HAL Id: hal-02711885
https://hal.inrae.fr/hal-02711885
Submitted on 1 Jun 2020
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Isolation and sequence of the murine Fgf6 cDNA
Vincent Ollendorff, Olivier Rosnet, Irène Marics, Daniel Birnbaum, Odile
Delapeyrière
To cite this version:
Vincent Ollendorff, Olivier Rosnet, Irène Marics, Daniel Birnbaum, Odile Delapeyrière. Isolation and
sequence of the murine Fgf6 cDNA. Biochimie, Elsevier, 1992, 74 (11), pp.1035-1038.
�10.1016/0300-9084(92)90025-A�. �hal-02711885�
S h o r t c o m m u n k a f i o n
Isolation and sequence of the m u ~ n e Fgf6 cDNA*
V O l l e n d o r f f , O RosneK I M a f i c s , D B i m b a u m , O
deLapeyfi~**
~ ~ ~ Molecular On~logy, ~ ! 9 INSERM, 27 Bd Le~ R o u t , 13009 Marseille, France(Rec~ved 16 S ~ m ~ r 199~ ~ ~ Se~emb~ 1992)
Summary - We have ~u~ed the ~mcm~ of ~ e routine Fg/6 gene enco~ng a fibm~ast growth factor wi~ ~e purpo~ of ~oking for p~ative ~ g ~ a ~ r y sequences ~ ~e $' and 3' nomcod~g m~on~ The Fgf6 cDNA contains a yew ~ng 3' untransla~d portion of 4015
n u c l i d e s .
f l b ~ M ~ t g r o w ~ factor / o n c o ~ n e / nucleotide sequence / ~ g u M ~ ~emen~
~ t ~ d u ~ n
The fibrobla~ growth factor ( F G ~ ~ m i ~ comprises ~ date seven memb~s ~ ~ e ~ v ~ v e d ~ ~ v e m l ~ o g ~ processes [!1. We h ~ e characterized ~ e ~ n e enco~ng ~ e f i ~ h member of t ~ s ~mily ~ man [2-4] m d in mouse ~]. The transcripts of the
FGF
genes ~ general, and of Fgf6 ~ p a n ~ , comprise a ~ n g 3' tail co~esponding m an untranslated ~ o n . The m ~ of ~ s e q u e ~ present in humm as wall as in mouse ~ m ~ n s unknown. Howeve~ several ~eces ~ evidence ~ d ~ a t e that t ~ s ~ p e of ~ q u ~ m ~ ~ some role. A d d ~ o n ~ open ~ading flames (ORF~ have been ~ u n d ~ 5' or 3' reg~ns ~
FGF
genes D, 7]. M o ~ impo~anfl~ Curat~a and B a ~ o [ ~ have e~a~ished ~ e p~sence of r e g ~ m o ~ ~ e - meres w i ~ enhancer activ~y in ~ e 3' nomco~ng reg~n ~ the c ~ s d y related
Fgf4/K-Fgf
~ n e . T ~ s enhancer ~ n d s nuc~ar factors [9]. For ~ e pu~ose of stud~ng the stru~ure and posfi~e ~ g ~ a m ~ ro~ of ~ e 5' m d 3' untranflated ~ o n s of Fg/6, we c~ned and sequenced the mufine Fgf6 cDNA.M a t e r , s and m ~ h o d s cDNA cloning and screening
~o~sDtr~NcAt~nar~ gf~t~. "~h~Nc~I~ 15s~n~hd~sT~ioenm~7~c~ *The nucleotide sequence data presented in thh paper have been submitted to the EMBL-Genbank Data Libraries under the accession numbers M92415 (550 first nucleotides, of geno- ,m~x~n~edncMe92an4 ~ n ~ s ~ t of the sequence).
fink~s and l~ation m ~ ~10 arms wen performed us~g Amersham syntheMs and c ~ n g ~ts as ~commended by ~e manufacm~ The l~a~d mastiff was packaged us~g Gigapack GAd exffac~ (S~a~gene) and the phages wen ~ ~d u~ng NMSI4 as a bac~fi~ host swan. T~s I~ra~ was ~ e n e d with genomic (EcoRV-Na~ ~agme~) or cDNA (Psd-Nc~ fragment) mouse Fgf6 probes ~ h p~v~u~y de- scribed ~ ~1).
Isolation of cDNA c~nes by PCR amplification
An ~iqu~ ~ Bg) of totfl RNA from 1 5 ~ a y mouse emb~o was used to genemm a flint g~nd eDNA. The RNA was heamd to 90°C for 5 min and the cDNA w ~ syn~etized by incubating 10 m ~ ~ room ~ m p ~ u m and 1 h at 42°C ~ a 20-~ fin~
~ ' a c l t ~ t ~ g ~ atRnaNn95°C for(Pr°mega)5 rain, hNf of and l X~e cDNA PCR buffe~action After mix was succeed ~ PCR a m ~ c a t i o n ~ a fin~ vo~me of 50 ~ of I x PCR buffer containing 25 pm~ of each ~igonuc~o- tide pfim~ and 1 U of Taq p~ymera~ (CereS. The same PCR buffer ~ 0 mM KCI, 20 mM Tr~-HCI pH 8.4, 1 ~ mM MgCI>
1 mg ml -~ BSA) was used ~ both ~v~se tran~ripmse and ~n~7~tt~£h~twe~, a ~ c i ~ i t ~ l d ~ t ~ ~ i~t wg~cCh ~efOr anne~gcyde from~mperatu~65°C to 55°C. was ~ e ~ s i ~ m ~ r ~ e 1 ; ~ ; ~
where ~e eg~n~on time was I0 m~. The PCR pmdu~s wen then Num~nded uNng the K~now p N y m e ~ and subcloned into a SmaI-d~gested Bluescd~ (Stra~gene) ~asmid.
cDNA sequencing
Nuc~m~e sequenc~g was performed by ~e d~eoxy~h~n ~ r m ~ n m~hod u~ng single-s~anded and/or dou~e-swan- ded Blue~fi~ (S~agene) ~ m ~ e s and ~ e mottled T7 p ~ y m ~ e (Sequenase) protocol (US Biochemicals~
1 ~ 6
A.
/ / /; / / / / / / ; !,/
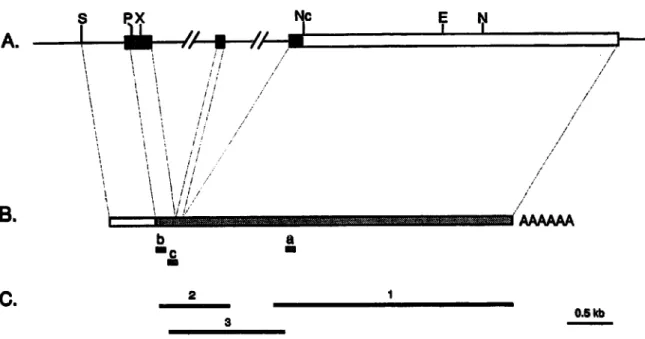
E I . / / B . ~ ! ! ! i / / / ! / i / / / / / ~ ~ ~ C ~ • 5 ~ C . 2 1Fig 1. Clo~ng of Fgf6
cooing
and untran~ated region~ Mo~ of the coding regions (black boxes) and the tot~ity of the Y untrans~ted region (open box) of the Fgf6 gene (A) were obtained by cDNA cloning (B, dashed box~ 5' Sequences (B, open box) were derived from genomk clonin~ R e s t r i ~ n enzyme sites mentioned in the ~xt are showm abbreviations are ~s fob lows: E, EcoRV; N, Nae~ Nc, Ncol; ~ PstI; S, Sphl; X, Xhol. The insets of the two cDNA dories (1 and 2) and the pmdu~ of the PCR ~oning (3) discussed in the texL are shown (C). The portions of the oligonucleotide pfime~ (a, b, and c) are shown above. The sc~e ~ the same for the three panels.R e s ~ and d ~ c u s ~ o n
In figure 1 the ~ones ~ were ~ a t e d from a mouse e m b ~ o cDNA l~rary are describe~ A fi~t eDNA c ~ n e ( c ~ n e 1) c o m ~ n e d 2.8 kb of 3' untran~ated sequence. A second cDNA ~ o n e (clone 2) contained part of ~ e 3' r e ~ o ~ ~ e wh~e of ~ e coding ~quences of exons 3 and 2, and ex~nded 5' into the ~ g g e ~ part of exon 1. In b ~ w e e n these two cDNA~ a poRion of • e 3' sequence r a i s i n g in the cDNA clones was o ~ n e d by c ~ n g of a p o ~ m e ~ s e c h i n reaction (PCR) produ~ obtained a~er two PCR s~ps. The first ~ep used a 3' primer ~ ) d e i g n e d in ~ e 5' end of ~ e fi~t ~one and a 5' primer (b) chosen in the fi~t e x o ~ T h e e prime~ span two different exons. The~ nucleo- fide sequences were 5'-GGATTGTCTTGCAGGT- CAGG-T and 5'-AACACACGAGGAGAACCCCT- 3', respe~ively. The second step was performed in the same conditions e x c e ~ for the use of a 5~ AGAGTGCTCTCTTCATTGCC-3'- nes~d 5' primer ~ ) defigned in ~ e second exon. The combin~ion of • e cDNA c ~ n e s and PCR produ~ r e s ~ d in a 4 ~ - kb cDNA, m ~ n g the fi~t 54 n u c ~ i d e s of Fgf6 coding sequence. Further scree~ng of ~ t h e r the orig- in~ or newly made cDNA fibraries did not allow ~ ~ e eDNA ~ones ex~nded f u l ~ ~ ~ e 5' e n ~ pre-
sumably because of strong secondary structure~ Further u p . r e a m sequences corresponding to the Y coding end o f the Fgf6 gene were therefore obtained from genomic ~ones. The genomic sequence was ex~nded even more 5' to include putative regulatory e ~ m e n t ~ P o t i o n s o f p r e ~ o u ~ y ~olated cosmid clones from the Fgf6 ~ c u s were subcloned. One such fragment, named XX ~ ] , conesponded to the 5' por- tion of the Fgf6 gene. A SphI-XhoI subfragment (fig IA) derived from XX was sequenced.
As in human [3], a 624-nucleofide-long ORF ~ able to code for a protein of 208 residues (fig 2). However, there are three in-frame ATG codons and two sho~er proteins could pos~bly be made. The 5' region com~ns n ~ t h e r CAAT nor TATA boxes Putative regulatory d e m e n t s are ~ c a t e d about 300 bp upstream of the ATG (fig 2); they i n c ~ d e a decanu- ~eotide repe~ AGGGTGGTGA, and the palindromk TGACGTCA ~ n d i n g si~ for the CREB ~ e m e n t found in the 5' part of cAMP regulated genes [11]. The 3' untran~ated region, ~arting a~er the stop codon ~ position 1121, is 4015 n u c ~ o t ~ e s ~ n ~ The polyadenylation ~ ~ preceded by an unusual poly(A) ~ g n ~ sequence at position 5122. A poly(A) stre~h ~arting ~ position 1686 may represent the remnant of an arch~c polyadenylation sequence. ~ ~
1
CCC~CC~TCC~CCT~C~G&~TGCT~T~C~C~TTCCT~TAG ~ ~ ~ ~ ~ T ~ T C T ~ G121 ~ ~ ~ A ~ T ~ T ~ C ~
181 T ~ ~ ~ ~ T A ~ C T A T ~ T ~ T C
241 ~ ~ ~ ~ A ~ C ~ C ~ T G
301 ~ A T A ~ ~ ~ ~ T ~ T ~ C ~
351
~ T ~ ~ T ~ ~ T ~ C C ~ C ~ T421 ~ C ~ G ~ ~ A ~ A ~ T T C ~ C ~ C G ~
481
~ ~ T ~ G A ~ T ~ T ~ C ~ _ _ 5~ ~ ~ ~ T T ~ G T C C T ~ T ~ T _ _ ~ ~ ~ ~ C ~ T ~ C T C ~ 6~ ~ C C K ~ C T ~ T A ~ T ~ T ~ ~I ~ T ~ ~ ~ T C T ~ T ~ C ~7~
~ C ~ C T ~ T ~ C ~ C C ~ A 8~ ~ T ~ C ~ C ~ A ~ T ~ G C C ~ ~ G9~ ~ T C ~ T T ~ T ~ ~ C ~ T ~ C ~ G C T T C ~
9~ ~ T ~ G T ~ C G ~ ~ C T ~ C ~ C T A C ~
~21 G T ~ C C T ~ T T ~ T ~ C ~ T ~ G ~ G ~ T
10~ N
~
~
T
~
~
~
~
II~ C ~ ~ T G C G ~ ~ T ~ G ~ 1201 ~ T h ~ C ~ A ~ A T C ~ T ~ G T G ~ T ~ T 1~1 ~ ~ ~ ~ C ~ G ~ 1321 ~ ~ C ~ ~ C C T ~ T A ~ T T 1381 ~ ~ T ~ ~ T ~ G ~ ? ~ 1441 T A ~ ~ ~ ~ f f f f A ~ T I ~ T T 1501 ~ ~ ~ ~ G ~ C ~ 1561 I ~ ~ ~ ~ T ~ A ~ ~1621
~ A ~ A ~ ~ A ~ A T ~ T A T ~1681 A ~ ~ ~ ? ~ C ~ C ~ T T C ~ ~
1741 ~ T ~ A ~ T A ~ T ~ ~ T C C C C ~ A ~
1801 ~ ? ~ T A ~ ~ G ~ T ~ ~ T
1861 GTCCTGTAG~C~TT~C~~CTCC~ ~? ~T~G
1921 C ~ G C C ~ G T ~ C G ~ ~ ? ~
19~
~ ~ C ? ~ T ~ C C ? C C ~ ~ G T ~ T 20~ T A ~ G ~ ~ T ~ T ~ C T C T C ~ C T ~ 21~ TCU~CT~C~G~CCC~TA~C~T~CTCG~CT~G 21~ C T A ~ T C ~ T ~ T ~ ~ T G A C C T ~ A ~ 2221 C C T ~ C f A ~ G ? A ~ T ~ ~ G ~ G C ~ 22~ T C ~ ~ f ~ ~ A T ~ T ~ C ~ ? T C 23~ C ~ A ? ~ ~ A G ~ ~ T T ~ T ~2401
T C C T ~ T ~ C ~ C G T A ~ T A T ~ T A T ~~61
~ T ~ C C C T C ~ T A C C C ~ C ~ ~ T C C C C ~ 2521 ? C T ~ ? A C ~ ? ~ T A C C ~ A W C ~ A T A ~ C ~ A 2581 ~CAGATCTC~CTCACGGACTCTGAGCTGCAG~CG~TC~G~TTC~CGATGC~G~G 2641 ~ C ~ T T C ~ C T T ~ C C ~ T ~ G ~ T ~ T ~ A ~2701
A ~ G ~ C C G R ~ G A ~ G A ~ T ~ T ~ T ~ T ~ G A ~ T ~ C2761 A T ~ T C ~ T T ~ A ~ ~ C C ~ T ~ A G
2821
~ ~ T T ~ T C ~ T A T T ~ T C ~ T ~ G T ~ 288~ ~ ~ T ~ C T ~ T A T C ~ T A ~ T A T ~ T ~ T A T ~ G 294~ ~T~T~CTT~A~T~TGA~T~T~CTGATGT~CT~G 300~ ~ T ~ T ~ T ~ T A T C T G ~ T A ~ G T ~ T A C ~ C 306~ T A G A T ~ T ~ C ~ T ~ C C ~ C ~ C T ~ C ~ ~ 3~2~ ~ C C ~ C C ~ T C C T ~ T C C T T T 3~8~ CTA~TC~AT~TTA~T~CTTGA~TCTAGTG~ACT~GACCCTG 324~ ~T~TG~TCTTT~T~CTTT~T~TG~GT~TC~TCC~A~G 330~ GTTC~T~CCACTC~CCCTT~T~CG~T~G~G~GCCCCGG~G 336~ CCCTGAT~CGGCCTG~GT~CCT~GA~G~C~GTTCCT 342~ ATG~TGA~TT~TTGT~~GTA~T~CC~TTTTGCT~AC 348~ TGGTT~T~ATGGRT~TACT~CT~TTTCCTGT~G~T~TCTT~GA 354~ GGTTTCTCTTTGACC~GT~A~GTA~GT~CC~TC~C 3~0~ ~ G T T ~ G ~ A T ~ G A ~ T ~ G A G ~ T C C C T T 3 ~ CTCTCCT~TACTGT~AG~TT~TCCT~C~CACCT~CTCCTT 372~ G ~ G T ~ T T C G T ~ T ~ G T G T G T T C ~ T T ~ G ~ C 3781 ~AG~T~T~GT~GA~CT~GT~TCT~GAG~TCT~TT~GTTC 384~ CCTGTA~G~T~T~T~GT~C~T~TGTCT~GTCTCTCTCT 390~ C ~ T C ~ G ~ G ~ T ~ G A T A T G ~ C T ~ T T ~ T C 396~ ~C~CCTCGTGA~C~T~CTTTTGATA~TTTGAC~GT~ 4~2~ ~T~GAT~TAC~T~TACCTT~GA~T~CC~GTTCCT 408~ C T G ~ G A ~ C C T A K C ~ ~ T C T ~ G T ~ C ~ G T A T G G 4X4~ T T T ~ C C ~ T ~ C ~ C T ~ T ~ A ~ A ~ C ~ T C ~ C T C T T 420~ A ~ T G ~ C T C T k ~ C ~ C C T G ~ T ~ T ~ T G K C ~ C C ~ C 426~ TTGTT~TGA~T~C~AC~TTGA~CCCCTTTCCTGTC~C~C 432~ A ~ T ~ C ~ T ~ T ~ C T ~ G ~ A ~ G A G G ~ A ~ G ~ G ~ T C ~ 438~ TG~TCC~TC~CCT~G~T~T~TT~G~TAC~TGT~ 444~ ~TC~T~CCC~CTC~GTATA~T~CT~GATTCC~C~CCCTT 45~ TTTGT~GCT~X~TA~C~TCTAKCTTA~T~TCCC~G 456~ A T ~ T ~ C T ~ A ~ C ~ G A ~ T C ~ C T ~ G T ~ A T ~ A 46~ TGA~TA~CTACC~CC~C~GA~GAC~C~TTG~CTT~CTG~G 4~8~ ~ C ~ T ~ ~ T ~ C ~ A ~ T A C T ~ 4741 G T ~ A ~ T C T ~ G T ~ k T ~ C T C K C C ~ G A ~ T A C ~ T ~ 480~ A ~ G A C T C T ~ T ~ G T ~ T ~ C ~ T G T ~ G T A T A C C 48~I T ~ C T A C T G ~ C ~ C ~ T ~ C T ~ G T ~ G ~ A ~ C 492~ ~ T ~ T ~ T A ~ G ~ C T ~ T C T ~ T ~ A ~ C ~ T A ~ A ~ T ~ 498~ A~TG~T~TGT~TT~G~T~T~TTCT~TG~T~TAT~ATGTG 50~ ~ C ~ T ~ T ~ C C ~ ~ T ~ T ~ A T A C G ~ T ~~01 ACT~TTCACTCCGRCCTC~TT~C~~
Figlows:genomic
sequences2"threeNUC~°fideAT
G codonsSequence(1
to 550)(bold°fand
cDNAand
the underlined),m°USesequence
Fgf6 theC°ding(55
lstoptoandcodonthe
Theuntranflated(bold)
end~
' putativeCOdingreg~n~5,sequenceTheregulatorynUCk°fideis
boxed.dementsSequenCesped~(unded~ed),
featuresis
a compoSRear
e vafiantfol.Of
polyadenylation fign~ sequence (doubly underlined) and a putative alternative po~adenylation fign~ sequence (douNy unde~
lined, position 1664). Doubly underlined and in bald ~ a block of 15 tetranuckotide repe~m
1 ~ 8
p~ceded by a pmative po~adenylation ~gn~ sequence thin may be used ~ h o a e r Fgf6 ~anscfipts of about 1 ~ kb are indeed som~imes observed in some tissues). A block of 15 ~ a n u c ~ o f i d e r e p e l s is pres- ent from position 3099 to 3159. These r e p e l s identify a putative m ~ r o s m d f i ~ and could be used to reve~ a p d y m o r p ~ s m ~ mous~ and maybe ~ s o in human w h e ~ ~ e y c o ~ d be conserved [12].
Indee& in a more general wa~
comparison
of ~ e 5' and 3' non-co~ng sequences described h e ~ w i ~ the co~espond~g human sequences c o ~ d y i d d infor- m ~ n about impo~am conserved ~ m u ~ of the 5' and 3' ~ g ~ n s . Furthermore, the c ~ n ~ g and se- quencing of the 3' un~anslmed region of Fgf6 will h e ~ in deriving specific Fgf6 probes useful in other ~ u d ~ s such as ~ situ h y b r i d i z ~ n a n ~ y ~ s of Fgf6 e x p l a i n . Indeed, ~ e 5' ~agmem of clone 1 ob- tained by E c o R V d~estion (named dream) used as ~ m ~ e for anfisense fiboprobes gives a good fign~ in h y b r i ~ z m ~ n of mouse embryo sections (deLapeyri~re ~ al, ~ p~paration).Acknow~dgments
T~s wo~ was supposed by In~rm and grants fram ARC and AFM. We are g m ~ ! to F Coffer for h~p~l advice and J Hanche for mchn~fl assistance. VO is the ~dp~m of a ~l- ~wsh~ from the 'L~ue Nm~nfle Cont~ ~ Cancer'.
References
I Gospodarowicz D (1991) B ~ g ~ acfiv~es of fibro- ~a~ grow~ ~ c ~ . Ann NY Acid Sci 638, I-8
2 Mafics I, A d ~ d e J, Raybaud E Mauei MG, C o ~ r E ~anche J, deLapeyfi~ O, Birnbaum D (1989) Chamc- ~fizm~n of ~e Hst-related FGF-6 gen~ a new member of
t ~ f i b ~ g ~ h ~ gene ~ f l ~ O ~ e ~ ~ 5 ~ 3 C o ~ r E Bat~ ~ M~cs L ~ ~ ~ ~ m - baum D (1991) ~ t ~ e s ~ ~ ~ ~ ~ p~- duct ~ d ~ of ~e fign~ ~ d e . O ~ e ~ 1 ~ 4 ~ r E ~ n d ~ ~ M~cs L ~ s ~ t ~ B ~ M, ~ c h e L M ~ t o ~ ~ ~ , d ~ ~ ~ B ~ u m D (1991) ~ e FGF6 ~ n e ~ n the FGF multigene ~mily. Ann ~ A c a d Sci 638, 53~1
5 d e L a p e y ~ O, Rosn~ ~ B e n h ~ h ~ R~baud ~ M a ~ o S, ~ a n c ~ J, G ~ a ~ E M ~ M ~ Cop~ I~d ~ ~n~ns ~ Co~i~ E ~ m ~ u m D (19~) S ~ u r e , chromosome m ~ n g ~ d expre~ion of the routine F~-6 gene. Oncogene 5, 823-831
6 ~ffa ~ ~ s ~ ~ ~ g a w a ~ S~amo~ ~ ~rada ~ Su~mura T (1987) cDNA s~uence ~ a human t ~ n s ~ ~ ~ r ~ ~o ~ a ~ ~ f i c ~ t ~ f ~ t y ~ g ~ e ~ USA 8~ 298~2984
7 Zhan ~ Ba~s B, Hu X, G ~ d ~ b M 0988) ~ e hum~ FGF-5 gene encodes a novd pm~n ~ a ~ d to ~roblast growth factor~ Mol ~ H B~l ~ M ~ 5
8 Curato~ AM, Basfl~o C (199~ E x p l a i n of the K-f~ proto-onco~ne is co~ml~d by Y r e g ~ o ~ e~me~s wh~h ~e s ~ ~r emb~onal c~cinoma cel~ Mol ~ H B ~ l !~ 2475-24~
9 Schoodemm~ L ~ e r W (1~1) ~me~dependent ~g~m~n of ~e kFGF gene ~ e m ~ o n ~ c~inoma and emb~on~ s~m ceils. M e ~ Dev ~ ~ 6
10 Don ~ Co~ ~ W a ~ w r i ~ B, B~er K, M~6ck J (1991) '~ucMown' ~ R m c~umvem ~ o u s ~ d ~ ~ne am~cation. N u c ~ Acids Res 1~ 4 ~ 8
I1 Hyman S~ Comb M, Mn YS, ~ L G~en M ~ Goodman HM 0988) A common trans-ac~g ~ctor ~ ~v~ved ~ tr~scfi~on~ wgu|ation of neumtr~smitter ~nes ~ ~ d ~ AM~ Mol ~ H B ~ ~ ~ 2 5 ~ 3
12 Moo~ S, S~geant ~ ~ n g Z Matt~k J, Georges ~ He~el D (1991) ~ e c o n ~ n ~ d ~ u c ~ ~cro- s a ~ l ~ s among mamm~an genomes ~ w s ~e use ~ h ~ e ~ o u s PCR ~ m e r ~ ~ ~osdy ~ated s ~ e s . G e n o m ~ 1~ 6 5 ~ 6 0