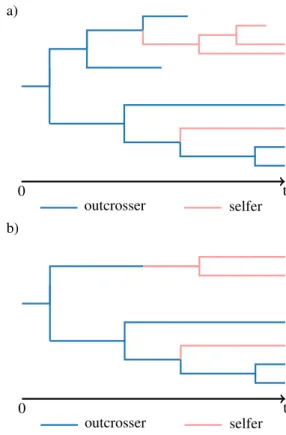
Modeling a trait-dependent diversification process coupled with molecular evolution on a random species tree
Texte intégral
Figure




Documents relatifs
Moreover, radial splitting of large tropical trees with high density wood is more prone to appear due to the decrease of the ratio between rupture energy and Young’s modulus when
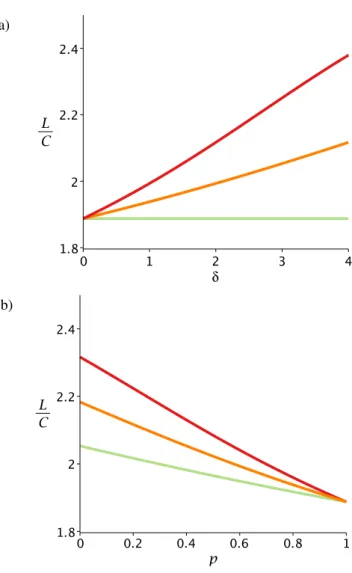
In agreement with simulation study 1, the results of simulation study 2 also showed that the estimated marginal posterior distributions covered the param- eter values used
By regarding the sampled values of the liabilities as observations from a linear Gaussian trait, the model parameters, with exception of thresholds, have fully conditional pos-
As an attempt to contribute to the urgently needed gap filling of leaf trait data for the Congo Basin, this paper presents a limited but unique dataset of leaf traits collected
The union of edges linking these points is called a branch of T (f). The proof is an easy application of Lemma 1.5.. We claim that ˜ F is the only element with this property.
By considering a continuous pruning procedure on Aldous’s Brownian tree, we construct a random variable Θ which is distributed, conditionally given the tree, according to
This pruning procedure allowed to construct a tree-valued Markov process [2] (see also [10] for an analogous construction in a discrete setting) and to study the record process
In this study, we used abundance data for 31 common tree species in 56,445 0,5-ha plots spread over more than 700,000 km 2 at the northern edge of the Congo basin and combined them