number of cfu⫾ SEM of at least four separate
experiments. *P< 0.05 comparatively to numbers of cfu for bacteria grown at low osmolarity. Relative values: fold variation of number of cell-associated bacteria LF82 or MG1655 after an overnight growth with increased concentrations of NaCl relative to those of LF82 or MG1655 after growth in the absence of NaCl.
B. Western immunoblot analysis of OmpC, OmpF and OmpA in LF82 and MG1655 after an overnight incubation of bacteria with increased concentrations of NaCl.
C. Fold variation of ratios OmpC/OmpA relative to that of K-12 strain MG1655 grown in LB without NaCl. Data are mean⫾ SEM of three separate experiments.
D. Fold variation of ompC and ompF mRNA levels in LF82 and MG1655 grown in LB with 20 g l-1of NaCl (white) relative to that of
each strain grown in LB without NaCl (black) using RT-PCR. 16S rRNA levels were measured as controls. Data are mean⫾ SEM of three separate experiments. *P< 0.05.
showed a 1.8-fold higher expression in bacteria grown at high osmolarity. The levels of ompF and ompC mRNA were measured by real-time reverse transcription polymerase chain reaction (RT-PCR) after growth of the bacteria at low and high osmolarities, 14.0-fold and 14.7-fold decreases in ompF mRNA levels were observed for strains LF82 and MG1655, respectively, with bacteria grown at high osmo-larity compared with low osmoosmo-larity (Fig. 1D). This con-firmed the results of protein analysis. In addition, 2.0-and 2.3-fold increases in the ompC mRNA levels were observed for strain LF82 and strain MG1655, respectively, with bacteria grown at high osmolarity compared with low osmolarity. Taken together, these results indicate that at high osmolarity OmpC expression is increased in the AIEC strain LF82 and in the E. coli K-12 strain MG1655. Like-wise, a significant increase in bacterial adhesion after growth of bacteria at high osmolarity was observed. This suggests that increased OmpC at high osmolarity could play a role in the interaction of bacteria with host cells.
Phenotype of LF82-DompC and LF82-DompF isogenic mutants
LF82 isogenic mutants with the ompC or the ompF gene deleted were constructed. Quantitative adhesion-invasion assays showed that the LF82-DompC mutant was consis-tently reduced in its abilities to adhere to and to invade Intestine-407 epithelial cells, having a 19.6%⫾ 8.6% residual adhesion level and a 15.5%⫾ 5.3% residual invasion level compared with wild-type strain LF82, taken as 100% (Fig. 2A). Transcomplementation experiments were performed with the entire ompC gene cloned into the arabinose inducible expression plasmid vector pBAD24, forming pPBI08. Increased adhesion and invasion levels of the LF82-DompC isogenic mutant transcomplemented with cloned ompC were observed with increased concen-trations ofL-arabinose (Fig. 2A). The arabinose-induced expression of OmpC in the LF82-DompC isogenic mutant transcomplemented with cloned ompC was confirmed by Western immunoblotting using anti-OmpC/F antibodies (Fig. 2B). Transcomplementation of the LF82-DompC isogenic mutant with the vector alone had no effect on its adhesion and invasion levels. In contrast, the adhesion level of the LF82-DompF isogenic mutant was 88.9%⫾ 6.8% of that of strain LF82 (Fig. 2A). Interest-ingly, the invasion level of the LF82-DompF isogenic mutant was increased, reaching 147.6%⫾ 29.2% of that of strain LF82 (Fig. 2A), and Western-blot analysis of whole bacteria extracts using antibodies raised against OmpC/F indicated an increased expression of OmpC in the LF82-DompF isogenic mutant compared with the wild-type strain (Fig. 2B). Thus, the increased invasion of the LF82-DompF mutant can be correlated with increased expression of OmpC in the absence of OmpF.
The decreased adhesion and invasion of LF82-DompC isogenic mutant are not due to decreased expression of type 1 pili and flagella
As type 1 pili and flagella have been previously shown to play a key role in the ability of strain LF82 to adhere to and to invade Intestine-407 cells (Boudeau et al., 2001; Barnich et al., 2003), we analysed whether the decreased adhesion and invasion of the LF82-DompC isogenic
Fig. 2. OmpC but not OmpF is involved in the adhesion-invasion of
AIEC strain LF82.
A. Effects of ompC or ompF disruption and effects of increased expression of ompC gene in the LF82-DompC isogenic mutant transcomplemented with the cloned ompC gene into pBAD24 vector in the presence of increased concentrations ofL-arabinose (0–2%) in supplemented minimum media M9 on adhesion to (black) and invasion (white) of Intestine-407 cells. Cell-associated bacteria were quantified after a 3 h infection period and invasion was determined after gentamicin treatment for an additional hour. The mean number of cell-associated LF82 bacteria was
2.9¥ 106⫾ 1.4 ¥ 106cfu per well. The mean number of intracellular
LF82 bacteria was 1.5¥ 104⫾ 0.9 ¥ 104cfu per well. Results are
expressed as cell-associated or intracellular bacteria relative to those obtained for strain LF82, taken as 100%. Each value is the mean⫾ SEM of at least four separate experiments. *P < 0.05 comparatively to the wild-type strain.
B. Western immunoblot analysis of OmpC and OmpF protein levels in LF82-DompC, LF82-DompF and LF82-DompC
transcomplemented with the cloned ompC gene into pBAD24 vector in the presence of increased concentrations ofL-arabinose (0–2%) in supplemented minimum media M9.
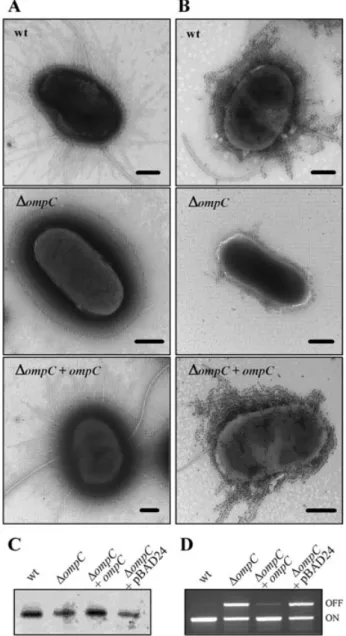
mutant were due to defects of type 1 pili and flagella expression. Electron microscopic examination of nega-tively stained bacteria indicated that only 7% of LF82-DompC bacteria were flagellated compared with 71% for the wild-type strain LF82 (Table 1). Concerning type 1 pili expression, 67% of the LF82-DompC bacteria were piliated, compared with 99% for the wild-type strain and we observed that when piliated the LF82-DompC bacteria produced only a few type 1 pili in comparison to the wild-type strain (Fig. 3A). These results were confirmed by gold immunolabelling (Fig. 3B) and Western immuno-blotting with whole-cell extracts (Fig. 3C) using polyclonal antibodies raised against purified type 1 pili. Interestingly, transcomplementation of the LF82-DompC isogenic mutant with the cloned ompC gene (plasmid pPBI08) restored flagella and type 1 pili expression to levels similar to those of the wild-type strain LF82 (Table 1, Fig. 3A–C), while transformation with the vector pBAD24 alone did not affect the phenotype. Thus, we further analysed whether defects in type 1 pili expression in the LF82-DompC isogenic mutant could be due to phase variation or to the absence of type 1 pili subunit polymerization on the surface of the bacteria. To check the phase variation, we performed PCRs using two sets of primers specific for the phase-ON and phase-OFF orientations of the invertible element (Schwan et al., 1992). The orientation of the invertible element in the wild-type strain LF82 was mostly in phase-ON (Fig. 3D). In contrast, PCR amplification of the LF82-DompC isogenic mutant DNA showed both phase-ON and -OFF orientations of the invertible element. Thus, the decrease in type 1 pili expression in the LF82-DompC isogenic mutant is not only correlated with a switch of the invertible element to the phase-OFF orientation.
Because the absence of OmpC can perturb outer mem-brane integrity and therefore the biogenesis of fimbriae,
we investigated whether the LF82-DompC isogenic mutant could express functional type 1 pili on the bacterial surface. The LF82-DompC isogenic mutant was trans-formed with plasmid pPBI01 harbouring the entire fim operon blocked in the phase-ON orientation to force the bacteria to express these pili. As a consequence of induced type 1 pilus expression, 98% of LF82-DompC/ pPBI01 bacteria were piliated (Table 1), indicating that the decrease in biogenesis of type 1 pili in the LF82-DompC isogenic mutant was not directly related to the absence of OmpC in the outer membrane. To investigate the respec-tive roles of type 1 pili, flagella and OmpC in the ability of strain LF82 to adhere to and to invade intestinal epithelial cells, quantitative adhesion and invasion assays were performed with induced type 1 pili expression and forced contact between bacteria and Intestine-407 epithelial cells. LF82-DompC isogenic mutant transformed with cloned fim operon still showed reduced abilities to adhere to and to invade intestinal epithelial cells, having, respectively, a 26.6%⫾ 4.0% residual adhesion and 15.6%⫾ 5.5% residual invasion levels compared with wild-type strain LF82 (Fig. 4). As the decrease in adhe-sion and invaadhe-sion of this mutant may be also related to the absence of bacterial motility, we added a centrifugation step to establish a close contact between bacteria and epithelial cells. We performed experiments with the LF82-DfliC isogenic mutant as a control, which is not able to express flagella and type 1 pili and has a greatly reduced ability to adhere to and to invade intestinal epithelial cells. After centrifugation, while the adhesion level of the LF82-DfliC transformed with pPBI01 was fully restored to the level of strain LF82, the adhesion levels of LF82-DompC/ pPBI01 were not higher than those in assays performed without centrifugation (Fig. 4A). In addition, while a cen-trifugation step did partially restore the invasion level of the LF82-DfliC/pPBI01 to the level of strain LF82/pPBI01,
Table 1. Expression of type 1 pili and flagella in wild-type strain LF82 compared with that in ompC or ompR mutants.
Strains
Percentage of bacteria expressinga
Flagella+ pili+ Flagella+ pili– Flagella– pili+ Flagella– pili–
LF82 71 0 28 1 LF82-DompC 7b 0 60b 33 LF82-DompC/pPBI08 (+ompC) 63 0 36 1 LF82-DompC/pBAD24 11b 0 49b 40 LF82-DompC/pPBI01 (+fim) 10 0 88 2 LF82-DompR 21b 0 18b 61 LF82-DompR/pPBI09 (+ompR) 70 0 24 6 LF82-DompR/pBAD24 16b 0 17b 67 LF82-DompR/pPBI01 (+fim) 20 0 70 10 LF82-DompR/pPBI08 (+ompC) 68 0 32 0 LF82-DompR/pPBI10 (+rpoE) 67 0 10 23 LF82-DompR/pPBI12 (+ompF) 69 0 30 1
a. Expression of flagella and type 1 pili was monitored by electron microscopic examination of negatively stained bacteria. For each strain,
100 bacteria were counted and data are the means of three separate experiments.
b. When piliated, the bacteria expressed very few type 1 pili.
it did not increase the invasion level of LF82-DompC/ pPBI01 (Fig. 4B). Thus, the decreases in adhesion and invasion levels observed for the LF82-DompC isogenic mutant were neither related to decreased type 1 pili expression nor to impaired bacterial motility.
The EnvZ/OmpR two-component system, which controls OmpC expression, is involved in the adherent-invasive phenotype of LF82
To better investigate the role of OmpC in strain LF82, we deleted the ompR gene, as OmpR is known to regulate OmpC and OmpF expression in E. coli strains (Hall and Silhavy, 1981a,b). As shown by Western-blot using anti-OmpC/F antibodies, the LF82-DompR isogenic mutant like the K-12 strain MG1655-DompR mutant had an absence of OmpC and OmpF expression (Fig. 5A). Transcomplementation of the LF82-DompR isogenic mutant with cloned ompR gene (plasmid pPBI09) restored OmpC and OmpF expression to levels similar to those observed in the wild-type strain LF82, and transcom-plementation of the wild-type strain LF82 with cloned ompR did not increase the expression of OmpC and OmpF (Fig. 5A), nor adhesion and invasion (Fig. 5B). Quantitative adhesion and invasion assays showed that the LF82-DompR isogenic mutant was consistently reduced in its ability to adhere to and to invade Intestine-407 cells, having 15.5%⫾ 2.7% residual adhe-sion and 13.0%⫾ 1.5% residual invasion levels com-pared with wild-type strain LF82, respectively (Fig. 5B).
Fig. 3. Analysis of type 1 pili and flagella expression in
LF82-DompC isogenic mutant.
A. Transmission electron micrographs of negatively stained LF82 bacteria, LF82-DompC isogenic mutant and LF82-DompC isogenic mutant transcomplemented with cloned ompC gene. Bars, 0.5mm. B. Gold immunolabelling of LF82 bacteria, LF82-DompC isogenic mutant and LF82-DompC isogenic mutant transcomplemented with cloned ompC gene using polyclonal antibodies raised against purified type 1 pili. Bars, 0.5mm.
C. Effects of ompC disruption on type 1 pili expression analysed by Western immunoblotting using anti-type 1 pili antibodies.
D. Regulation of type 1 pili in the LF82-DompC isogenic mutant. Determination by PCR analysis of the invertible element orientation of the fim operon in strain LF82, LF82-DompC isogenic mutant, LF82-DompC isogenic mutant transcomplemented with cloned ompC gene and LF82-DompC isogenic mutant transcomplemented with pBAD24 alone. A 450 bp product revealed ON orientation and a 750 bp product OFF orientation of the invertible element.
Fig. 4. Adhesion (A) and invasion (B) abilities of LF82-DompC, LF82-DfliC and these isogenic mutants transformed by pPBI01 harbouring the entire fim operon with intestinal epithelial cells I-407 without (black) or after centrifugation (white). See the legend to Fig. 2A. Each value is the mean⫾ SEM of at least four separate experiments. *P< 0.05 comparatively to the wild-type strain under the same conditions.
Transcomplementation of the LF82-DompR isogenic mutant with the cloned ompR gene (plasmid pPBI09) restored adhesion and invasion levels to levels similar to those of wild-type strain, while transcomplementation with pBAD24 alone did not restore the wild-type phenotype (Fig. 5B). Deletion of the ompR gene in strain LF82 resulted in a high decrease of type 1 pili expression as shown by gold immunolabelling assays and Western-immunoblotting using anti-type 1 pili antibodies (Fig. 6A and B). Electron microscopic examination of negatively stained bacteria indicated that only 39% of LF82-DompR bacteria were piliated compared with 99% for the wild-type strain (Table 1). This effect was specific for strain LF82, because deletion of ompR in K-12 strain MG1655 induced increased type 1 pili expression (Fig. 6A and B). Moreover, LF82-DompR isogenic mutant showed a loss of motility on swim agar plates (Fig. 6C) and electron micro-scopic examination of negatively stained bacteria showed that only 21% of LF82-DompR bacteria were flagellated (Table 1). In contrast, and as already reported by others for various E. coli K-12 strains (Shin and Park, 1995; Oshima et al., 2002), the MG1655-DompR mutant was more motile than the MG1655 wild-type strain (Fig. 6C). In order to investigate whether the decrease in type 1 pili and flagella expression were involved in the decreased adhesion and invasion of the LF82-DompR mutant, we performed experiments with induced type 1 pili expres-sion and forced contact between bacteria and Intestine-407 cells. The LF82-DompR transformed with the cloned fim operon (pPBI01) expressed type 1 pili and 90% of LF82-DompR/pPBI01 bacteria were piliated (Table 1). After centrifugation, the adhesion and invasion levels of LF82-DompR/pPBI01 were higher than those observed in assays performed without centrifugation, however, the adhesion and invasion levels reached only 27.9%⫾ 5.7% and 27.6%⫾ 6.8% of those of wild-type strain, respec-tively (Fig. 5B). Thus, the decreases in adhesion and inva-sion levels observed for the LF82-DompR isogenic mutant were neither related to decreased type 1 pili expression nor to a defect in bacterial motility, in contrast with a LF82-DfliC mutant for which, as shown above, induced type 1 pili expression and forced contact between bacteria and epithelial cells fully or partially restored the levels of adhesion and invasion, respectively (Fig. 4).
Induced expression of OmpC or of OmpF is able to restore adhesion, invasion, and flagella and type 1 pili expression in LF82-DompR mutant
Transcomplementation of the LF82-DompR isogenic mutant with the cloned LF82 ompC gene (plasmid pPBI08) restored type 1 pili expression as shown by gold immunolabelling assays and Western-immunoblotting using polyclonal antibodies raised against purified type 1
Fig. 5. Effects of ompR deletion in AIEC strain LF82 and in E. coli
K-12 strain MG1655.
A. Expression of OmpC and OmpF in AIEC strain LF82, in E. coli K-12 strain MG1655 and theDompR respective isogenic mutants. Outer membrane proteins preparations were separated by SDS-PAGE and analysed by Western-blot using antibodies raised against OmpC/OmpF.
B. Effects of ompR disruption and transcomplementation of LF82-DompR isogenic mutant with pPBI09 harbouring the LF82 ompR gene on adhesion to (black) and invasion (white) of Intestine-407 cells. See the legend to Fig. 2A. The mean number of cell-associated LF82 bacteria was 1.6¥ 107⫾ 0.2 ¥ 107cfu per
well. The mean number of intracellular LF82 bacteria was 4.2¥ 105⫾ 0.9 ¥ 105cfu per well. Each value is the mean⫾ SEM
of at least four separate experiments. *P< 0.05 comparatively to the wild-type strain.
C and D. Adhesion (C) and invasion (D) abilities of LF82-DompR transformed by pPBI01 harbouring the entire fim operon with intestinal epithelial cells I-407 without (black) or after centrifugation (white). See the legend to Fig. 2A. Each value is the mean⫾ SEM of at least four separate experiments. *P< 0.05 comparatively to the wild-type strain under the same conditions.
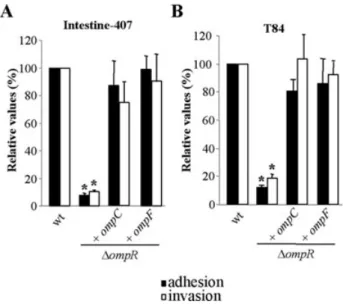
pili (Fig. 6A and B). As shown in Table 1, 100% of the LF82-DompR/pPBI08 bacteria expressed type 1 pili. Inter-estingly, the induced expression of the ompC gene in the LF82-DompR isogenic mutant also restored flagella expression and motility (Fig. 6C and Table 1). In addition, the induced expression of OmpC in the OmpR-negative mutant restored the ability to adhere to and to invade Intestine-407 cells. The adhesion and invasion levels of LF82-DompR transcomplemented with cloned ompC were 87%⫾ 18.2% and 75.0% ⫾ 15.0%, respectively, of those of strain LF82 (Fig. 7A) and similar results were also observed with polarized T84 intestinal epithelial cells (Fig. 7B). Thus, induced expression of OmpC in the LF82-DompR isogenic mutant restored a wild-type phenotype.
The induced expression of the ompF gene cloned in the pBAD33 expression vector (pPBI12) in the LF82-DompR isogenic mutant fully restored type 1 pili and flagella expression (Table 1) and the ability to adhere to and to invade (Fig. 7). This indicated that the adherent-invasive phenotype was linked to the expression of outer mem-brane proteins OmpC or OmpF. However, the role of OmpF in wild-type AIEC LF82 is limited because OmpF is not expressed at high osmolarity and because a LF82-DompF isogenic mutant was not affected in its ability to adhere to and to invade intestinal epithelial cells.
Fig. 6. Type 1 pili and flagella expression in LF82-DompR and MG1655-DompR.
A. Transmission electron micrographs of gold immunolabelling of MG1655 bacteria, MG1655-DompR isogenic mutant, LF82 bacteria, LF82-DompR isogenic mutant and LF82-DompR isogenic mutant transcomplemented with cloned ompC gene using polyclonal antibodies raised against purified type 1 pili. Bars, 0.5mm. B. Expression of type 1 pili, analysed by Western immunoblotting using polyclonal rabbit antiserum raised against purified type 1 pili. C. Motility assay of LF82, LF82-DompR isogenic mutant and LF82-DompR isogenic mutant transcomplemented with pPBI08 harbouring the cloned AIEC LF82 ompC gene and MG1655 and MG1655-DompR isogenic mutant on 0.3% agar after 6 h at 37°C. Motility was visualized as a halo radial diffusion of bacteria around the primary inoculum.
Fig. 7. Adhesion and invasion abilities of LF82-DompR isogenic mutant transcomplemented with cloned ompC or ompF gene. Adhesion (black) and invasion (white) ability with I-407 (A) and T84 (B) intestinal epithelial cells of LF82-DompR isogenic mutant and this isogenic mutant transformed with cloned ompC or ompF gene. See the legend to Fig. 2A. The mean number of cell-associated LF82 bacteria was 4.8¥ 106⫾ 0.1 ¥ 106and
5.3¥ 106⫾ 0.7 ¥ 106cfu per well with I-407 and with T84
respectively. The mean number of intracellular LF82 bacteria was 3.9¥ 105⫾ 1.6 ¥ 105and 1.8¥ 105⫾ 0.8 ¥ 105cfu per well with
I-407 and with T84 respectively. Each value is the mean⫾ SEM of at least four separate experiments. *P< 0.05.
The role of OmpC in adhesion and invasion of AIEC LF82 is indirect and involves thesEregulatory pathway
In E. coli, overproduction of outer membrane protein or misfolding proteins in the outer membrane or in the peri-plasm activate the alternative sigma factorsEor the
two-component regulatory system CpxRA (Raivio and Silhavy, 1999). We checked the involvement of these regulatory systems in the restoration of virulence of the LF82-DompR isogenic mutant transcomplemented with cloned ompC
gene (plasmid pPBI08). After growth of the bacteria in Luria–Bertani (LB) medium in presence of L-arabinose, the cpxR and rpoE mRNA levels were measured by real-time RT-PCR using primers described in Table 2, because it is well known that CpxR and RpoE upregulate their own transcription. The levels of cpxR mRNAs in the LF82-DompR isogenic mutant with induced expression of the ompC gene were not different from those of the wild-type strain LF82 or those of the LF82-DompR isogenic mutant (Fig. 8A). In addition, as shown in Fig. 8B, the levels of
Table 2. Oligonucleotides used for PCR and RT-PCR experiments.
Primer Oligonucleotide sequence (5′-3′) PCR product size (bp) Use
A2GBL-3 B2GBLnp5 AAAGCCACGTTGTGTCTCAA TTAGAAAAACTCATCGAGCA 957 Kanamycin resistance cassette amplification FIME INV GCAGGCGGTTTCTTACGGGG GAGGTGATGTGAAATTAATTTAC 750 OFF-oriented invertible element FIMA INV GATGCGGTACGAACCTGTCC GAGGTGATGTGAAATTAATTTAC 450 ON-oriented invertible element MI-ompC R ATATCAATCGAGATTAGAAAAACTCATCGAGCAG AAAACAATGAAAAAAGGGCCCGCAGGCCCTTTGTTCG 1057 LF82-DompC isogenic mutant construction MI-ompC F GCAGTGGCATAAAAAAGCAAATAAAGGCATATAA CAGAGGGTTAATAACAAAGCCACGTTGTGTCTCAA OmpC1 OmpC2 ACGTTTGGAGCTGGAGATCGC TTCTTCGGTCTGGTTGACGGC 200 Isogenic mutant verification OmpC3 OmpC4 CTCATGCGAACGGTCGCAAG TGGGGAGAATGGACTTGCCG 1298 Isogenic mutant verification
OmpC SalI AGCCGTCGACCAGAGGGTTAATAACATGAAAG 1142 Cloning of ompC
OmpC HindIII CCCAAGCTTGGGTTAGAACTGGTAAACCAGACCC OmpC RT1 OmpC RT2 CAGCCAGGTAGATGTTGTTAG AATTCGGTGGTGACACCTACG 380 RT-PCR MI-ompR R GAGCAATAACGTACGGGCAAATGAACTTCGTGGC GAGAAGCGGAATCGCCTTAGAAAAACTCATCGAGCA 1057 DompR isogenic mutants construction MI-ompR F TTTAAGAATACACGCTTACAAATTGTTGCGAACCT TTGGGAGTACAAACAAAAGCCACGACGTTGTGTCTCAA OmpR1 OmpR2 AACCCGCGTGAACTGCTGGC GCGGCATCGGCTCGTCTTCG 159 Isogenic mutant verification, RT-PCR OmpR3 OmpR4 CCAGATAAGTCGTCACCAGG AGATTTAGCTGGTGACGAACG 1191 Isogenic mutant verification OmpR SalI OmpR HindIII AGCCGTCGACGGGAGTACAAACAATGCAAG CCGAAGCTTCGGTCATGCTTTAGAGCCGTC 754 Cloning of ompR MI-ompF R AAACAGGACCAAAGTCCTGTTTTTTCGGCATTTA ACAAAGAGGTGTGCTATTAGAAAAACTCATCGAGCA 1057 LF82-DompF isogenic mutant construction MI-ompF F ATTGACGGCAGTGGCAGGTGTCATAAAAAAAACC ATGAGGGTAATAAATAAAAGCCACGTTGTCTCAA OmpF1 OmpF2 GATCGGCGTAGCGTTACGGG GGCGGCGTTGCTACCTATCG 320 Isogenic mutant verification, RT-PCR OmpF3 OmpF4 AGGGAAGTCCGCTATCAGGG GCGTCTTCAAGAGCCAGCGC 1845 Isogenic mutant verification
OmpF SalI ACGCGTCGACCTTATTGACGGCAGTGGCAGGTGTC 1149 Cloning of ompF
OmpF HindIII CCCAAGCTTGGGTTAGAACTGGTAAACGATACCCACAGC
CpxR1 GCGCAGATGACTATCTCCCG 187 RT-PCR
CpxR2 GTTTGCCCGTCGAAGCTGGC
RpoE3 GAGGGACTCAATAGTTCGGA 368 RT-PCR
RpoE4 AGAAGGGAGATCAGAAAGCC
RpoE SalI ACGCGTCGACTACCTCGGATGAGCGAGCAG 615 Cloning of rpoE
RpoE HindIII CCCAAGCTTGGGCCCGCTATCGTCAACGCCTG
DegS1 CAGCCTTAACCCGCTTTCCA 220 RT-PCR
DegS2 TCTGATCGGCGTCGTTGATG
adhesion and invasion of a LF82-DcpxR isogenic mutant were not significantly different from those of the wild-type strain LF82. These results indicate that the two-component regulatory system CpxRA is not involved in the adhesion and invasion abilities of AIEC strain LF82. In contrast, the levels of rpoE mRNAs in the LF82-DompR isogenic mutant with induced expression of the ompC gene were greater than those of the wild-type strain LF82 or those of the LF82-DompR isogenic mutant (Fig. 8A), indicating that the sE-regulatory pathway was activated
in the LF82-DompR mutant when OmpC was over-expressed. As previously reported, the RpoE factor may be involved in LF82 bacterial viability, since repeated attempts to isolate LF82-DrpoE isogenic mutant failed (Bringer et al., 2005). In order to analyse the role of RpoE in the virulence of AIEC strain LF82, we induced in the LF82-DompR isogenic mutant increased expression of the rpoE gene cloned in the pBAD24 expression vector, because we speculated that all RpoE overexpressed pro-teins would not be sequestered to the inner membrane and therefore could activate sE-gene transcription.
Interestingly, the increased expression of RpoE in the LF82-DompR isogenic mutant obtained with increased concentrations ofL-arabinose led to restoration of adhe-sion and invaadhe-sion (Fig. 8C), irrespective of the OmpC levels because no OmpC expression was still observed with increasedsEexpression (Fig. 8D). The adhesion and
invasion levels of the LF82-DompR isogenic mutant were restored with induced expression of RpoE but not in the presence of similar concentrations of L-arabinose (Fig. 8C). Hence, different genes may be involved in adhesion or in invasion and their expression is variously controlled by RpoE. In addition, we observed that the induced expression of the rpoE gene in the OmpR-negative mutant restored the type 1 pili and flagella expression to levels similar to those observed for the wild-type strain (Table 1). Thus, in the absence of OmpC, overproduction of RpoE in the LF82-DompR isogenic mutant fully restored the adherent-invasive phenotype.
High osmolarity activates thesEregulatory pathway
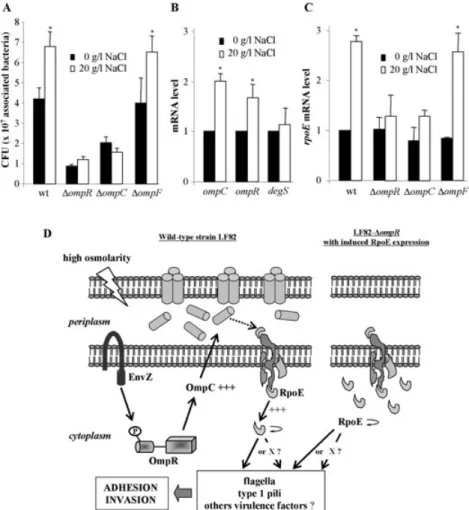
Comparative analysis of the ability of strain LF82 and the related omp isogenic mutants to interact with intestinal epithelial cells was performed after growth of bacteria in LB broth at low or high osmolarity (Fig. 9A). At low osmo-larity, the ompR and ompC mutants display low adher-ence rates and the ompF mutant behaves like the wild-type strain. At high osmolarity, a significant 1.6-fold increase in the number of associated bacteria, similar to that seen in the wild-type strain LF82, was observed for the LF82-DompF isogenic mutant. Such increases were not observed for LF82-DompR and -DompC mutants. Growth of LF82 bacteria at high osmolarity induced, in
Fig. 8. Involvement ofsEregulatory pathway in adherent-invasive
phenotype of strain AIEC LF82.
A. Fold variation of cpxR and rpoE mRNA levels in LF82-DompR isogenic mutant (white) and LF82-DompR isogenic mutant transcomplemented with pPBI08 harbouring the cloned ompC gene (grey) relative to that of wild-type strain LF82 (black) using RT-PCR. 16S rRNA levels were measured as controls. Data are mean⫾ SEM of three separate experiments.
B. Effects of cpxR disruption on adhesion to (black) and invasion (white) of Intestine-407 cells. Each value is the mean⫾ SEM of at least four separate experiments. See the legend to Fig. 2A. C. Effects of increased expression of rpoE gene in the LF82-DompR isogenic mutant obtained with increased
concentrations ofL-arabinose (0–2%) in supplemented minimum media M9 on adhesion to (black) and invasion (white) of
Intestine-407 cells. Each value is the mean⫾ SEM of at least four separate experiments. The mean number of cell-associated LF82 bacteria was 3.7¥ 106⫾ 0.5 ¥ 106cfu per well. The mean number
of intracellular LF82 bacteria was 2.3¥ 104⫾ 0.6 ¥ 104cfu per well.
See the legend to Fig. 2A. *P< 0.05.
D. Western immunoblot analysis of OmpC, OmpF and OmpA expression in LF82, LF82-DompR isogenic mutant and this mutant transcomplemented with the cloned rpoE gene (pPBI10).
addition to the increase in ompC mRNA levels as shown in Fig. 1D, a significant (1.7-fold) increase in ompR mRNA levels (Fig. 9B). Similarly the rpoE mRNA level increased 2.8-fold after growth of the LF82 bacteria at high osmo-larity (Fig. 9C). In contrast, the degS mRNA levels were similar after growth of the LF82 bacteria at low and high osmolarities (Fig. 9B). The determination of rpoE mRNAs levels in the LF82-DompC, -DompF and -DompR isogenic mutants after growth of bacteria at low and high osmolari-ties indicated that at low osmolarity the levels of rpoE mRNAs in the various omp mutants were not different
from those of the wild-type strain LF82 (Fig. 9C). Thus, decreases in adhesion and invasion levels of the ompR and ompC mutants at low osmolarity can not be explained by a decrease in RpoE expression. This suggests that under low osmolarity conditions, OmpC plays a direct or indirect role in AIEC adhesion and invasion, indepen-dently of the RpoE regulatory pathway. In contrast, at high osmolarity, a 3.1-fold significant increase in the rpoE mRNA levels was observed for the LF82-DompF mutant, while no increase was observed for LF82-DompC and -DompR mutants.
Fig. 9. Effects of high osmolarity in AIEC strain LF82.
A. Effects of bacterial growth in LB broth without NaCl or with 20 g l-1NaCl on adhesion to Intestine-407 cells. Each value is the mean number
of cfu⫾ SEM of at least four separate experiments. *P < 0.05 comparatively to numbers of cfu for bacteria grown at low osmolarity. B. Fold variation of ompC, ompR and degS mRNA levels in wild-type strain LF82 grown in LB broth with 20 g l-1of NaCl (white) relative to
that of wild-type strain LF82 grown in LB broth without NaCl (black) using RT-PCR. 16S rRNA levels were measured as controls. Data are mean⫾ SEM of three separate experiments. *P < 0.05.
C. Fold variation of rpoE mRNA levels in wild-type strain LF82 and the related omp mutants grown in LB broth with 20 g l-1of NaCl (white)
relative to that of wild-type strain LF82 grown in LB broth without NaCl (black) using RT-PCR. Data are mean⫾ SEM of three separate experiments. *P< 0.05 comparatively to mRNA level for bacteria grown at low osmolarity.
D. Model for the involvement of OmpC in the adherence-invasiveness phenotype of AIEC strain LF82 under high osmolarity encountered in gastrointestinal tract. High osmolarity is sensed by the EnvZ/OmpR two component system and induce OmpC overexpression. This overexpression should directly activates the complex DegS-RseA-RseP that frees RpoE into the cytoplasm to function as a sigma factor to activatesEregulon expression as previously reported (De Las Penas et al., 1997; Missiakas et al., 1997; Ades et al., 1999; Alba et al., 2002;
Kanehara et al., 2002; Walsh et al., 2003). RpoE induces in turn its own expression and directly or indirectly via one or several intermediates (X) the expression of flagella, type 1 pili and others factors involved in adhesion and invasion process of AIEC strain LF82. However, in a LF82-DompR mutant, induced RpoE overexpression bypasses the need for OmpC.
Discussion
The aim of the present study was to investigate the OmpC expression and the role of OmpC in CD-associated adherent-invasive E. coli strain LF82 under conditions of high osmolarity encountered by bacteria in the gastrointestinal tract (Fordtran and Ingelfinger, 1968; Chowdhury et al., 1996) because it has been reported that passage of bacteria from the stomach into the small intestine represents an osmotic upshift that in many food-borne pathogens, serves to trigger the expression of genes that are necessary for survival and colonization (Nikaido and Rosenberg, 1983; Foster and Spector, 1995). Increased concentrations of NaCl increased OmpC expression in AIEC LF82. This was also observed for the non-pathogenic E. coli strain MG1655, confirming previous reports with various K-12 strains, for reviews (Forst and Inouye, 1988; Mizuno and Mizushima, 1990; Pratt et al., 1996). The high OmpC expression in AIEC strain at high osmolarity, together with the abnormal ileal colonization with AIEC bacteria as previously reported (Darfeuille-Michaud et al., 1998), could explain in part why high levels of anti-E. coli OmpC are observed in CD patients (Landers et al., 2002; Mow et al., 2004). Increased bacterial adhesion to intestinal epithelial cells was also observed when AIEC LF82 bacteria were grown at high osmolarity, and therefore we investigated whether OmpC could be involved in the interaction of AIEC LF82 bacteria with host cells.
A LF82-DompC mutant showed decreases in its abilities to adhere to and to invade, indicating a possible role of OmpC as an adhesin or an invasin in AIEC bacteria. Conflicting reports exist regarding the role of OmpC in the virulence of pathogenic bacteria. A Shigella flexneri DompC mutant was considerably impaired in its ability to invade HeLa and Caco-2 intestinal epithelial cells (Ber-nardini et al., 1993) and mutants of an enterotoxigenic E. coli strain deleted for ompC and ompF, in addition to deletion of aroC, are attenuated in virulence in humans (Turner et al., 2001). However, in Salmonella enterica serovar Typhimurium, no significant difference in binding to intestinal epithelial cells was observed between an OmpC-negative mutant and the wild-type strain (Hara-Kaonga and Pistole, 2004). The decreased adhesion and invasion levels of the OmpC-negative LF82 mutant could result from the decrease in type 1 pili expression, as already reported for LF82 mutants deficient for flagellar biogenesis or lipoprotein NlpI expression (Barnich et al., 2003; 2004). Induced expression of type 1 pili in the LF82-DompC isogenic mutant did not restore the ability of the OmpC null mutant to adhere to and to invade intestinal epithelial cells, even after a forced contact between bac-teria and host cells. This indicates that OmpC acts inde-pendently of type 1 pili and flagella expression, and
perhaps has a role per se or regulates the expression of other factors involved in adhesion and invasion processes.
Co-regulated expression of the outer membrane pro-teins OmpC and OmpF exists in E. coli and involves the two-component OmpR–EnvZ regulatory system (Hall and Silhavy, 1981a,b). We observed that deletion of ompR gene in AIEC strain LF82, as in non-pathogenic E. coli K-12 strain MG1655, induced absence of OmpC and OmpF expression. Interestingly, the phenotypes of the negative OmpR mutants engineered from AIEC strain LF82 or K-12 strain MG1655 were different. The LF82-DompR mutant synthesized very small amounts of flagella and type 1 pili, and deletion of ompR in K-12 strain MG1655 resulted in increased motility and type 1 pili expression. This indicates that OmpR functions differently in pathogenic AIEC and commensal bacteria. It has been previously reported that in E. coli K-12 OmpR negatively regulates the master flagellar regulatory operon flhDC (Shin and Park, 1995; Pruss, 1998; Oshima et al., 2002) but in Salmonella enterica serovar Typhimurium inactivation of ompR does not affect flhDC expression (Kutsukake, 1997). The LF82-DompR isogenic mutant showed decreased abilities to adhere to and to invade intestinal epithelial cells Intestine-407 and T84. As in results obtained with an OmpC null mutant, induced type 1 pili expression and forced contact between bacteria and intestinal epithelial cells did not restore the ability of the OmpR mutant to adhere to and to invade. Interestingly, the expression of the cloned ompC or ompF gene in the LF82-DompR isogenic mutant restored flagella and type 1 pili expression, as well as the ability of the bacteria to adhere to and to invade intestinal epithelial cells. One explanation could be that overexpression of OmpC or OmpF in aDompR mutant context could transduce outer membrane or periplasmic stress signals to the cytoplasm. However, in a wild-type strain context and at high osmo-larity, we can speculate that the expression of flagella, type 1 pili and interaction with host cells is linked to high OmpC levels because we observed a quasi absence of OmpF and an increase in OmpC. This hypothesis is rein-forced by the fact that an OmpF null mutant showed an increased invasion level, together with increased OmpC expression. However, the phenotypes of the OmpC mutant expressing OmpF and the OmpR mutant lacking OmpC with induced overexpression of OmpF were different. A possible explanation is that the induced over-expression of OmpF in the ompR mutant can lead to envelope perturbations and thus to outer membrane or periplasmic stress signals sufficient to promote AIEC adhesion and invasion via RpoE activation.
The Cpx and sE extracytoplasmic stress responses
sense and respond to misfolded or overexpressed pro-teins in the bacterial envelope (Mecsas et al., 1993; © 2007 The Authors
Danese et al., 1995; Raina et al., 1995; Raivio and Silhavy, 1999). Overexpression of OmpC in the LF82-DompR mutant did not modify the level of cpxR mRNA. As activation of the Cpx regulatory pathway is known to induce cpxRA operon transcription (Raivio and Silhavy, 1999), we hypothesized that the CpxRA regulatory pathway is not involved in the OmpC overexpression-induced responses in AIEC strain LF82. This was con-firmed because a CpxR-negative mutant of strain LF82 had a similar phenotype as the wild-type strain. In E. coli K-12, overproduction of the various outer membrane pro-teins including OmpF and OmpC causes an increase insE
activity (Mecsas et al., 1993; Grigorova et al., 2004). Like-wise, the overproduction of OmpC activates the sE
pathway, because we observed that induced expression of OmpC in LF82-DompR isogenic mutant led to increased rpoE mRNA level. Experiments performed to create a LF82-DrpoE isogenic mutant to verify the role of RpoE in AIEC LF82 have failed. Interestingly, induced expression of RpoE increased the abilities of the LF82-DompR mutant to interact with host cells and restored flagella and type 1 pili expression. In addition, an increase in rpoE mRNA levels was observed in AIEC strain LF82 after growth of bacteria at high osmolarity together with increases in ompR and ompC mRNA levels. It has been reported that overexpression of OmpC directly activates protease DegS, which then cleaves RseA and thereby activates RpoE regulatory pathway (Ades et al., 1999; Alba et al., 2002; Walsh et al., 2003). No increase in degS mRNA levels were observed when LF82 bacteria were grown at high osmolarity, indicating that activation of the RpoE regulatory pathway in strain LF82 does not need any increase in the DegS protease level. After growth of the bacteria at high osmolarity, no increase in rpoE mRNA levels was observed for LF82 OmpR and OmpC null mutants, as well as no increase in the ability of these mutants to adhere to intestinal epithelial cells. All together this indicates a link between the OmpC level, the RpoE level and bacterial interaction with host cells. This hypoth-esis is reinforced by the fact that at high osmolarity the behaviour of the LF82 OmpF null mutant, in which we observed an increase in OmpC expression, was similar to that of the wild-type strain LF82.
ThesEregulon encodes many pathogen-related
func-tions and this explains why cells lackingsEare defective
in pathogenesis. Rhodius et al. performed a promoter pre-diction model for E. colisE, which enabled them to predict
a total of 89 unique sE-controlled transcription units in
E. coli K-12 and eight related genomes (Rhodius et al., 2006). This study and others have shown that in addition to transcribe genes encoding chaperones and proteases targeted to the cell envelope that will degrade overex-pressed proteins, sE transcribes genes that ensure the
synthesis, assembly and homeostasis of
lipopolysaccha-ride and outer membrane proteins (Dartigalongue et al., 2001; Rezuchova et al., 2003; Rhodius et al., 2006). In addition, the sE regulon encodes multiple functions
related to pathogenesis (Humphreys et al., 1999; Cano et al., 2001; Manganelli et al., 2001; Kovacikova and Skorupski, 2002; Testerman et al., 2002; Bang et al., 2005; Rhodius et al., 2006). Interestingly among the genes regulated or predicted to be regulated by sE in
E. coli, the yfgL gene is upregulated when RpoE is over-expressed (Onufryk et al., 2005; Rhodius et al., 2006). We previously reported that, in the AIEC strain LF82, YfgL is required for invasive ability in relation to outer mem-brane vesicle release (Rolhion et al., 2005). In addition, it has been recently reported that in E. coli DH5a a link between outer membrane vesiculation and sE-pathway
could exist (McBroom et al., 2006). But among the sE-regulated genes predicted in the various genomes
(Rhodius et al., 2006), we found none that could be involved in type 1 pili or flagella expression. Restoration of type 1 pili and flagella expression in LF82-DompR
isogenic mutant was observed when RpoE was
overexpressed. This may indicate that one or several intermediates, whose transcription is sE-dependent, are
involved in the regulation of type 1 pili and flagella in AIEC strain LF82. The whole genome of strain LF82 is currently sequenced and in order to identify all AIECsE-regulated
genes, we will subsequently search for such intermedi-ates by performing a promoter prediction model forsE.
In summary, we propose a model for the involvement of OmpC in the interaction of AIEC bacteria with intes-tinal epithelial cells under conditions of high osmolarity similar to that of the gastrointestinal tract (Fordtran and Ingelfinger, 1968; Chowdhury et al., 1996). At high osmolarity, increased expression of OmpC in AIEC LF82 bacteria and activation of the sE regulatory pathway
were observed. This can modulate flagella and/or type 1 pili encoding gene expression but also the expression of genes encoding other yet unidentified virulence factors also involved in AIEC interactions with host cells (Fig. 9D). In addition, activation of the RpoE regulatory pathway can bypass the effect of OmpC, as shown in the LF82 OmpR mutant. Our results rose also the ques-tion of whether OmpC is really involved as adhesin or invasin in the interaction of some pathogenic bacteria with host cells as already reported by others, or whether its role is linked to the sE regulatory pathway. We also
showed that flagella and/or type 1 pili encoding gene regulation involving the EnvZ/OmpR two-component system is opposite in AIEC strain LF82 and in a K-12 strain. This could indicate that AIEC bacteria have evolved from non-pathogenic to pathogenic bacteria by elaborating intestinal environment adaptation mecha-nisms for which high osmolarity constitutes key signals to activate the expression of virulence genes.
Experimental procedures
Bacterial strains, plasmids and cell lines
Strain LF82 was isolated from chronic ileal lesion of a patient with CD and belongs to E. coli serotype O83:H1. It adhered to and strongly invaded Intestine-407, HEp-2 and Caco-2 cells (Boudeau et al., 1999). E. coli strain JM109 was used as host strains for cloning experiments. Bacterial strains and plas-mids used in this study are listed in Table 3. Bacteria were grown routinely in LB broth and in M9 minimal media supple-mented with 1 mM MgSO4, 0.2% carbon source (glucose or
L-arabinose) and 0.00005% thiamine without shaking or on LB agar plates (Institut Pasteur Production) overnight at 37°C. Plasmid vectors pBAD24 and pBAD33 were used for cloning procedures (Guzman et al., 1995). Antibiotics were added to media at the following concentrations: ampicillin (50mg ml-1
), kanamycin (50mg ml-1
) and chloramphenicol (25mg ml-1). Intestine-407 cells (derived from human
intesti-nal embryonic jejunum and ileum) and T84 (derived from human colon carcinoma) were purchased from Flow Labora-tories and cultured according to the manufacturer’s protocols.
Adhesion and invasion assay
The bacterial invasion was performed using the gentamicin protection assay as described previously (Boudeau et al., 1999). Briefly, Intestine-407 and T84 cells were seeded in 24-well tissue culture plates with 4¥ 105and 2¥ 105cells per
well respectively. Monolayers were then infected at a multiplic-ity of infection of 10 bacteria per cell in 1 ml of the cell culture medium without antibiotics and with heat-inactivated fetal calf serum (FCS) (Biowhittaker Cambrex Compagny, Verviers, Belgium). After a 3 h incubation period at 37°C, monolayers were washed three times in phosphate-buffered saline (PBS,
pH 7.2). The epithelial cells were then lysed with 1% Triton X-100 (Euromedex, Mundolsheim, France) in deionized water. Samples were diluted and plated onto Muller-Hinton agar plates to determine the number of colony-forming units (cfu) corresponding to the total number of cell-associated bacteria (adherent and intracellular bacteria). To determine the number of intracellular bacteria, fresh cell culture medium containing 100mg ml-1gentamicin was added for 1 h to kill extracellular
bacteria. Monolayers were then lysed with 1% Triton X-100, and bacteria were quantified as described above. When needed, the infected monolayers were centrifuged for 10 min at 1000 g before the 3 h infection period.
Motility assay
Bacterial strains were grown overnight at 37°C without agi-tation on LB broth and 4ml of the culture were inoculated into the centre of a 0.3% LB agar plates. The plates were incu-bated at 37°C for 6 h, and motility was assessed qualitatively by examining the circular swim formed by the growing motile bacterial cells.
Transmission electron microscopy
Negative staining. Bacteria were grown overnight in LB broth
containing 10 g l-1NaCl at 37°C without shaking, placed for
1 min on carbon-Formvar copper grids (Electron Microscopy Sciences, Hatfield, England) and negatively stained during 1 min with acid phosphotungstic pH 6.0. Grids were exam-ined with Hitachi H-7650 transmission electron microscope.
Immunolabelling. Gold immunolabelling was performed by
the method of Levine (Levine et al., 1984). A drop of bacteria Table 3. Bacterial strains and plasmids used in this study.
Strain or plasmid Relevant characteristics Source or reference
Strains
LF82 E. coli isolated from an ileal biopsy of a CD patient Darfeuille-Michaud et al. (1998) LF82-DompC LF82 isogenic mutant with the ompC gene deleted This study
LF82-DompF LF82 isogenic mutant with the ompF gene deleted This study LF82-DompR LF82 isogenic mutant with the ompR gene deleted This study LF82-DcpxR LF82 isogenic mutant with the cpxR gene deleted Bringer et al. (2005) LF82-DfliC LF82 isogenic mutant with the fliC gene deleted Barnich et al. (2003)
MG1655 E. coli K-12 serotype OR:H48/K- Laboratory stock
MG1655-DompR MG1655 isogenic mutant with the ompR gene deleted This study Plasmids
pKOBEG pBAD cloning vector harbouringl phage redgba operon, Chaveroche et al. (2000) chloramphenicolr
pBAD24 E. coli cloning vector, ampicillinr Guzman et al. (1995)
pBAD33 E. coli cloning vector, chloramphenicolr Guzman et al. (1995)
pPBI01 pHSG575 harbouring the 11.2 kb SalI fragment with the entire fim operon of K-12 E. coli strain J96
Boudeau et al. (2001) pPBI08 pBAD24 harbouring the 1.1kb HindIII–SalI fragment with
the entire ompC gene of strain LF82
This study pPBI09 pBAD24 harbouring the 0.7kb HindIII–SalI fragment with This study
the entire ompR gene of strain LF82
pPBI10 pBAD24 harbouring the 0.6kb HindIII–SalI fragment with the entire rpoE gene of strain LF82
This study pPBI12 pBAD33 harbouring the 1.1kb HindIII–SalI fragment with
the entire ompF gene of strain LF82
This study
grown overnight in LB broth at 37°C without shaking was placed on carbon-Formvar copper grids. Excess liquid was removed and the grid was placed face down on a suitable dilution of antiserum raised against type 1 pili for 15 min. After 30 washings in wash solution (PBS+ 1% bovine serum albumin and 1% Tween 20), the grid was placed on a drop of gold-labelled goat anti-rabbit serum (BB International, Cardiff, UK) for 15 min. After a further thorough washing, the grid was negatively stained with acid phosphotungstic pH 6.0.
Construction of isogenic mutants and transcomplementation assays
Isogenic mutants were generated with a PCR product using the method described by Chaveroche et al. (Chaveroche
et al., 2000). Briefly, the strategy was to replace a
chromo-somal sequence with a selectable antibiotic resistance gene (kanamycin) generated by PCR. This PCR product was gen-erated by using primers with 50-nt extensions that are homologous to regions adjacent to the gene to delete and template E. coli strain carrying a kanamycin resistance gene (Table 2). In addition, strain LF82 was transformed with pKOBEG, a plasmid encoding Red proteins from phagel under the control of a promoter inducible by L-arabinose. These proteins protect linear DNA from degradation in bacteria. The plasmid was maintained in bacteria at 30°C with 25mg ml-1
of chloramphenicol and was suicided at 42°C. Strain LF82/pKOBEG was grown at 30°C with 1 mM L-arabinose to induce Red protein expression. When OD620
reached 0.5, the bacterial culture was incubated for 10 min at 42°C in order to suicide the plasmid. Bacteria were washed three times with 10% glycerol, and PCR products were electroporated. The isogenic mutants were then selected on LB agar containing 50mg ml-1kanamycin. The replacement
of the gene by the kanamycin resistance cassette in each isogenic mutant was confirmed by PCR.
A 1142, 754 and 615 bp PCR product obtained using, respectively, primers OmpC SalI/OmpC HindIII, OmpR SalI/ OmpR HindIII and RpoE SalI/RpoE HindIII containing, respectively, the entire ompC, ompR and rpoE gene were cloned into the pBAD24 vector and designated, respectively, pPBI08, pPBI09 and pPBI10 (Tables 2 and 3). A 1149 bp PCR product obtained using OmpF SalI/OmpF HindIII con-taining the entire ompF gene was cloned into the pBAD33 vector and designed pPBI12 (Tables 2 and 3). These construct were used to transform the wild-type strain LF82, LF82-DompC or LF82-DompR isogenic mutant.
Growth of the wild-type strain LF82, DompC, LF82-DompF or LF82-DompR isogenic mutant was performed at 37°C in the bacterium-cell incubation medium used for adhe-sion and invaadhe-sion experiments (Eagle minimal essential medium cell culture medium supplemented with 10% heat-inactivated FCS). The growth curves for wild-type strain LF82 and the three isogenic mutants were similar at all time points (data not shown).
Protein preparation, SDS-PAGE and Western immunoblotting analysis
Expression of OmpC, F and A was analysed with whole-cells extracts. After an overnight incubation at 37°C in LB broth
with or without NaCl, bacteria were centrifugated and resus-pended in SDS-PAGE loading buffer (2% SDS, 50 mM Tris-HCl pH 6.8, 12.5% glycerol, 400 mMb-mercaptoethanol and 0.01% Bromophenol Blue). For outer membrane prepara-tions, cells were recovered and treated as previously described (Pugsley and Schnaitman, 1979). Protein prepara-tions were heated for 5 min at 95°C and separated by SDS-10% or -12% PAGE in presence or not of urea 6 M. Expression of type 1 pili was analysed with whole-cell extracts. After on overnight incubation at 37°C in LB broth or M9 without shaking, bacteria at the same OD were centri-fuged and resuspended in SDS-PAGE loading buffer and heated for 5 min at 95°C. Protein preparations were acidified with HCl, heated for 5 min at 95°C and separated by SDS-12% PAGE. Western immunoblotting was performed accord-ing to the procedure of Towbin (Towbin et al., 1979). Proteins were electroblotted onto nitrocellulose membranes (Amer-sham International) and the membranes were reacted with the rabbit antiserum against OmpC/F (diluted 1:1000), OmpA (diluted 1:10 000), E. coli type 1 pili (diluted 1:750). Immu-noreactants were detected using horseradish peroxydase anti-rabbit IgG antibody (diluted 1:10 000), ECL reagents (Amersham International) and autoradiography. OmpC and A were quantified by densitometry (Scion Image).
RNA manipulations, reverse transcription and RT-PCR
Cultures were grown at 37°C in LB with or without NaCl to OD 0.2 at 620 nm and when needed, L-arabinose (0.2%) was added to induce the overexpression of OmpC. Total RNAs were extracted from bacteria at OD 0.4 at 620 nm and treated with DNase (Roche Diagnostics, Manheim, Germany) to remove any contaminating genomic DNA. The RNAs were reverse transcribed and amplified using specific primers to
cpxR, rpoE, ompR, ompC, ompF and degS mRNAs or 16S
rRNA (Table 2). Amplification of a single expected PCR product was confirmed by electrophoresis on a 2% agarose gel. RT-PCR was performed using a Light Cycler (Roche Diagnostic), and quantification of the cpxR, rpoE ompR,
ompC, ompF and degS mRNA levels or 16S rRNA (as a
control) was performed using RNA master SYBR Green I (Roche Diagnostic) with 0.5mg of total RNA.
Statistical analysis
For analysis of the significance of differences in adhesion and invasion levels, Student’s t-test was used. All experiments were repeated at least three times. A P-value less than or equal to 0.05 was considered statistically significant.
Acknowledgements
This study was supported by the Ministère de la Recherche et de la Technologie (EA3844), by the INRA (USC 2018) and by grants from the Association F. Aupetit (AFA), and Institut de Recherche des Maladies de l’Appareil Digestif (IRMAD, Laboratoire Astra France). We are grateful to Jean Michel Betton (CNRS URA 2185, Institut Pasteur, Paris, France) for his interest in our work and helpful discussions. We thank Roland Lloubès (CNRS UPR 9027, Institut de Biologie
Structurale et Microbiologie, Marseille, France) for OmpC/F antibodies, Nico Nouwen (Departement of Molecular Micro-biology, University of Groningen, Haren, the Netherlands) for OmpA antibodies and Maryvonne Moulin-Schouleur (Pathogénie Bactérienne, UR86, INRA, Nouzilly, France) for
E. coli type 1 pili antibodies. We also thank Christelle
Drégneaux and Claire Szczepaniak (CICS, Université d’Auvergne, Clermont-Ferrand, France) for technical assis-tance with electron microscopy.
References
Ades, S.E., Connolly, L.E., Alba, B.M., and Gross, C.A. (1999) The Escherichia coli sigma (E)-dependent extracy-toplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev 13: 2449– 2461.
Alba, B.M., Leeds, J.A., Onufryk, C., Lu, C.Z., and Gross, C.A. (2002) DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma (E)-dependent extracytoplasmic stress response. Genes Dev 16: 2156– 2168.
Arnott, I.D., Landers, C.J., Nimmo, E.J., Drummond, H.E., Smith, B.K., Targan, S.R., and Satsangi, J. (2004) Sero-reactivity to microbial components in Crohn’s disease is associated with disease severity and progression, but not NOD2/CARD15 genotype. Am J Gastroenterol 99: 2376– 2384.
Bang, I.S., Frye, J.G., McClelland, M., Velayudhan, J., and Fang, F.C. (2005) Alternative sigma factor interactions in
Salmonella: sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol Microbiol 56: 811–823.
Barnich, N., Boudeau, J., Claret, L., and Darfeuille-Michaud, A. (2003) Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn’s disease.
Mol Microbiol 48: 781–794.
Barnich, N., Bringer, M.A., Claret, L., and Darfeuille-Michaud, A. (2004) Involvement of lipoprotein NlpI in the virulence of adherent invasive Escherichia coli strain LF82 isolated from a patient with Crohn’s disease. Infect Immun 72: 2484–2493.
Beaven, S.W., and Abreu, M.T. (2004) Biomarkers in inflam-matory bowel disease. Curr Opin Gastroenterol 20: 318– 327.
Bernardini, M.L., Sanna, M.G., Fontaine, A., and Sansonetti, P.J. (1993) OmpC is involved in invasion of epithelial cells by Shigella flexneri. Infect Immun 61: 3625–3635. Boudeau, J., Glasser, A.L., Masseret, E., Joly, B., and
Darfeuille-Michaud, A. (1999) Invasive ability of an
Escherichia coli strain isolated from the ileal mucosa of a
patient with Crohn’s disease. Infect Immun 67: 4499–4509. Boudeau, J., Barnich, N., and Darfeuille-Michaud, A. (2001) Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn’s disease is involved in bacterial invasion of intestinal epithelial cells. Mol Microbiol 39: 1272–1284.
Bringer, M.A., Barnich, N., Glasser, A.L., Bardot, O., and Darfeuille-Michaud, A. (2005) HtrA stress protein is involved in intramacrophagic replication of adherent and
invasive Escherichia coli strain LF82 isolated from a patient with Crohn’s disease. Infect Immun 73: 712–721. Burke, D.A., and Axon, A.T. (1988) Adhesive Escherichia coli
in inflammatory bowel disease and infective diarrhoea.
BMJ 297: 102–104.
Cano, D.A., Martinez-Moya, M., Pucciarelli, M.G., Groisman, E.A., Casadesus, J., and Garcia-Del Portillo, F. (2001)
Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect Immun 69: 6463–6474.
Chaveroche, M.K., Ghigo, J.M., and d’Enfert, C. (2000) A rapid method for efficient gene replacement in the filamen-tous fungus Aspergillus nidulans. Nucleic Acids Res 28: E97.
Chowdhury, R., Sahu, G.K., and Das, J. (1996) Stress response in pathogenic bacteria. J Biosci 21: 149–160. Danese, P.N., Snyder, W.B., Cosma, C.L., Davis, L.J., and
Silhavy, T.J. (1995) The Cpx two-component signal trans-duction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic pro-tease, DegP. Genes Dev 9: 387–398.
Darfeuille-Michaud, A., Neut, C., Barnich, N., Lederman, E., Di Martino, P., Desreumaux, P., et al. (1998) Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 115: 1405–1413. Darfeuille-Michaud, A., Boudeau, J., Bulois, P., Neut, C.,
Glasser, A.L., Barnich, N., et al. (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127: 412– 421.
Dartigalongue, C., Missiakas, D., and Raina, S. (2001) Char-acterization of the Escherichia coli sigma E regulon. J Biol
Chem 276: 20866–20875.
De Las Penas, A., Connolly, L., and Gross, C.A. (1997) The sigmaE-mediated response to extracytoplasmic stress in
Escherichia coli is transduced by RseA and RseB, two
negative regulators of sigmaE. Mol Microbiol 24: 373–385. Duchmann, R., Lochs, H., and Kruis, W. (1999) [Crohn disease, ulcerative colitis. When bacteria attack the intes-tinal wall{]. MMW Fortschr Med 141: 48–51.
Elson, C.O. (2000) Commensal bacteria as targets in Crohn’s disease. Gastroenterology 119: 254–257.
Fordtran, J.S., and Ingelfinger, F.J. (1968) Absorption of water, electrolytes, and sugars from the human gut. In
Handbook of Physiology. Heidel, W., and Code, C.F. (eds).
Baltimore, MD: Waverly Press, pp. 1457–1490.
Forst, S., and Inouye, M. (1988) Environmentally regulated gene expression for membrane proteins in Escherichia
coli. Annu Rev Cell Biol 4: 21–42.
Foster, J.W., and Spector, M.P. (1995) How Salmonella survive against the odds. Annu Rev Microbiol 49: 145–174. Grigorova, I.L., Chaba, R., Zhong, H.J., Alba, B.M., Rhodius, V., Herman, C., and Gross, C.A. (2004) Fine-tuning of the
Escherichia coli sigmaE envelope stress response relies
on multiple mechanisms to inhibit signal-independent pro-teolysis of the transmembrane anti-sigma factor, RseA.
Genes Dev 18: 2686–2697.
Guzman, L.M., Belin, D., Carson, M.J., and Beckwith, J. (1995) Tight regulation, modulation, and high-level expres-sion by vectors containing the arabinose PBAD promoter.
J Bacteriol 177: 4121–4130.