HAL Id: hal-01975970
https://hal.archives-ouvertes.fr/hal-01975970
Submitted on 9 Jan 2019
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Towards a congruent reclassification and nomenclature
of the thermophilic species of the genus
Pseudothermotoga within the order Thermotogales
Hassiba Belahbib, Zarath Summers, Marie-Laure Fardeau, Manon Joseph,
Christian Tamburini, Alain Dolla, Bernard Ollivier, Fabrice Armougom
To cite this version:
Hassiba Belahbib, Zarath Summers, Marie-Laure Fardeau, Manon Joseph, Christian Tamburini, et
al.. Towards a congruent reclassification and nomenclature of the thermophilic species of the genus
Pseudothermotoga within the order Thermotogales. Systematic and Applied Microbiology, Elsevier,
2018, 41 (6), pp.555-563. �10.1016/J.SYAPM.2018.04.007�. �hal-01975970�
Towards
a
congruent
reclassification
and
nomenclature
of
the
thermophilic
species
of
the
genus
Pseudothermotoga
within
the
order
Thermotogales
Hassiba
Belahbib
a,
Zarath
M.
Summers
b,
Marie-Laure
Fardeau
a,
Manon
Joseph
a,
Christian
Tamburini
a,
Alain
Dolla
c,
Bernard
Ollivier
a,
Fabrice
Armougom
a,∗aAixMarseilleUniv,UniversitédeToulon,CNRS,IRD,MIO,Marseille,France
bExxonMobilResearchandEngineeringCompany,1545Route22East,Annandale,NJ08801,UnitedStates cAixMarseilleUniv,CNRS,LCB,Marseille,France
a
r
t
i
c
l
e
i
n
f
o
Articlehistory:
Received15December2017 Receivedinrevisedform20April2018 Accepted27April2018 Keywords: Thermotogae Pseudothermotoga Taxonomy Classification
a
b
s
t
r
a
c
t
ThephylumThermotogaegathersthermophilic,hyperthermophic,mesophilic,andthermo-acidophilic anaerobicbacteriathataremostlyoriginatedfromgeothermallyheatedenvironments.Themetabolic andphenotypicpropertiesharboredbytheThermotogaespeciesquestionstheevolutionaryevents driv-ingtheemergenceofthisearlybranchoftheuniversaltreeoflife.RecentreshapingoftheThermotogae taxonomyhasledtothedescriptionofanewgenus,Pseudothermotoga,asistergroupofthegenus ThermotogawithintheorderThermotogales.ComparativegenomicsofbothPseudothermotogaand Ther-motogaspp.,including16S-rRNA-basedphylogenetic,pan-genomicanalysisaswellassignatureindel conservation,providedevidencethatThermotogacaldifontisandThermotogaprofundaspeciesshouldbe reclassifiedwithinthegenusPseudothermotogaandrenamedasPseudothermotogacaldifontiscomb.nov. (typestrain=AZM44c09T)andPseudothermotogaprofundacomb.nov.(typestrain=AZM34c06T),
respec-tively.Inaddition,baseduponwhole-genomerelatednessindicesandDNA–DNAHybridizationresults, thereclassificationofPseudothermotogalettingaeandPseudothermotogasubterraneaaslatterheterotypic synonymsofPseudothermotogaelfiiisproposed.Finally,potentialgeneticelementsresultingfromthe distinctevolutionarystoryoftheThermotogaandPseudothermotogacladesarediscussed.
©2018TheAuthors.PublishedbyElsevierGmbH.ThisisanopenaccessarticleundertheCC BY-NC-NDlicense(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
Thermotogamaritima[19]embodiestheoriginalmemberofthe phylum Thermotogae,whichcurrently harborsat least65 culti-vatedspecies distributedwithin 13genera [23].Although their truephylogeneticpositionremainsamatterofdebate[6,13],the Thermotogales,whicharecloselyrelatedtoAquificales[36],are com-monlydepictedas slowlyevolving organismsdefining an early branchinglineagewithintheBacteria[1].Thephylogenetic relat-ednessamongtheThermotogalesspeciesdonotreflecthoweverthe geneticvariabilityoftheirgenomeswhichundergopervasive hor-izontalgenetransfer(HGT),includinggeneinfluxfromFirmicutes andArchaea[32,34,58].
∗ Correspondingauthorat:AixMarseilleUniv,UniversitédeToulon,CNRS,IRD, MediterraneanInstituteofOceanography(MIO),163AvenueLuminy,13288, Mar-seille,France.
E-mailaddress:fabrice.armougom@mio.osuphytheas.fr(F.Armougom).
The phylum Thermotogaewas originallydescribed from sin-gular characteristicscarried by itsrepresentativeswhich thrive most often at high-temperature and exhibit a single-unit lipid membrane as well as a unique loose-fitting sheath structure (called“toga”)surroundingthecell[40].Untilrecently,the phy-lum Thermotogae was believed to mainly comprise anaerobic, thermophilic (Topt≥70◦C) and hyperthermophilic (Topt≥80◦C),
fermentativerod-shapedbacteriawiththehighestgrowth temper-ature(65–90◦C)undernear-neutralpHconditions[14].Thisview wasdispelledwiththediscoverywithintheorderThermotogales ofmesophilic[18,33]andmoderatelythermophilicacidophilic lin-eagescomprisingthegenusMesotogaandgeneraMesoaciditogaand Athalassotoga[21,45],respectively.
ThephylumThermotogaeinitiallyconsistedwithinasingleclass Thermotogae;a singleorder,Thermotogales;and asinglefamily, Thermotogaceae[44].Recentlyhowever,BhandariandGuptahave revisitedthetaxonomyoftheThermotogaebyidentifying molec-ularsignaturesfromwholegenome-basedcomparisons[5].The newtaxonomicproposalresultedfromtheidentificationof
Con-servedSignatureIndels(CSIs)atseveraltaxonomiclevelsthatwas fairlycongruentwiththeinferenceof16SrRNA-based phyloge-neticrelatednessoftheThermotogaespecies[5].Accordingtotheir findings,thephylumThermotogaewasredefinedanddividedinto fourseparateorders(Thermotogales,Kosmotogales,Petrotogalesand Mesoaciditogales)andfivefamilies(Thermotogaceae, Fervidobacte-riaceae,Kosmotogaceae,PetrotogaceaeandMesoaciditogaceae).At thegenuslevel,theoriginalgenusThermotogawasproposedto besplitintotwogeneraThermotogaandPseudothermotoga.This conclusionwassupportedby16SrRNA-basedphylogenetic recon-structionandidentificationofclade-specificCSIsthatindicatedtwo distinctevolutionaryhistoriesofcultivatedmembersoftheoriginal genusThermotoga[5].Todate,thegenusPseudothermotoga con-tainsP.elfii,P.lettingae,P.subterranea,P.hypogeaandP.thermarum
[35].BhandariandGupta’soriginalwork[5]echoesthemodern taxonomypracticethatreferstotheuseofthecompletegenome DNAsequenceasthereferencestandardtodetermineboth phy-logenyandtaxonomyofspecies[48].Withtherapidadvancesin affordablefullgenomesequencing,thecurrentviewoftaxonomy therebyintegrateswholegenome-basedcomparisons[42,53]with aninflowofOverallGenomeRelatednessindices(OGRI)[8] includ-ingdigitalDNA–DNAHybridization(DDH)[30],AverageNucleotide Identity(ANI)[15,26],ortetranucleotideregression[52].TheANI measureiscurrentlythemostacceptedOGRIbythescientist com-munity[25].
Recently,twonovelthermophilicanaerobicspecieswere iso-lated from terrestrial hot springs in Japan and were classified as belonging to the genus Thermotoga as T. caldifontis and T. profunda [31]. Using multiphasic molecular approaches based on16S rRNA,core-genomephylogeny,and identificationofthe Pseudothermotoga-specificCSIs,weprovideevidencefor reclassify-ingT.profundaandT.caldifontiswithinthegenusPseudothermotoga asPseudothermotogaprofundacomb.nov.,andPseudothermotoga caldifontiscomb.nov.,respectively.Wealsodemonstrateby exper-imentalDDHandOGRImeasuresthatPseudothermotogalettingae andPseudothermotogasubterraneashouldbereclassifiedaslatter heterotypicsynonymsofPseudothermotogaelfii.
Methods
Genomefeatureanalysis
Thermotoga and Pseudothermotoga genomes were retrieved fromtheNCBIftpsite(ftp://ftp.ncbi.nlm.nih.gov/)underthe acces-sion numbers listed in Table 1. The coding sequences (CDS) information wasextracted from the gbkand GFF files. The GC content was estimated by the geecee function of the EMBOSS package 6.6.0.0 [46]. The hypothetical protein fractions were
deducedfromtheproteinannotationfiles(faa).TheT.caldifontis andT.profundagenomeswereincludedinthegenus Pseudother-motoga for the genome feature analysis. Mean comparison of Thermotogaceae clades was performed by the non-parametric Mann–Whitney–Wilcoxon test. The Mann–Whitney–Wilcoxon testcanbeappliedonsmallsamplesizeanddoesnotrequireany assumptionaboutthevarianceequalityandpopulation distribu-tions.TheMann–Whitney–Wilcoxontests,genomesize,geneand GCcontentswereperformedbyGraphPadprismSoftwareversion 5.0.M,SDindicatemeanandstandarddeviation,respectively. SSUrRNA-basedphylogeny
Thenear-fulllength16SrDNAsequences(>1300pb)ofthe Ther-motogaespecieswereretrievedfromtheNCBIdatabase(https:// www.ncbi.nlm.nih.gov/) and aligned with the MAFFT software v7.123b[24].Amaximumlikelihood(ML)phylogenetictreewas constructedfrom1624nucleotidepositionsusingthePhyML3.1 algorithm [17] implemented in the Seaview package v4.6 [16]
withGTR+I+Gsubstitutionmodeland1000bootstrapsreplicates. The phylogenetic tree was rooted using the Coprothermobac-terproteolyticusIT3(GU363592)andCoprothermobacterplatensis (Y08935.1)16SrDNAsequencessincethegenusCoprothermobacter wastheclosestbranchingoutgroupinupdateSSUrRNA phyloge-netictree[57].
Core-genome-basedphylogeny
The genome sequences of cultivated Thermotogaceae were retrievedfromtheNCBIaccessionnumberssummarizedinTable1. A core/pan-genome analysis was performed using the Roary pangenomepipelinev3.6.1[37].TheRoarypipelinegenerated a coregenealignmentfromPRANKv.140110[29]witha50% iden-titycut-off level, as wellasa gene presence/absence matrix. A maximum-likelyhood(ML)core-genometreewasinferredusing PhyML 3.1 algorithm implemented in Seaview v4.6 with gen-eralized time-reversible (GTR) and gamma distribution. PhyML approximatedlikelihoodratio(aLRT)SH-likevalueswereindicated inaconsensustree.Theroaryplots.pyscriptswereusedto com-binethecore-genome-basedtreeandthegenepresence/absence matrix.
MolecularsignatureswithinthegenusPseudothermotoga
TheCSIs identifiedasuniquetothegenusPseudothermotoga wereoriginatedfromreferenceproteinsincludingthe Glycerol-3-phosphatedehydrogenase,thehypotheticalproteinTheth0845, the hypothetical protein TM1140 and the hypothetical protein
Table1
GenomicandgrowthfeatureswithintheThermotogaceae.
Strains Genomesize(bp) GCcontent(%) PredictedCDS CDSwithoutpredicted
function Optimalgrowth temperature Accessionnumber P.elfiiDSM9442T 2,169,860 39 2063 497(24%) 66 AP014507 P.lettingaeTMOTDSM14385 2,135,342 39 2047 414(20%) 65 CP000812
T.profundaAZM34c06T 2,187,612 39 2073 600(29%) 60 AP014510
P.hypogeaDSM11164T 2,165,416 49 2073 663(32%) 70 CP007141
T.caldifontisAZM44c09T 2,014,912 51 1940 564(29%) 70 AP014509
P.thermarumDSM5069T 2,039,943 40 1965 428(21%) 70 CP002351 T.naphthophilaRKU-10T 1,809,823 46 1805 333(19%) 80 CP001839 T.petrophilaKU-1T 1,823,511 46 1772 335(19%) 80 CP000702 Thermotogasp.Cell2 1,749,904 46 1632 321(18%) – CP003409 Thermotogasp.RQ2 1,877,693 46 1832 337(18%) 80 CP000969 Thermotogasp.RQ7 1,851,618 47 1812 347(19%) 80 CP007633 T.neapolitanaDSM4359T 1,884,562 47 1829 354(19%) 80 CP000916 Thermotogasp.2812B 1,843,731 46.2 1808 361(20%) – CP003408 T.maritimaMSB8T 1,869,612 46 1858 369(20%) 80 AE000512
Tlet0907[5].Thereferenceproteinsequenceswereretrievedfrom theircorrespondinggenomes,P.lettingaeTMOT(Tlet907),T.
mar-itimaMSB8T (TM1140), P.thermarumLA3T (Therth0845)and P.
lettingaeTMOT(Glycerol-3-phosphatedehydrogenase).The
iden-tificationofthehomologousproteinsintheotherThermotogaceae genomeswasperformedbyBLASTpsimilaritysearch[2]usingeach ofthefourreferenceproteinsasseed.Aproteinsetwasbuiltwith eachreferenceproteinandtheirhomologswithinThermotogaceae spp.Themultiplesequencealignmentsoftheproteinsetswere per-formedbyMAFFTv.7123b[24].Themultiplesequencealignment wasvisualizedbyJalview2.9.0b2[54]usingClustalXaminoacid colors.OnlythealignmentpartcontainingtheCSIregionwas dis-played.TheCSIpositionswerereportedaccordingtothereference sequenceineachCSIalignment.
DNA–DNAHybridizationdissimilaritymeasures
DNA–DNAHybridization(DDH)studieswereperformedusingP. lettingaeTMOT(DSM14385)[4]andP.elfiiSEBR6459T(DSM9442)
[43].TheDDHstudieswerecarriedoutbytheDSMZasdescribed byDeLeyetal.[27]withsomemodifications[11,20]andusinga model2600spectrophotometerequippedwitha2527-R thermo-programmerandplotter(GilfordInstruments).Renaturationrates werecomputedwiththeTRANSFER.BASprogram[22].
Whole-genomerelatednessindices
TheAverageNucleotideIdentity(ANI)andAverageAminoAcid Identity(AAI)indiceswerededucedfrompairwisegenome[26]
and conserved coding proteins [47] comparisons, respectively, usingtheavailablefullThermotogaceaegenomes.TheANIb mea-surewas preferredto ANImsince it was more appropriate for thecomparison of distantrelated genomes and forThermotoga spp.[28].TheANIbreliedonthesequencealignmenttoolMUMer
[9] and was implemented in theJSpecies 1.2.1 software pack-age(www.imedea.uib.es/jspecies).TheANIbrangevalues(95–96%)
correspondtothethresholdforspeciesdelineation(32).TheAAI calculationwasperformedbyCompareMv0.0.21software(https:// github.com/dparks1134/CompareM),whichusesDIAMOND0.8.14
[7]forbestreciprocalhits.Homologywasdefinedwithsequence similarityabove50%,sequence coverageabove75%and a mini-mumE-valueof 1e-05.TheresultingANIb andAAIvalues were transformedindistancematrixandthehierarchicalclusteringwas performedbyhclust(Rpackage)usingacompletelinkagemethod. Theheatmap.2function(Rggplotspackage)wasusedtoshowthe ANI/AAIheatmapwithacolorschemereliedonANIvalues.
Resultsanddiscussion
GenomiccharacteristicswithintheThermotogaceae
Previous work revealed that the former genus Thermotoga consistedoftwodistinctgroupsofspeciesaccordingtotheir opti-malgrowthtemperature(Topt)higherthan77◦Candlowerthan
70◦CforThermotogaspp.andPseudothermotogaspp.,respectively
[14].WithaToptcomprisedbetween60◦Cand70◦C,Thermotoga
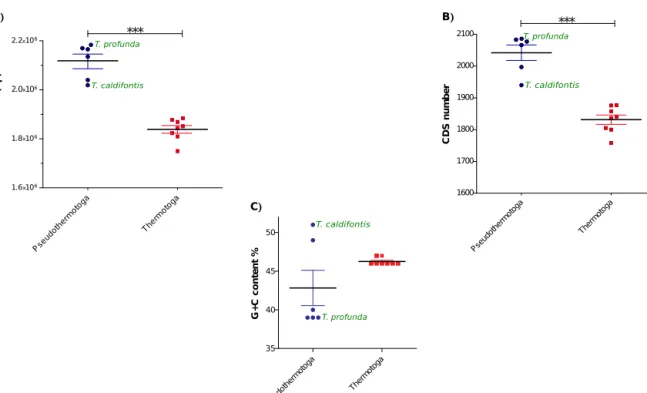
caldifontisandThermotogaprofunda sharedgrowthtemperature characteristicsofPseudothermotogaspp.Inadditiontothis phe-notypic feature, T. caldifontis and T. profunda carried genomic attributesmoresimilartothePseudothermotogathantothe Ther-motoga representatives(Table1,Fig.1).Themeangenomesize reached2.12Mb(SD=0.073)and1.84Mb(SD=0.044)withinthe generaPseudothermotogaandThermotoga,respectively(Fig.1A). ThegenomesizeofPseudothermotogaspp.wastherefore signifi-cantlylarger(Mann–Whitney–Wilcoxontest,p=0.0007)andmore variablethantheThermotogagenomes.Moreover,thegenomesize and growth temperature were negatively associated (r=−0.95) withinThermotogaceae(Table1).Recently,the“streamlining” con-cept has been proposed, i.e. that thermal adaptation involves reductioninthegenomesize[50].Asexpected,themeangene con-tentofPseudothermotogaspeciescomparedtoThermotogaspecies consisted ofmore coding sequences (Mann–Whitney–Wilcoxon
Fig.1. GenomefeatureswithinThermotogaceaefamily.
(A)Genomesizevariability.(B)CDSnumberdistribution.(C)GCcontentdistribution.Blacklineindicatesmean±SD.T.caldifontisandT.profundaspeciesareincludedwithin thegenusPseudothermotoga.
Fig.2.16SrRNA-basedphylogenetictreeoftheThermotogae.
MLtreebuiltfrom6516SrRNAsequencesand1624positions.Bootstrapvaluesfrom1000replicatesareindicatedonlywhen>50%.Thetreeisrootedwith16Sof CoprothermobacterproteolyticusIT3andCoprothermobacterplatensis.Barscale,9%nucleotidesubstitutions.Orderandfamilytaxonomiclevelsareindicatedinblackandblue, respectively.ThePseudothermotogaandThermotogageneraareboxedinred;theredcircleindicatestheirexclusivecommonancestor.Detailsofthebranchsupportvalues ofphylogeneticrelatednesswithinPseudothermotogaandThermotogaisshownbyacladogramonthetopright.
test,p=0.0007)(Fig.1B),withoutaffectinggenomecompactness (similargenedensity,datanotshown).Thehypotheticalprotein fractiondeducedfromtheCDSfunctionalannotationwaslarger withinthePseudothermotogaspp.thanwithintheThermotogaspp. (Table1).ComparedtotheGC contentofthePseudothermotoga spp.(M=42.83,SD=5.601),thoseofThermotogaspp.wereslightly higher(notsignificant,p=0.3308),andmoresimilartoeachother (M=46.25,SD=0.463).T.caldifontisandP.hypogeaspecieshadthe highest GC content (Table1, Fig. 1C), were closely related but slightlydifferedfromtherestofthePseudothermotogaspp.because oftheirlowergenomesizeandahigherGCcontent.
ThephylogeneticrelatednessofThermotogae
Theevolutionaryrelationshipsof 65species affiliatedtothe phylumThermotogaewereestablishedby16SrRNA-based phyloge-neticinference(Fig.2).Therecentdescriptionofthermoacidophilic MesoaciditogalescomprisingonlyMesoaciditogalauensisand Atha-lassotoga saccharophila formed the deepest diverging lineage within this phylum [21,45]. The next well-supported branch separatedbothPetrotogalesandKosmotogalesordersfromthe Ther-motogales.The Petrotogales clade consists of Marinitoga spp. as wellastheGeotogaandPetrotogasistergroups.Withintheorder Kosmotogales,thethermophilicKosmotogalineagenotablyharbors theK.olearia and therecent K.pacifica species, which demon-strateanoutstandinglywidegrowthtemperaturerange,20–79◦C and 33–78◦C, respectively [10,23]. The closest relative of Kos-motogawasthemesophilicMesotogalineage,withlowergrowth temperaturerange(30–40◦C).However,thelowbootstrap
sup-port(60,Fig.2)oftheKosmotogalesandPetrotogalesdichotomy broughtintoquestiontheirtruephylogeneticplacement[41,45]
despitearecentcore-gene-basedphylogenywith17Thermotogae species[23].Finally,Thermotogalesappearedtobethemostdiverse lineage,encompassingthegeneraThermotoga,Pseudothermotoga, Thermosipho,OceanotogaandFervidobacterium.Withintheorder Thermotogales,theThermotogaandPseudothermotogacladesshared anexclusivecommonancestor(redcircle,Fig.2);theywere sis-tergroupsandwerethereforemorecloselyrelatedtooneanother thantoThermosiphospp.Fromtheirmostrecentancestor,more evolutionarychangewasobservedwithinthePseudothermotoga thanwithintheThermotogalineage.However,theweaknessofthe phylogeneticsignalfromthe16SrRNA-basedphylogenydidnot offeranacceptablemeanstodeterminetherelationshipsbetween Thermotogaspecies(Fig.2).Thepoorlyresolvedphylogenyofthe Thermotogarepresentativesindicatesaneedtoinvestigate addi-tionalphylogeneticmarkers(RpoB),core-genesphylogeny[23]or concatenatedhousekeepingproteins[5].Additionally,afull com-parativegenomicsstudyofthegenefloweventsundergonebythe membersofthetwogenerawouldshedlightontheirevolutionary history.ThisfirstphylogeneticapproachsuggestedthatThermotoga profundaandThermotogacaldifontisbelongtothe Pseudothermo-togacladeratherthantoThermotoga(Fig.2).
Thecore-genome-basedphylogenyofThermotogaceae
ToconfirmthetaxonomicaffiliationofThermotogacaldifontis andThermotogaprofundawithinthegenusPseudothermotoga,we investigatedlarge-scalefeaturesoftheirgenomes.Theinferenceof
Fig.3. Core-genome-basedphylogenyoftheThermotogaceaefamily.
Thecore-genesconsensusmidpointtree(left)isdeducedfrom603,752positionsof614sharedgenes.PhyMLbranchsupportvaluesareindicatedinblackrespectively.The genepresence/absencematrixisdeducedfrom5052totalgenes(right).Bluetileshowsthepresenceofgeneinagenome.Alignedbluetilesaregenessharedbyaleasttwo genomes.Pseudothermotogagenusisboxedinred.Thepan-genomicprofileforthePseudothermotogaandThermotogacladesareshownasbarcharts.
core-genome-basedphylogenyofthefamilyThermotogaceaerelied ontheanalysisof thegenecontentofeightThermotogaandsix Pseudothermotogaspecies.Theidentificationofthegenesshared byalltheavailableThermotogaceaegenomes (thecore-genome) wasdeducedbygeneorthologyandresultedin614genesusedfor theThermotogaceaephylogenyreconstruction.The core-genome-basedphylogenypreservedP.hypogeaastheclosestrelativeofT. caldifontis,andthesistertaxaP.elfiiastheclosestrelativesofT. pro-funda(Fig.3),aswasobservedinthe16SrRNA-basedphylogeny (Fig.2).ThelackofconsistencyinthetreetopologyofThermotoga spp.between16S-rRNA-basedandcore-genome-basedphylogeny canbeexplainedbythelowbootstrapbranchsupportsfor this clade(Fig.2).InFig.3,primarygeneticindicatorsofthedichotomy ofgeneraPseudothermotogaandThermotogaweredenotedwitha
genepresence(bluetile)/absencematrix,wherethegenesshared bytwoormorespeciesareshownbyalignedbluetilesorblocks. SimilargenecontentprofilesofThermotogaspeciesresultedina considerablecore-genomesize(numberofgenessharedbyallthe members)tothedetrimentoftheaccessorygenome(sharedby atleasttwomembersbutnotbyall)andspecies-specificgenes (Fig.3).Incontrast,thePseudothermotogaspeciesshowedmore variabilityintheirgenecontentprofiles,whichwasexhibitedbya lowercoregenomesizebutagreateraccessorygenomeandmore species-specificgenes(Fig.3).ThegenecontentshowedthatT. pro-fundaandT.caldifontisweregeneticallymorecloselyrelatedtothe PseudothermotogathantoThermotogaspecies.Asdiscussedabove, decipheringthedistinctevolutionaryhistoryoftheThermotogaand Pseudothermotogalineageswillrequiremorecomplex
compara-Fig.4. PartialsequencealignmentsofthePseudothermotoga-specificConservedSignatureIndels.
Partialalignmentsextractedfromfullproteinsequencealignments.Thepositionsaddedtosequencelabelsmeanstart/endpositionsaccordingtothefullalignedsequences. PositionsonthetopoftheCSIsboxedindashedredlinestargetthereferencesequence(labeledinbold).T.caldifontisandT.profundaholdthefourPseudothermotoga-specific CSIs.(A)Glycerol-3-phosphatedehydrogenase.ALeucineinsertionconservedinallPseudothermotogamembers.(B)HypotheticalproteinTlet0907.Pseudothermotoga membersshowaspecificfive-aminoaciddeletion.(C)HypotheticalproteinTheth0845.Afive-amino-acidinsertionisconservedwithinthePseudothermotogaclade.(D) HypotheticalproteinTM1140.ALeucinedeletionconservedwithinthePseudothermotogaclade.
Fig.5.Whole-genomerelatednesswithintheThermotogaceaefamily.
HierarchicalclusteringfromAverageNucleotideIdentity(vertically)andAverageAminoAcidIdentity(horizontally)measuresusing19Thermotogaceaegenomes.Valuescale isdepictedinred,yellowandgreencolorsforANIgenomepairwisecomparison.Thevaluescalerangesfrom0to100%accordingtosequenceidentitylevel.Theverticalline atthevalue96indicatestheANIspeciesdemarcationthreshold.
tivegenomicsincludinggenefamilyreconstruction,identification ofspecies-specificgenes,functionalsystems,andhorizontalgene transfer,adominantforceofThermotogalesevolution[34]. MolecularsignaturesofthePseudothermotogaclade
Previousgenomic insightsontheThermotogae havebrought outthediscoveryoffourConservedSignatureIndels(CSIs) spe-cific to the members of the Pseudothermotoga clade [5]. The Pseudothermotoga-specific CSIs have beenidentified from Ther-motogalesproteinfamilyalignmentsoftheGlycerol-3-phosphate dehydrogenase (one amino-acid insertion), the hypothetical Theth0845(5/6amino-acidinsertion),thehypotheticalTM1140 (oneamino-acid deletion) and thehypothetical Tlet0907(five amino-aciddeletion)[5].Presenceofthefour Pseudothermotoga-specificCSIswithinThermotogaprofundaandThermotogacaldifontis offersadditionalevidenceoftheirreclassificationproposalas Pseu-dothermotogaspecies.Themultiplesequencealignment(MSA)of thehomologstotheGlycerol-3-phosphatedehydrogenase(Fig.4A) and tothehypothetical protein TM1140 (Fig. 4D)showed that thetwoPseudothermotoga-specificCSIs,aLeucineinsertionanda
Leucinedeletion,respectively,werealsoconservedinT.caldifontis andT. profunda(Fig.4A, D).ThenextPseudothermotoga-specific CSI,afive-amino-aciddeletionderivedfromtheMSAofhomologs tothe Tlet0907protein, wasalso sharedbyboth T. caldifontis andT.profunda(Fig.4B).However,BhandariandGuptareported asix-amino-acid deletionintheirMSAofhomologs toTlet907
[5]. In thesame way,the MSA ofhomologs to theTheth0845 proteinexhibitedalsoaslightmodifiedpatternfromtheoriginal Pseudothermotoga-specificCSIdescribedinsupplementaryFig.S39 ofBhandariandGuptawork[5](Fig.4C).
Our analysis led us to identify two slight modified Pseudothermotoga-specificCSIs(Fig.4B,C)comparedtotheoriginal work[5],thatcouldbeexplainedbytheMSAmethodsusedand bythedivergenceoftheselectedsequencesintheMSA.Usingthe T-COFFEEMSAmethod[3](datanotshown),theCSIsidentified were congruent with our MAFFT-based analysis. The accuracy evaluation of theMSA methods (benchmark)commonly shows that consistency-based programssuchas T-COFFEE and MAFFT exceedtheothermethods[38].Itisalsoimportanttoemphasize thatthepreviousidentificationofPseudothermotoga-specificCSIs
thermarum) whereas all Pseudothermotoga spp. were included in ouranalysis (Fig.4). Finally, thepairwise identity matrix of theTheth0845proteinfamilyalignment(Fig.4C)provided low sequenceidentityvalues(<35%)closetothetwilightzone[49]that couldstimulatedebateaboutthetrueevolutionaryrelatednessof theseproteinsandespeciallythebiologicalrelevanceofthisCSI. The occurrenceof the fourPseudothermotoga-specific CSIs in T. caldifontisandT. profundaisthereby anadditionalargumentto reclassifythesetwoorganismswithinthegenusPseudothermotoga asP.caldifontiscomb.nov.andP.profundacomb.nov.,respectively. SpeciesdelineationwithinthegenusPseudothermotoga
Fardeauetal.[12],previouslyreportedthatDNA–DNA homolo-giesbetweenP.elfiistrainG1andP.subterraneaDSM9912T,P.
lettingaeDSM14385TandP.elfiiDSM9442Twere83%,71.4%and
86%,respectively.ThisstudyalsoindicatedaDNA–DNAhomology of83%betweenP.subterraneaDSM9912TandP.elfiiDSM9442T,
andof78%betweenP.subterraneaDSM9912TandP.lettingaeDSM
14385T[12].Inourstudy,complementaryDDHexperimentswere
carriedoutinduplicatewithP.lettingaeDSM14385TandP.elfii
DSM9442T,theyindicatedDNA–DNAhomologyof94%(91.5%and
96.7%)betweenthesetwospecies.AllDDHvaluesofthepairwise speciescomparisonweretherebyabove70%.Consequently,allthe Pseudothermotogastrainscouldnotbedifferentiatedatthespecies levelbyDDHwhentakingintoaccounttherecommendationofthe adhoccommittee[55],whichadvisedathresholdvalueof70%DDH similarityforthedefinitionofbacterialspecies.
ThecloserelationshiplinkingDDHandANIvalues[15,25]led ustocalculateANIindicesforestimatingoverallgenomesimilarity and clarifying thegenetic relationshipswithin thegenus Pseu-dothermotoga.InadditiontoANIindices,theAminoAcidIdentity (AAI)measurewasalsoconsideredforitsrobustnessfordivergent species[47].WithintheThermotogaceaegenomes,theestimated ANIandAAIvaluesrangedfrom64.19to99.96%andfrom63.32to 99.98%,respectively.ThedoubleANI–AAIclusteringdistinguished thePseudothermotogaandThermotogagenera(Fig.5).The Thermo-togaclusteringshowedintra-speciesrelationshipsforThermotoga sp.Xyl54, Thermotoga.spCell2,Thermotoga.sp. TBGT1765and Thermotoga.spTBGT1766(AAIandANIvalues>98).Strikingly,both ANIandAAIvaluesrevealedintra-speciesrelationshipsbetweenP. elfiiandP.lettingaewith99.12%and99.5%ofidentitylevel, respec-tively.Based uponDDHand ANI–AAIresults, P.elfii,P.lettingae andP.subterraneaspeciesshouldbeconsideredasheterotypic syn-onyms.Hereinweproposetounitethesethreespecies.According torules38,42and24b(2)oftheBacteriologicalCode[39],thename Pseudothermotogaelfiihaspriorityandhenceshouldbeusedforthe unifiedtaxon.
Conclusions
With the significant advances in full genome sequencing, modern taxonomy practice promotes the useof gene marker-based/core-genome-based phylogenetic inferences, comparative genomics, and whole genome relatedness indices as reliable methods for taxonomic classification [8,42,51,53]. As a result, investigationoftheThermotogaandPseudothermotogageneraby moderntaxonomypracticeandclassicalDDHmeasuresputs for-wardcongruentclassificationandnomenclatureofthesegenera withintheThermotogaceaefamily.OurdatashowthatThermotoga caldifontisandThermotogaprofundaspeciessharesimilarfeatures (genome size, CDS number) withthe Pseudotermotoga species. TheyarealsophylogeneticallyrelatedtoPseudothermotogaspecies, and carry the fourPseudothermotoga-specific CSIs. Moreover, a preliminarypan-genomicanalysisshows thatT.profundaandT.
caldifontisaregeneticallymorecloselyrelatedtothe Pseudothermo-togaspp.WetherebyproposetoreclassifyThermotogacaldifontis and Thermotoga profunda asPseudothermotoga caldifontis comb. nov.andPseudothermotogaprofundacomb.nov.,respectively.We alsodemonstratethat Pseudothermotogaelfii,P.subterraneaand P. lettingae are heterotypic synonyms and should therefore be unitedwiththeprioritynameofPseudothermotogaelfii.Despite theconsiderableeffortneededtounderstandtheThermotogae phy-logenetic relationships[13,58], the evolutionary eventsdriving Thermotogaesub-groups remainblurred. Furthercomplex com-parativegenomicsmayassistthedecipheringoftheevolutionary mechanismswithinThermotogae.
DescriptionofPseudothermotogaprofundacomb.nov. Pseudothermotogaprofunda(pro.fun’daL.fem.adj.profunda deep,pertainingtothesourceofisolationofthetypestrain) Basonym:ThermotogaprofundaMorietal.2014.
Thedescriptionofthistaxonisthesameasthatgivenpreviously
[31].
ThetypestrainisAZM34c06T=NBRC106115T=DSM23275T.
Thesequenceaccessionnumber(16SrRNAgene)forthetypestrain isNR133904.1.
TheaccessionnumberofthegenomesequenceisAP014510. DescriptionofPseudothermotogacaldifontiscomb.nov. Pseudothermotogacaldifontis(cal.di.fon’tis.L.adj.caldushot;L. masc.n.fons,fontisaspring,N.L.gen.n.caldifontisofahot spring,pertainingtothesourceofisolationofthetypestrain) Basonym:ThermotogacaldifontisMorietal.2014.
Thedescriptionofthistaxonisthesameasthatgivenpreviously
[31].
ThetypestrainisAZM44c09T=NBRC106116T=DSM23272T.
Thesequenceaccessionnumber(16SrRNAgene)forthetypestrain isNR133903.1.
TheaccessionnumberofthegenomesequenceisAP014509. EmendeddescriptionofthegenusPseudothermotogaBhandari andGupta2014
Themorphologicalandphysiologicaldescriptionofthisgenus, whosetypespecies isPseudothermotogathermarum[5],remains thesame asdescribed byWindbergeret al.[56].Otherspecies belongingthisgenusareP.profunda(Basonym:T.profunda)and P.caldifontis(Basonym:Thermotogacaldifontis)(Morietal.[31]). EmendeddescriptionofthespeciesPseudothermotogaelfii BhandariandGupta2014
ThecharacteristicsofthisspeciesareasdescribedbyRavotetal.
[43]withthefollowingamendments.Thefollowingsubstratesmay beused:methanol,lactate,pyruvate,galactose,cellobiose,glycerol, starch, xylan, pectin, methylamine, dimethylamine, trimethy-lamine,2-oxoglutarate,serine,yeastextract,peptone,gelatinand casaminoacids.Acetate,betaine,leucine, isoleucine,valine, for-mate and H2/CO2 (80/20, v/v) may be oxidized either in the
presence of thiosulfate or a hydrogenotrophic methanogen as electronacceptor.Elementalsulfur,Fe(III)and anthraquinone-2,6-disulfonatemayserveaselectronacceptors.ThetypestrainisDSM 9442T=ATCC51869T.
Acknowledgement
We thank the Professor William B. Whitman, University of Georgia,fortherevisionofthemanuscript.
Funding
ThisprojecthasbeenfundedwithsupportfromExxonMobil ResearchandEngineering.
AppendixA. Supplementarydata
Supplementarydataassociatedwiththisarticlecanbefound, intheonlineversion,athttps://doi.org/10.1016/j.syapm.2018.04. 007.
References
[1]Achenbach-Richter,L.,Gupta,R.,Stetter,K.O.,Woese,C.R.(1987)Werethe originaleubacteriathermophiles?Syst.Appl.Microbiol.9,34–39.
[2]Altschul,S.F.,Gish,W.,Miller,W.,Myers,E.W.,Lipman,D.J.(1990)Basiclocal alignmentsearchtool.J.Mol.Biol.215,403–410.
[3]Armougom,F.,Moretti,S.,Poirot,O.,Audic,S.,Dumas,P.,Schaeli,B.,Keduas,V., Notredame,C.(2006)Expresso:automaticincorporationofstructural informa-tioninmultiplesequencealignmentsusing3D-Coffee.NucleicAcidsRes.34, 600–603.
[4]Balk,M.,Weijma,J.,Stams,A.J.M.(2002)Thermotogalettingaesp.nov.,anovel thermophilic,methanol-degradingbacteriumisolatedfromathermophilic anaerobicreactor.Int.J.Syst.Evol.Microbiol.52,1361–1368.
[5]Bhandari,V.,Gupta,R.S.(2014)Molecularsignaturesforthephylum(class) Thermotogaeandaproposalforitsdivisionintothreeorders(Thermotogales, Kosmotogalesord.nov.andPetrotogalesord.nov.)containingfourfamilies (Thermotogaceae,Fervidobacteriaceaefam.nov.,Kosmotogaceaefam.nov.and Petrotogaceaefam.nov.)andanewgenusPseudothermotogagen.nov.withfive newcombinations.AntonievanLeeuwenhoekInt.J.Gen.Mol.Microbiol.105, 143–168.
[6]Brochier,C.,Philippe,H.(2002)Phylogeny:anon-hyperthermophilicancestor forBacteria.Nature417,244.
[7]Buchfink,B.,Xie,C.,Huson,D.H.(2015)Fastandsensitiveproteinalignment usingDIAMOND.Nat.Methods12,59–60.
[8]Chun,J.,Rainey,F.A.(2014)Integratinggenomicsintothetaxonomyand sys-tematicsoftheBacteriaandArchaea.Int.J.Syst.Evol.Microbiol.64,316–324.
[9]Delcher,A.L.(2002)Fastalgorithmsforlarge-scalegenomealignmentand com-parison.NucleicAcidsRes.30,2478–2483.
[10]DiPippo,J.L.,Nesbø,C.L.,Dahle,H.,Doolittle,W.F.,Birkland,N.K.,Noll,K.M. (2009)Kosmotogaoleariagen.nov.,sp.nov.,athermophilic,anaerobic het-erotrophisolatedfromanoilproductionfluid.Int.J.Syst.Evol.Microbiol.59, 2991–3000.
[11]Escara,J.F.,Hutton,J.R.(1980)ThermalstabilityandrenaturationofDNAin dimethylsulfoxidesolutions:accelerationoftherenaturationrate. Biopoly-mers19,1315–1327.
[12]Fardeau,M.-L.,Goulhen,F.,Bruschi,M.,Khelifi,N.,Cayol,J.-L.,Ignatiadis,I., Guyot,F.,Ollivier,B.(2009)ArchaeoglobusfulgidusandThermotogaelfii, Ther-mophilicisolatesfromdeepgeothermalwateroftheParisBasin.Geomicrobiol. J.26,119–130.
[13]Forterre,P.(2015)Theuniversaltreeoflife:anupdate.Front.Microbiol.6,1–18.
[14]Frock,A.D.,Notey,J.S.,Kelly,R.M.(2010)ThegenusThermotoga:recent devel-opments.Environ.Technol.31,1169–1181.
[15]Goris,J.,Konstantinidis,K.T.,Klappenbach,J.A.,Coenye,T.,Vandamme,P., Tiedje,J.M.(2007)DNA-DNAhybridizationvaluesandtheirrelationshipto whole-genomesequencesimilarities.Int.J.Syst.Evol.Microbiol.57,81–91.
[16]Gouy,M.,Guindon,S.,Gascuel,O.(2010)SeaViewversion4:amultiplatform graphicaluserinterfaceforsequencealignmentandphylogenetictreebuilding. Mol.Biol.Evol.27,221–224.
[17]Guindon,S.,Dufayard,J.F.,Lefort,V.,Anisimova,M.,Hordijk,W.,Gascuel,O. (2010)Newalgorithmsandmethodstoestimatemaximum-likelihood phylo-genies:assessingtheperformanceofPhyML3.0.Syst.Biol.59,307–321.
[18]BenHania,W.,Ghodbane,R.,Postec,A.,Brochier-Armanet,C.,Hamdi,M., Fardeau,M.L.,Ollivier,B.(2011)Cultivationofthefirstmesophilic represen-tative(“mesotoga”)withintheorderThermotogales.Syst.Appl.Microbiol.34, 581–585.
[19]Huber,R.,Langworthy,T.A.,König,H.,Thomm,M.,Woese,C.R.,Sleytr,U.B., Stetter,K.O.(1986)Thermotogamaritimasp.nov.representsanewgenusof uniqueextremelythermophiliceubacteriagrowingupto90◦C.Arch.Microbiol.
144,324–333.
[20]Huss,V.A.R.,Festl,H.,Schleifer,K.H.(1983)Studiesonthe spectrophotomet-ricdeterminationofDNAhybridizationfromrenaturationrates.Syst.Appl. Microbiol.4,184–192.
[21]Itoh,T.,Onishi,M.,Kato,S.,Iino,T.,Sakamoto,M.,Kudo,T.,Takashina,T., Ohkuma,M.(2016)Athalassotogasaccharophilagen.nov.,sp.nov.,isolated
fromanacidicterrestrialhotspring,andproposalofMesoaciditogalesord.nov. andMesoaciditogaceaefam.nov.inthephylumThermotogae.Int.J.Syst.Evol. Microbiol.66,1045–1051.
[22]Jahnke,D.(1992)BASICcomputerprogramforevaluationofGILFORDSYSTEM 2600spectrophotometeronaPC/XT/ATtypepersonalcomputer.System15, 61–73.
[23]Jiang,L.,L’Haridon,S.,Jebbar,M.,Xu,H.,Alain,K.,Shao,Z.(2017)Complete genomesequenceandwhole-genomephylogenyofKosmotogapacificatype strainSLHLJ1TfromanEastPacifichydrothermalsediment.Stand.Genom.Sci. 12,3.
[24]Katoh,K.,Standley,D.M.(2013)MAFFTmultiplesequencealignmentsoftware version7:improvementsinperformanceandusability.Mol.Biol.Evol.30, 772–780.
[25]Kim,M.,Oh,H.S.,Park,S.C.,Chun,J.(2014)Towardsataxonomiccoherence betweenaveragenucleotideidentityand16SrRNAgenesequencesimilarity forspeciesdemarcationofprokaryotes.Int.J.Syst.Evol.Microbiol.64,346–351.
[26]Konstantinidis,K.T.,Tiedje,J.M.(2005)Genomicinsightsthatadvancethe speciesdefinitionforprokaryotes.Proc.Natl.Acad.Sci.U.S.A.102,2567–2572.
[27]DeLey,J.,Cattoir,H.,Reynaerts,A.(1970)Thequantitativemeasurementof DNAhybridizationfromrenaturationrates.Eur.J.Biochem.12,133–142.
[28]Li,X.,Huang,Y.,Whitman,W.B.(2015)Therelationshipofthewholegenome sequenceidentitytoDNAhybridizationvariesbetweengeneraofprokaryotes. AntonievanLeeuwenhoekInt.J.Gen.Mol.Microbiol.107,241–249.
[29]Löytynoja,A.(2014)Phylogeny-awarealignmentwithPRANK.MethodsMol. Biol.(CliftonN.J.)1079,155–170.
[30]Meier-Kolthoff,J.P.,Klenk,H.P.,Göker,M.(2014)TaxonomicuseofDNAG+C contentandDNA-DNAhybridizationinthegenomicage.Int.J.Syst.Evol. Micro-biol.64,352–356.
[31]Mori,K.,Yamazoe,A.,Hosoyama,A.,Ohji,S.,Fujita,N.,Ishibashi,J.I.,Kimura,H., Suzuki,K.I.(2014)Thermotogaprofundasp.nov.andThermotogacaldifontissp. nov.,anaerobicthermophilicbacteriaisolatedfromterrestrialhotsprings.Int. J.Syst.Evol.Microbiol.64,2128–2136.
[32]Nesbø,C.L.,Bapteste,E.,Curtis,B.,Dahle,H.,Lopez,P.,Macleod,D.,Dlutek, M.,Bowman,S.,Zhaxybayeva,O.,Birkeland,N.-K.,Doolittle,W.F.(2009)The genomeofThermosiphoafricanusTCF52B:lateralgeneticconnectionstothe FirmicutesandArchaea.J.Bacteriol.191,1974–1978.
[33]Nesbø,C.L.,Bradnan,D.M.,Adebusuyi,A.,Dlutek,M.,Petrus,A.K.,Foght,J., Doolittle,W.F.,Noll,K.M.(2012)Mesotogaprimagen.nov.,sp.nov.,thefirst describedmesophilicspeciesoftheThermotogales.Extremophiles16,387–393.
[34]Nesbø, C.L.,Swithers,K.S.,Dahle,H., Haverkamp,T.H.A., Birkeland,N.-K., Sokolova,T.,Kublanov,I.,Zhaxybayeva,O.(2015)Evidenceforextensivegene flowandThermotogasubpopulationsinsubsurfaceandmarineenvironments. ISMEJ.9,1532–1542.
[35]Oren,A.,Garrity,G.M.(2014)Listofnewnamesandnewcombinations pre-viouslyeffectively,butnotvalidly,published.Listno.158.Int.J.Syst.Evol. Microbiol.64,2184–2187.
[36]Oshima,K.,Chiba,Y.,Igarashi,Y.,Arai,H.,Ishii,M.(2012)Phylogeneticposition ofAquificalesbasedonthewholegenomesequencesofsixAquificalesspecies. Int.J.Evol.Biol.2012,1–9.
[37]Page,A.J.,Cummins,C.A.,Hunt,M.,Wong,V.K.,Reuter,S.,Holden,M.T.G., Fookes,M.,Falush,D.,Keane,J.A.,Parkhill,J.(2015)Roary:rapidlarge-scale prokaryotepangenomeanalysis.Bioinformatics31,3691–3693.
[38]Pais,F.S.-M.,Ruy,P.deC.,Oliveira,G.,Coimbra,R.S.(2014)Assessingthe effi-ciencyofmultiplesequencealignmentprograms.AlgorithmsMol.Biol.9,4.
[39]Parker,C.T.,Tindall,B.J.,Garrity,G.M.(2015)Internationalcodeof nomencla-tureofprokaryotes.Int.J.Syst.Evol.Microbiol.
[40]Petrus, A.K.,Swithers,K.S.,Ranjit,C.,Wu,S.,Brewer,H.M.,Gogarten,J.P., Pasa-Tolic,L.,Noll,K.M.(2012)Genesforthemajorstructuralcomponentsof Thermotogalesspecies’togasrevealedbyproteomicandevolutionaryanalyses ofOmpAandOmpBhomologs.PLoSOne7,1–9.
[41]Pollo,S.M.J.,Zhaxybayeva,O.,Nesbø,C.L.(2015)Insightsintothermoadaptation andtheevolutionofmesophilyfromthebacterialphylumThermotogae.Can.J. Microbiol.61,655–670.
[42]Ramasamy,D.,Mishra,A.K.,Lagier,J.C.,Padhmanabhan,R.,Rossi,M.,Sentausa, E.,Raoult,D.,Fournier,P.E.(2014)Apolyphasicstrategyincorporatinggenomic dataforthetaxonomicdescriptionofnovelbacterialspecies.Int.J.Syst.Evol. Microbiol.64,384–391.
[43]Ravot,G.,Magot,M.,Fardeau,M.L.,Patel,B.K.,Prensier,G.,Egan,A.,Garcia,J.L., Ollivier,B.(1995)Thermotogaelfiisp.nov.,anovelthermophilicbacteriumfrom anAfricanoil-producingwell.Int.J.Syst.Bacteriol.45,308–314.
[44]Reysenbach,A.L.(2001)PhylumBII.Thermotogaephy.nov.Bergey’sManual ofSystematicBacteriology.In:Boone,D.R.,Castenholz,R.W.(Eds.), Springer-Verlag,pp.369–387.
[45]Reysenbach,A.L.,Liu,Y.,Lindgren,A.R.,Wagner,I.D.,Sislak,C.D.,Mets,A., Schouten, S. (2013) Mesoaciditoga lauensis gen. nov., sp.nov., a moder-atelythermoacidophilicmemberoftheorderThermotogalesfromadeep-sea hydrothermalvent.Int.J.Syst.Evol.Microbiol.63,4724–4729.
[46]Rice,P.,Longden,I.,Bleasby,A.(2000)EMBOSS:TheEuropeanMolecular Biol-ogyOpenSoftwareSuite.TrendsGenet.16,276–277.
[47]Rosselló-Móra,R.(2005)Updatingprokaryotictaxonomy.J.Bacteriol.187, 6255–6257.
[48]Rosselló-Móra,R.,Amann,R.(2015)Pastandfuturespeciesdefinitionsfor BacteriaandArchaea.Syst.Appl.Microbiol.38,209–216.
[49]Rost,B.(1999)Twilightzoneofproteinsequencealignments.ProteinEng.12, 85–94.
[50]Sabath,N.,Ferrada,E.,Barve,A.,Wagner,A.(2013)Growthtemperatureand genomesizeinbacteriaarenegativelycorrelated,suggestinggenomic stream-liningduringthermaladaptation.GenomeBiol.Evol.5,966–977.
[51]Schleifer,K.H.,Amann,R.,Rosselló-Móra,R.(2015)Taxonomyintheageof genomics.Introduction.Syst.Appl.Microbiol.38,207–208.
[52]Teeling,H.,Meyerdierks,A.,Bauer,M.,Amann,R.,Glöckner,F.O.(2004) Appli-cationoftetranucleotidefrequenciesfortheassignmentofgenomicfragments. Environ.Microbiol.6,938–947.
[53]Vandamme,P.,Peeters,C.(2014)Timetorevisitpolyphasictaxonomy.Antonie vanLeeuwenhoekInt.J.Gen.Mol.Microbiol.106,57–65.
[54]Waterhouse,A.M.,Procter,J.B.,Martin,D.M.A.,Clamp,M.,Barton,G.J.(2009) JalviewVersion2-Amultiplesequencealignmenteditorandanalysis work-bench.Bioinformatics25,1189–1191.
[55]Wayne,L.G.,Brenner,D.J.,Colwell,R.R.,Grimont,P.A.D.,Kandler,O.,Krichevsky, M.I.,Moore,L.H.,Moore,W.E.C.,Murray,R.G.E.,Stackebrandt,E.,Starr,M.P.,
Tru-per,H.G.(1987)Reportoftheadhoccommitteeonreconciliationofapproaches tobacterialsystematics.Int.J.Syst.Bacteriol.37,463–464.
[56]Windberger,E.,Huber,R.,Trincone,A.,Fricke,H.,Stetter,K.O.(1989) Ther-motogathermarumsp.nov.andThermotoganeapolitanaoccurringinAfrican continentalsolfataricsprings.Arch.Microbiol.151,506–512.
[57]Yarza,P.,Ludwig,W.,Euzéby,J.,Amann,R.,Schleifer,K.H.,Glöckner,F.O., Rosselló-Móra,R.(2010)Updateoftheall-specieslivingtreeprojectbased on16Sand23SrRNAsequenceanalyses.Syst.Appl.Microbiol.33,291–299.
[58]Zhaxybayeva,O.,Swithers,K.S.,Lapierre,P., Fournier,G.P.,Bickhart,D.M., Deboy,R.T.,Nelson,K.E.,Nesbø,C.L.,Doolittle,W.F.,Gogarten,J.P.,Noll,K.M. (2009)Onthechimericnature,thermophilicorigin,andphylogenetic place-mentoftheThermotogales.Proc.Natl.Acad.Sci.U.S.A.106,5865–5870.