Plastid Proteome Assembly without Toc159: Photosynthetic
Protein Import and Accumulation of
N-Acetylated Plastid
Precursor Proteins
C WSylvain Bischof,
aKatja Baerenfaller,
aThomas Wildhaber,
aRaphael Troesch,
aPierre-Alexandre Vidi,
b,1Bernd Roschitzki,
cMatthias Hirsch-Hoffmann,
aLars Hennig,
a,2Felix Kessler,
bWilhelm Gruissem,
a,cand Sacha Baginsky
a,3,4aDepartment of Biology, Eidgeno¨ssische Technische Hochschule Zurich, 8092 Zurich, Switzerland bLaboratoire de Physiologie Ve´ge´tale, 2007 Neuchaˆtel, Switzerland
cFunctional Genomics Center Zurich, 8057 Zurich, Switzerland
Import of nuclear-encoded precursor proteins from the cytosol is an essential step in chloroplast biogenesis that is mediated by protein translocon complexes at the inner and outer envelope membrane (TOC). Toc159 is thought to be the main receptor for photosynthetic proteins, but lacking a large-scale systems approach, this hypothesis has only been tested for a handful of photosynthetic and nonphotosynthetic proteins. To assess Toc159 precursor specificity, we quantitatively analyzed the accumulation of plastid proteins in two mutant lines deficient in this receptor. Parallel genome-wide transcript profiling allowed us to discern the consequences of impaired protein import from systemic transcriptional responses that contribute to the loss of photosynthetic capacity. On this basis, we defined putative Toc159-independent and Toc159-dependent precursor proteins. Many photosynthetic proteins accumulate in Toc159-deficient plastids, and, surprisingly, several distinct metabolic pathways are negatively affected by Toc159 depletion. Lack of Toc159 furthermore affects several proteins that accumulate as unprocessedN-acetylated precursor proteins outside of plastids. Together, our data show an unexpected client protein promiscuity of Toc159 that requires a far more differentiated view of Toc159 receptor function and regulation of plastid protein import, in which cytosolic Met removal followed by N-terminal acetylation of precursors emerges as an additional regulatory step.
INTRODUCTION
Assembly of the chloroplast proteome requires transport com-plexes that recognize plastid precursor proteins in the cytosol and mediate their translocation into the chloroplast. Most of the nuclear-encoded chloroplast proteins contain a cleavable N-terminal transit peptide and are imported by the TOC/TIC (for translocon at the outer/inner membrane of the chloroplast)
import machinery (Agne and Kessler 2009; Balsera et al., 2009). In Arabidopsis thaliana, recognition and selection of chloroplast-imported proteins are mediated by GTP binding proteins that belong to two small families: Toc34/33 and Toc159/132/120/90. Members of the Toc159 family share a GTP binding domain (G-domain) and a membrane anchoring domain (M-domain). They are distinguished by differently sized acidic domains (A-domain), with Toc90 entirely lacking the A-domain. The differently sized A-domains in Toc159, Toc132, and Toc120 are located at the N-terminal side of the G- and M-domains and largely account for the molecular mass differ-ences between the different members of the Toc159 family (Agne and Kessler, 2009). The receptors are required for photosyn-thetic growth because their loss-of-function produces plants with a pale green or albino phenotype (Jarvis et al., 1998; Bauer et al., 2000). The analysis of Toc33 mutants (ppi1 for plastid protein import 1) suggested that this receptor may be preferen-tially involved in the import of photosynthetic proteins (Kubis et al., 2003). Similarly, the loss of Toc159 function (ppi2) results in albino plants that do not grow beyond the cotyledon stage on soil because the accumulation of photosynthetic proteins is signif-icantly reduced (Bauer et al., 2000).
Reverse genetic studies and precursor binding assays sug-gested two different classes of receptors with specialized functions:
1 Current address: Basic Medical Sciences, Purdue University, West
Lafayette, IN 47907.
2 Current address: Department of Plant Biology and Forest Genetics,
Uppsala BioCenter, Swedish University of Agricultural Sciences, Box
7080, 75007 Uppsala, Sweden.
3 Current address: Martin-Luther-University Halle-Wittenberg, Institute
of Biochemistry and Biotechnology, Plant Biochemistry, Weinbergweg
22 (Biozentrum), 06120 Halle (Saale), Germany.
4 Address correspondence to sacha.baginsky@biochemtech.uni-halle.de.
The authors responsible for distribution of materials integral to the
findings presented in this article in accordance with the policy
described in the Instructions for Authors (www.plantcell.org) are:
Wilhelm Gruissem (wgruissem@ethz.ch) and Sacha Baginsky (sacha.
baginsky@biochemtech.uni-halle.de).
C Some figures in this article are displayed in color online but in black
and white in the print edition.
one class comprising Toc159/90 and the other class Toc132/120 (Hiltbrunner et al., 2004; Ivanova et al., 2004; Kubis et al., 2004; Smith et al., 2004; Infanger et al., 2011). The loss of either Toc132, Toc120, or Toc90 did not result in a visible phenotype, but Toc132
Toc120 double mutants appeared pale green (Kubis et al., 2004) or
were even embryo lethal (Ivanova et al., 2004), suggesting func-tional overlap between Toc132 and Toc120. Neither full-length Toc132 nor Toc120 could complement ppi2 (Ivanova et al., 2004; Kubis et al., 2004). Based on these data, it was proposed that Toc132 and Toc120 are specific for the import of nonphotosyn-thetic proteins (Kubis et al., 2004). In contrast with Toc132 and Toc120, the loss of Toc90 function in the ppi2 background resulted in a more severe phenotype, suggesting functional overlap be-tween Toc159 and Toc90 (Hiltbrunner et al., 2004). Indeed, partial complementation of the ppi2 phenotype was achieved by the overexpression of Toc90 (Infanger et al., 2011).
Although it is currently unclear how the different Toc receptors recognize their substrates, it is conceivable that amino acids in the N-terminal transit peptide of plastid precursor proteins are in-volved in the recognition and in establishing different import specificities. Canonical chloroplast targeting requires transit pep-tides that are typically 20 to 100 amino acids long and enriched in hydroxylated amino acids but low in acidic amino acids (Bruce, 2001). After import, most transit peptides are cleaved by a stromal processing peptidase (SPP; Richter and Lamppa, 1998, 1999). Processing of imported precursors is essential, and the lack of SPP results in embryo lethality in Arabidopsis (Tro¨sch and Jarvis, 2011). Because the SPP cleavage site sequence is not highly conserved, it was proposed that recognition requires physico-chemical properties of the transit peptide rather than a particular amino acid sequence (Emanuelsson et al., 2000; Zhang and Glaser, 2002; Rudhe et al., 2004). Following transit peptide removal, the newly generated protein N terminus may be acety-lated (Ferro et al., 2003; Kleffmann et al., 2007; Zybailov et al., 2008). N-acetylation is catalyzed by N-terminal acetyltransferases in the cytosol and in the plastid (Sherman et al., 1985; Meinnel et al., 2006; Goetze et al., 2009). It is therefore possible that a chloroplast-targeted protein may be acetylated as a precursor in the cytosol and after import and processing as the mature protein. N-terminal acetylation is thus a useful modification to map the different N termini of proteins in different compartments. Interest-ingly, the lack of a cytoplasmic N-acetyltransferase affects plastid biogenesis suggesting a functional crosstalk between cytosolic
N-acetylation and chloroplast function (Pesaresi et al., 2003).
The assembly and maintenance of the chloroplast prote-ome are highly regulated processes and involve not only the organellar import machinery but also multiple anterograde and retrograde signaling events that affect the transcription of nuclear- and chloroplast-encoded genes (Mochizuki et al., 2001, 2008; Strand et al., 2003; Larkin et al., 2003; Koussevitzky et al., 2007; Moulin et al., 2008; Kakizaki et al., 2009). Similar transcript patterns observed under different environmental con-ditions or genetic modifications identified key regulators that affect entire sets of plastid proteins and biochemical networks (Richly et al., 2003; Biehl et al., 2005). In order to understand chloroplast proteome assembly, it is therefore necessary to investigate the complexity and quantitative nature of the pro-teome together with the regulation of the corresponding genes.
Here, we report an integrated approach to investigate the qualitative and quantitative contribution of the Toc159 receptor to the assembly of the plastid proteome. We analyzed the proteome and the transcriptome of Toc159-deficient plants to discern the consequences of impaired protein import from systemic transcriptional responses resulting from the loss of photosynthetic capacity. The analysis of N-terminal protein acetylation showed that some chloroplast-targeted precursor proteins accumulate outside of plastids in Toc159-deficient plants. Interestingly, these precursors were not exclusively pho-tosynthetic proteins but have diverse metabolic functions. Sev-eral of these proteins were acetylated at position 2, indicating that they entered the common modification pathway for cyto-solic proteins. Our results show that Toc159 has broad speci-ficity, which is different from the proposed role as a receptor for import of abundant photosynthetic proteins. Together, our com-bined proteome and transcriptome analysis of mutants lacking Toc159 provides new insights into nuclear–organelle interactions during chloroplast proteome assembly.
RESULTS
Quantitative Analysis of Protein Accumulation in Wild-Type and Toc159-Deficient Leaves
To investigate the contribution of Toc159 to the assembly of the plastid proteome, we measured the quantitative protein accu-mulation in two Arabidopsis mutant lines deficient in Toc159 in comparison to the wild type and the wild type grown on Suc (wtS) (Figures 1A to 1D). The Toc159 null mutant ppi2 (Bauer et al., 2000) can grow beyond the cotyledon stage only on medium supplemented with Suc (Figure 1A). Elevated levels of soluble sugars interfere with developmental processes and affect gene expression (Bla¨sing et al., 2005; Gibson, 2005). To bypass the requirement of exogenous sugar supply, we generated a Toc159 cosuppression line (Toc159cs) that develops silencing of the
Toc159 gene only during later stages of development, resulting in
normal appearing seedlings and older plants with chlorophyll-deficient white rosette leaves (Figure 1B). Toc159cs lines were originally constructed to overexpress the M- and G-domains of Toc159 in a wild-type background (see Supplemental Figure 1 online) but developed the cosuppression phenotype shown in Figure 1A. Several independent Toc159cs lines were selected containing only one single transgene insertion. Thermal asym-metric interlaced PCR was used to identify lines in which the transgene was inserted in a noncoding region (see Supplemental Figure 1D online). Immunoblotting analyses with Toc159 anti-bodies confirmed the complete loss of Toc159 in ppi2 and in white leaves of Toc159cs (Figure 1E).
Using tandem mass spectrometry (MS/MS), we analyzed the leaf proteome composition of wild-type, wtS, ppi2, and
Toc159cs plants in three biological replicates each. In total,
4582 different proteins were identified (wild type, 2760; wtS, 2882; ppi2, 3150; Toc159cs, 3119) (Figure 1F; see Supplemental Data Set 1 online). Quantitative information was obtained for all identified proteins using normalized spectral counting (nSpC) (Baerenfaller et al., 2008). Since our experiment was designed to
analyze the consequences of Toc159 depletion for the assembly of the quantitative plastid proteome, we grouped the proteomics data of the wild type (wild type and wtS) and the Toc159-deficient plants (ppi2 and Toc159cs) and compared the protein identifi-cation and accumulation within and between these two groups. In total, 2347 proteins were detected in the wild type and wtS and 2496 in ppi2 and Toc159cs. The Spearman rank correlation coefficients obtained from the mean nSpC within these two groups are 0.85 for the wild type and wtS and 0.69 for ppi2 and
Toc159cs. Although a large number of proteins were identified in
all four plant types (1755 proteins), their quantitative accumula-tion was considerably different between the wild-type and the Toc159-deficient plants with Spearman rank correlation coeffi-cients between 0.43 and 0.47.
Plastid Proteome Assembly in the Absence of Toc159 To investigate the changes in the abundance of plastid pro-teins, we extracted these from the leaf data sets using a
high-confidence chloroplast proteome reference table comprising 1155 proteins (Baginsky and Gruissem, 2009; see Supplemental Data Set 2 online). We identified in total 926 plastid proteins in our data sets. In Toc159-deficient plants (the ppi2 mutant and
Toc159cs), we identified 767 plastid proteins and 569 were
identified in all four profiled plant types (Figure 2A). The quanti-tative accumulation of plastid proteins was very similar between wild-type and wtS plastids and between ppi2 and Toc159cs, with Spearman rank correlation coefficients of 0.945 and 0.813, re-spectively, and statistical tests support this conclusion. Out of 737 plastid proteins identified with more than 10 spectra, only 27 were significantly different between wild type and wtS and only 22 between Toc159cs and the ppi2 (t test, P value < 0.05). Only two proteins are present in both sets of significantly changed proteins: ferritin (AT5G01600) andb-amylase (AT4G17090); thus, we have no indication for a systematic influence of Suc on quantitative plastid protein accumulation. These results indicate that effects of exogenous Suc supply on plastid proteome assembly are minor under our experimental conditions and that both growth
Figure 1. Quantitative Proteome Profiling of ppi2, Toc159cs, Wild-Type, and wtS Leaves.
(A) to (D) Phenotypes of 35-d-old plants used for large-scale proteome profiling grown in short-day conditions (8 h light/16 h dark) on soil or on half-strength Murashige and Skoog medium supplemented with 100 mM Suc for ppi2 and wtS. ppi2 (A), Toc159cs (B), wild type (C), and wtS (D). Bars = 5 mm.
(E) Protein immunoblotting using antibodies directed against Toc159 confirming the lack of Toc159 in ppi2 and Toc159cs plants. Coomassie blue staining shows an equivalent protein loading for each protein extract separated by SDS-PAGE. wt, wild type.
(F) Proteins identified by MS/MS in ppi2, Toc159cs, wild-type, and wtS leaves. Total proteins were extracted from leaves, separated by gel electrophoresis, digested with trypsin, and analyzed by liquid chromatography MS/MS using a linear trap quadrupole Fourier transform–ion cyclotron resonance mass spectrometer.
conditions, with and without Suc, can be used for a quantitative comparison of the plastid proteomes in wild-type and Toc159-depleted plastids. Furthermore, statistical testing (t test, P value < 0.05) revealed that the abundances of the different components of the protein import machinery were not significantly affected by Suc, suggesting that similar constraints exist for the assembly of
the plastid proteome in ppi2 and Toc159cs (see Supplemental Figure 2 online).
Next, we used the quantitative data for 737 plastid proteins of which 680 are nuclear encoded that were identified with at least 10 spectra across all samples to identify groups of plastid proteins whose accumulation is significantly affected by the
Figure 2. Quantitative Accumulation of Plastid Proteins Identified in Leaves.
(A) Venn diagram of the plastid proteins identified in leaves by MS/MS. Plastid localization is based on a chloroplast proteome reference table with 1155 proteins. wt, wild type.
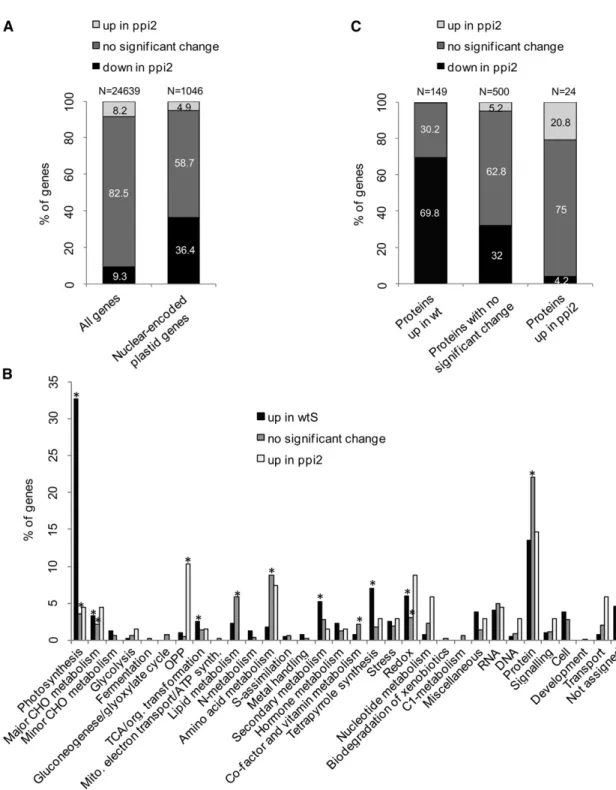
(B) Differential accumulation of nuclear-encoded plastid proteins in wild-type and Toc159-deficient leaves. Numbers have been rounded to one decimal. (C) Metabolic annotation of the nuclear-encoded genes destined to the plastid based on their distribution in MapMan bins. Categories indicated with an asterisk are significantly overrepresented, showing P values < 1e-05 in Fisher’s exact test. CHO, carbohydrate; OPP, oxidative pentose phosphate.
absence of Toc159. For statistical analysis, we omitted proteins identified with <10 spectra because low-abundance proteins are not accurately quantified (Zybailov et al., 2009). We performed a
t test and defined proteins that accumulated to higher levels in
wild-type or in Toc159-deficient plants as those proteins that had a P value < 0.05 both in the comparisons of the wild type against
Toc159cs and wtS against ppi2, and no significant difference in
the comparisons of the wild type against wtS and Toc159cs against ppi2 to exclude those proteins from the analysis whose accumulation is affected by Suc. In addition, we required a minimum fold change above 1.5 to exclude spurious small changes that are statistically significant from the interpretation. To these proteins we added those that had been identified in at least two replicates in each wild-type condition and not at all in both Toc159-deficient conditions and vice versa. Altogether, 184 plastid proteins accumulated to higher levels in wild-type and 24 in Toc159-deficient plants, of which 155 and 24, respectively, are nuclear encoded (Figure 2B; see Supplemental Data Set 3 online). The vast majority of the identified plastid proteins are not significantly affected by Toc159 depletion (74%). Two groups of plastid proteins (“no significant change” and “up in Toc159-deficient”) include proteins whose accumulation is not signifi-cantly affected by the absence of Toc159. Functional annotation of these putative Toc159-independent proteins and Fisher’s exact test revealed a significant enrichment of the categories
photosynthesis (“no significant change”) and tricarboxylic acid
(TCA) cycle TCA/org. transformation (“up in Toc159-deficient”) (Figure 2C). By contrast, the third group of proteins, “up in wild type,” comprises 155 nuclear-encoded plastid proteins that accumulate to higher levels in wild-type plastids and thus com-prises putative Toc159-dependent substrates (Figures 2B and 2C). Functional annotation showed that these 155 proteins belong to various functional categories (Figure 2C) with a signif-icant enrichment in photosynthesis.
Proteins for Photosynthetic Functions Are Downregulated at the Transcriptional Level inppi2
To distinguish proteins whose accumulation is directly affected by a loss of Toc159 function from those that are downregulated at the transcriptional level, we profiled the transcriptome of wild-type and ppi2 plants using plants grown on Suc to account for a possible effect of Suc on gene expression. After RNA hybridiza-tion to AGRONOMICS1 genome tiling arrays (Rehrauer et al., 2010), signals above background in at least one of the samples were detected for 24,639 of the
;
30,000 probe sets corre-sponding to 24,378 genes. Of these genes, 1998 (8.2%) were significantly upregulated in the wild type and 2268 (9.3%) in ppi2 (Figure 3A; see Supplemental Data Set 4 online). Our transcrip-tomics data confirmed the trend that photosynthetic proteins are expressed at lower levels in the import mutant as previously reported in published microarray, SAGE and RT-PCR data (Kubis et al., 2003; Kakizaki et al., 2009; Lee et al., 2009b). Notably, 84% of the downregulated genes reported in the SAGE data set were also found downregulated under our experimental conditions (see Supplemental Data Set 4 online). Similarly, we could confirm the reported upregulation of several heat shock–related proteins (Kakizaki et al., 2009). Of the 1046 nuclear-encoded genescoding for plastid proteins for which we had array data, 381 (36.4%) had significantly reduced RNA levels in ppi2, with a significant overrepresentation of genes encoding proteins for photosynthetic functions (Figure 3B; see Supplemental Data Set 4 online). Thus, the decreased accumulation of photosynthetic proteins in ppi2 is most likely a composite effect of reduced protein import and downregulation of genes for photosynthetic proteins, possibly as a systemic response to the loss of photo-synthetic capacity.
We observed that 104 (expected in a random distribution are 59, P value in Fisher’s exact test = 2.2e-07) of the set of 155 nuclear-encoded plastid proteins with decreased protein accu-mulation in Toc159-deficient plants for which we had transcript levels were also downregulated at the transcriptional level in ppi2 (69.8%; Figures 3B and 3C; see Supplemental Data Set 3 online). For the 24 proteins that accumulated to higher levels in Toc159-deficient plants (Figure 3C), 75% showed no significant change in RNA expression level, 20.8% were upregulated in ppi2 com-pared with the wild type, and only one was downregulated. From the 529 proteins (Figure 2B; i.e., the 74% plastid proteins from the category “no significant change”), more than 80% were either unchanged or downregulated at the transcriptional level in ppi2, thus indicating that they are efficiently imported into plastids de-spite the absence of Toc159. These Toc159-independent pro-teins also include propro-teins involved in photosynthesis (Figure 2C). In conclusion, the comparison between protein and transcript accumulation therefore confirms that Toc159 is not exclusively involved in the import of photosynthetic proteins and that many photosynthetic proteins enter the plastids in the absence of Toc159 (Figure 2C).
Peptide Detection Incidence Indicates Plastid Precursor Protein Accumulation outside of Plastids in Toc159-Deficient Plants
The accumulation of plastid proteins in Toc159-deficient leaves does not necessarily indicate their correct import into the chlo-roplast. We therefore analyzed the peptide detection incidence along the amino acid sequence of each protein to distinguish between mature plastid proteins and unprocessed precursors. For each identified protein, the most N-terminal detected peptide was considered and placed into the indicated amino acid bin corresponding to its location in the protein (Figure 4). After import, transit peptides of plastid proteins are usually cleaved off and degraded. Therefore, peptides from the transit peptide region should not be detectable from mature plastid proteins. The bin distribution of peptide detection reveals a detection gap at the N-terminal region of plastid proteins, thus suggesting that the transit peptides of most of the plastid proteins were cleaved (Figure 4A). The distribution was similar between the wild type, wtS, ppi2, and Toc159cs, suggesting that almost all precursor proteins were imported into plastids and processed in the absence of Toc159. As a control, we analyzed the peptide detection incidence for nonplastid proteins (Figure 4B). In this case, peptide detections from the N-terminal region were more prevalent, indicating that the low peptide detection in the N terminus of plastid proteins is most likely a consequence of N-terminal processing.
Figure 3. Transcriptional Response in ppi2 Compared with wtS.
(A) Expression data for 24,639 genes of ppi2 compared with wtS grown on Suc (all genes) and for the subset of 1046 nuclear-encoded plastid proteins. The percentage of downregulated genes in ppi2 highly increases when only nuclear-encoded genes destined to the plastid are taken into account. (B) Metabolic annotation of the nuclear-encoded genes destined to the plastid based on their distribution in MapMan bins. Categories indicated with an asterisk are significantly overrepresented, showing P values < 1e-05 in Fisher’s exact test.
(C) Expression data for the genes for which the proteins were found significantly upregulated in wild type (wt)/wtS, unchanged, or significantly upregulated in toc159cs/wtS in the proteomics data. CHO, carbohydrate; OPP, oxidative pentose phosphate.
We observed small peaks at positions 1 to 10 for plastid proteins in ppi2 and Toc159cs that were not present in the wild type (Figure 4A). This detection bin contains 22 proteins, 14 of which have plastid transit peptides that TargetP predicts to be processed and removed after protein import. We retrieved information about the localization of these proteins from the Plant Proteomics Database (PPDB) (Sun et al., 2009) and marked all proteins whose localization is ambiguous (see Supplemental Data Set 5 online). The remaining proteins include established chloroplast proteins, such as a pho-tosystem II oxygen-evolving complex protein (AT5G66570), RNA binding protein 29 (AT2G37220), or phosphoglycerate kinase (AT3G12780), which have predicted transit sequences of 29, 47, and 75 amino acids, respectively. Peptides within the cleaved transit peptide sequences were not found in the wild type or in the Arabidopsis proteome map (Baerenfaller et al., 2008), indicating that normally the transit sequences are efficiently
cleaved and degraded. Their detection in ppi2 and Toc159cs therefore provides a first hint that precursor proteins accumulate in these two genotypes (Table 1). To gain further insights into processing of chloroplast-targeted proteins in the wild type and in the mutants, we systematically analyzed N-terminal protein acetylation.
Plastid Precursor Proteins Accumulate outside of Plastids in Toc159-Deficient Plants Where They Are Modified by N-Terminal Met Excision and N-Terminal Acetylation The detection of peptides within the N-terminal transit peptide sequence of several plastid proteins suggests their accumulation as unprocessed precursors in Toc159-deficient plants. How-ever, the exact size of the transit peptide is difficult to predict and therefore could be erroneous. For further information about precursor processing, we therefore mapped the in vivo N ter-mini of proteins by analyzing N-terminal protein acetylation (N-acetylation) (see Supplemental Data Set 6 online). Since
N-acetylation occurs in the cytosol and in plastids, it allows us
to distinguish between plastid precursor and mature proteins (Figure 5A). Cytosolic proteins are predominantly acetylated at position 2, which corresponds to the detection of N-acetylated peptides in the 1-10 amino acid bin (Figure 5B). This distribution was similar among the wild type, wtS, ppi2, and Toc159cs, indicating that cytosolic N-terminal Met excision (NME) and subsequent N-acetylation is functional in all plant lines (Meinnel et al., 2006).
By contrast, differences were found in the distribution of
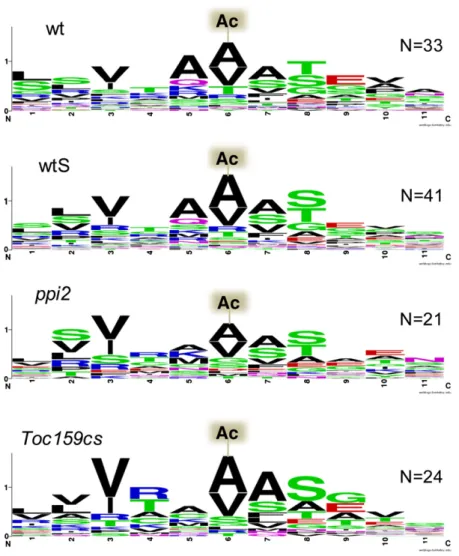
N-acetylation sites in plastid proteins. In the wild type, peaks of N-acetylated peptides were detected between amino acid
po-sitions 30 and 80, with a minor peak in the 1-10 amino acid bin (Figure 5A). N-acetylation between positions 30 and 80 corre-sponds to the position of the mature N terminus after removal of transit sequences and is therefore characteristic for imported and processed proteins. The minor peak at positions 1 to 10 comprises four nuclear-encoded proteins, a dynamin-like pro-tein (AT5G42080), a propro-tein of unknown function (AT5G01750), a malate dehydrogenase (AT5G11670), the Fe-superoxide dismu-tase 1 (AT4G25100), and three plastid-encoded proteins (see Supplemental Data Set 7 online). The nuclear-encoded proteins do not have predicted transit sequences, which is supported by our data.
For ppi2 and Toc159cs plastid proteins, two N-acetylation peaks were visible: a major peak at positions 1 to 10 and a second, minor peak at positions 30 to 80 (Figure 5A). As discussed above, the peak at positions 30 to 80 is indicative for precursor process-ing after import in ppi2 and Toc159cs plastids. The major peak at positions 1 to 10 in Toc159-deficient plants comprises three plastid-encoded proteins and 16 nuclear-encoded proteins (see Supplemental Data Set 7 online). Ten of these nuclear-encoded plastid proteins have a canonical transit sequence of a predicted length between 29 and 75 amino acids (Table 1). The accumulated proteins are not restricted to abundant photosynthetic proteins but also include minor constituents of the chloroplast proteome, such as a 29-kD ribonucleoprotein, the chaperonin 20, and a thiazole-requiring enzyme. Notably, the sequence context around the identified N-acetylation sites is consistent with established
Figure 4. Peptide Detection Incidence and Transit Peptide Cleavage in Leaves.
(A) Peptide minimal position of plastid proteins identified in leaves. The positions of the most N-terminal detected peptide of each identified protein were grouped into bins. The reduced detection of peptides at the N terminus of plastid proteins supports the removal of transit peptides after import in the wild type and in Toc159-deficient plants. The number of proteins for each blot (wild type, wtS, Toc159cs, and ppi2) is 763, 771, 645, and 659.
(B) Peptide minimal position of nonplastid proteins identified in leaves indicates that the N terminus of most nonplastid proteins is not cleaved and readily detectable by MS. The number of proteins for each blot (wild type, wtS, Toc159cs, and ppi2) is 1936, 2051, 2427, and 2444.
requirements for the cytosolic NME/acetylation pathway (i.e., small, uncharged residues, such as Ala following the initiator Met) (Figures 5C and 5D) (Sherman et al., 1985; Meinnel et al., 2006).
The detection of unprocessed plastid precursor proteins in the import-deficient plants could result either from a cleavage and processing defect in the mutant plastids or from precursor protein accumulation outside of plastids, most likely in the cytosol. The data presented in Figure 6 show that the amino acid context of the identified N-acetylation sites of processed plastid proteins was similar in the wild type and mutants. Although it is possible that a defect in the plastid protease network could potentially affect precursor processing in plastids (Zybailov et al., 2009), our data show that the accumulation of proteins involved in the posttrans-lational protein homeostasis network is not negatively affected by a defect in Toc159 (see Supplemental Data Set 8 online). Most of the N-acetylated mature plastid proteins that we identified here start with Ala or Val, and Ile and Val are overrepresented at the position three amino acids N-terminal to the N-acetylation site in the precursor protein. This is true for the mature proteins in all genotypes tested and confirms previous data (Zybailov et al.,
2008). Together, these data argue against a cleavage and pro-cessing defect in ppi2 and Toc159cs lines. We therefore tested whether the detection of unprocessed precursors results from their accumulation outside of plastids using isolated plastids. Proteome Analysis with Isolated Plastids Confirms the Accumulation of Precursor Proteins outside Plastids in Toc159-Deficient Mutants
To confirm the accumulation of N-acetylated plastid precursors outside of plastids in Toc159-deficient plants, we isolated plas-tids from wild-type and ppi2 plants because unprocessed cyto-solic precursors should be removed during plastid isolation. We analyzed quantitative protein accumulation in isolated wild-type and ppi2 plastids using MS/MS in three biological replicates and identified 3231 unique proteins (see Supplemental Figure 3 and Supplemental Data Set 9 online). We focused our analysis on 856 proteins that were present in the chloroplast proteome reference table (Baginsky and Gruissem, 2009). Of these proteins, 708 were identified in the wild type, 737 in ppi2, and 589 in both
Table 1. Unprocessed Precursor Proteins Accumulate outside Plastids in Toc159-Deficient Plants
Accession Description
N-Acetylation
Position (Leaves)
Minimal Peptide Position in Leaves TP Length Metabolic Function toc159 (Wild
Type) toc159 Wild Type AtProteome
N-acetylated plastid precursor proteins in Toc159-deficient plants
AT1G10960 ATFD1 (FERREDOXIN1) 2 2 NF 59 69 Photosystem
AT1G60950 FED A (FERREDOXIN2) 2 2 130 59 52 Photosystem
AT2G37220 29-kD ribonucleoprotein,
chloroplast
2 2 63 68 47 RNA met.
AT2G39730 RUBISCO ACTIVASE 2 73 73 73 58 Photosystem
AT3G12780 PGK1 2 2 80 80 75 Photosystem
AT1G61520 LHCA3; chlorophyll binding 2 14 14 49 48 Photosystem
AT3G54050 Fructose-1,6-bisphosphatase, putative 2 (60) 2 60 60 57 Photosystem
AT5G20720 CPN20 (CHAPERONIN20) 2 2 59 59 50 Protein
AT5G54770 THI1 (THIAZOLE REQUIRING) 2 2 47 46 45 Not assigned
AT5G66570 PSBO-1 2 2 107 107 29 Photosystem
Putative plastid precursor proteins identified in Toc159-deficient plants
AT1G62640 3-Ketoacyl-acyl carrier protein
synthase III
NF 44 92 72 74 Lipid met.
AT1G80030 DNAJ heat shock protein, putative NF 72 133 118 92 Stress
AT2G04030 CR88 (embryo defective 1956) NF 8 69 62 60 Abiotic stress
AT2G34460 Flavin reductase-related NF 35 36 48 51 Unknown
AT3G63140 CSP41A NF 50 102 80 72 RNA met.
AT4G13430 Isopropyl malate isomerase NF 48 56 56 76 TCA/org.
AT5G06290 2-Cys peroxiredoxin B NF 20 104 104 90 Redox
AT5G08280 Hydroxymethylbilane synthase NF 70 81 81 86 Tetrapyrrole
AT5G16390 Acetyl coenzyme A carboxylase 1 NF 64 67 78 82 Lipid met.
AT5G20250 DIN10 (DARK INDUCIBLE10) NF 9 20 20 – Minor CHO met.
AT5G26742 EMB1138 NF 27 61 61 60 RNA met.
AT5G49910 cpHSC70-2 NF 45 152 152 92 Abiotic stress
AT5G62790 1-Deoxy-D-xylulose 5-phosphate
reductoisomerase
NF 14 109 54 86 Secondary met.
Cytosolic location of plastid proteins was assessed by detection of N-acetylation within the predicted transit peptide (TP). Provided is also the minimal position of detected peptides in wild-type leaves and in AtProteome (Baerenfaller et al., 2008). Data “Wild Type” encompass data from wild type and wtS, and “toc159” data from ppi2 and Toc159cs, respectively. CHO, carbohydrate; NF, not found; met., metabolism; org., organic acid; -, no transit peptide predicted.
genotypes (see Supplemental Figures 3A and 3B online). The high number of proteins common to both plastid types provides additional evidence that most plastid proteins accumulate in both wild-type and ppi2 plastids despite the lack of Toc159. The N-terminal peptide detection incidence of isolated wild-type and
ppi2 plastids was very similar, indicating that the transit sequence
of most plastid proteins was cleaved after import (see Supple-mental Figure 3C online). Also, the amino acid context around the identified N-acetylation sites of wild-type and ppi2 plastid pro-teins was in agreement with the previous observation for all leaf data sets, confirming that processing of imported plastid proteins is functional in the wild type and in ppi2 plastids (Figure 6; see Supplemental Data Set 9 and Supplemental Figure 3D online).
Analysis of acetylated N termini supports the import and correct processing of plastid precursor proteins in isolated wild-type and ppi2 plastids (Figure 7A) because peptide detec-tion peaked between posidetec-tions 30 and 80 (cf. Figure 5A). Com-parison of the N-acetylation site distribution in plastid proteins between leaves and isolated plastids from wild-type plants confirmed that precursor proteins do not accumulate in wild-type plants (Figure 7B; see Supplemental Data Sets 9 and 10 online). By contrast, the distribution of N-acetylation sites was different for leaves and isolated plastids of ppi2 (Figure 7C). The
major peak in bin 1-10 in leaves was absent in isolated plastids, suggesting that cytosolic precursor proteins were removed during plastid isolation (see Supplemental Data Set 10 online). Since peptide detection in data-dependent acquisition may result in false-negative results, we searched for the peptide masses of N-terminal peptides from precursor proteins in the leaf data set and in isolated plastids. The absence of detectable peptide masses in MS spectra provides a strong argument against its presence in the sample because the mass spectro-metric measurement is extremely sensitive. Most peptides of a complex sample will give rise to a data point (i.e., peak) in the spectrum, even though only a very small fraction of these are identified in data-dependent acquisition experiments (Beck et al., 2011). While the precursor peptide masses were clearly identified in leaf extracts of Toc159-deficient plants, no masses matching the precursor peptide were identified in isolated plas-tids (exemplified for At3g12780 and At1g61520 in Figure 8 and Supplemental Figure 4 online). If nuclear-encoded plastid precursor proteins were present in plastids, the isolation pro-cedure should have enriched these compared with the leaf extract and, thus, they should be more readily detectable. Based on these data, we therefore conclude that the unprocessed precursor proteins in ppi2 and Toc159cs accumulate outside of
Figure 5. Met Removal and N-Terminal Acetylation in Leaves.
(A) N-acetylation sites of plastid proteins identified by MS/MS. N-acetylated proteins were grouped according to the position of their N-acetylation site.
N-acetylation sites of plastid proteins identified at positions 30 to 80 reflect the correct cleavage and processing of transit peptides after import in
wild-type and in Toc159-deficient plants. The increased percentage of N-acetylated plastid proteins at positions 1 to 10 in Toc159-deficient leaves suggests their accumulation outside plastids as a consequence of partially impaired import. The number of proteins for each blot (wild type, wtS, Toc159cs, and
ppi2) is 42, 48, 37, and 33.
(B) Distribution of N-acetylation sites of nonplastid proteins indicates that most of these undergo Met removal and N-acetylation in the cytosol. The number of proteins for each blot (wild type, wtS, Toc159cs, and ppi2) is 121, 140, 191, and 168.
(C) Schematic representation of the cytosolic two-step Met removal/acetylation pathway. (1) N-terminal start Met is usually removed from newly synthesized precursor proteins containing a transit peptide (TP) if the second residue is small and uncharged. (2) Addition of N-acetylation at the second residue by N-acetyltransferases.
plastids. The typical processing pattern of cytosolic proteins that is also observed for the detected precursor proteins (i.e., NME and N-terminal acetylation) (Figure 5) furthermore provides a strong argument for their cytosolic localization.
Identification of Toc159-Dependent and Toc159-Independent Plastid Proteins
Using the quantitative protein and transcript data presented above, we defined substrates that depend on Toc159 for import into the chloroplast and searched for features that distinguish such Toc159-dependent and Toc159-independent plastid pro-teins. We defined as Toc159-dependent those proteins that accumulated to significantly higher levels in the wild type and wtS compared with ppi2 and Toc159cs leaves as described above (see Supplemental Data Set 3 online). To exclude those proteins whose decreased abundance in ppi2 could be
attrib-uted to lower transcript levels, we additionally excluded those from the list of Toc159-dependent proteins that were signifi-cantly downregulated in ppi2 compared with wtS. As a final criterion, plastid proteins annotated by PPDB and AT_CHLORO to be localized in the outer envelope were removed (Sun et al., 2009; Ferro et al., 2010). Together, 44 proteins fulfilled these requirements and constitute the set of Toc159-dependent pro-teins, whose accumulation is affected at the protein and not at the transcript level (see Supplemental Data Set 11 online). By contrast, Toc159-independent proteins are those proteins that have increased or equal protein abundance in ppi2 or Toc159cs compared with the wild type and wtS, while their expression levels are similar in ppi2 and wtS. Together, 308 Toc159-independent proteins matched these criteria (see Supplemental Data Sets 1 and 3 online). Most of these were also identified in isolated ppi2 plastids, further supporting their Toc159-independent import (see Supplemental Data Set 9 online).
Figure 6. Sequence Context around N-Acetylation Sites of Plastid Proteins Identified in Leaves.
Most plastid proteins are N-acetylated (Ac) on Val or Ala. The similar sequence context in wild-type and in Toc159-deficient plants indicates that precursor processing of imported proteins is also functional in the mutant lines. N-acetylation sites ranging between positions 25 and 90 were used for the alignment.
Among the Toc159-independent proteins was plastoglobulin 35 (PGL35) (At4g04020) that we selected as an additional control for Toc159-independent protein import. The PGL35 protein can be considered representative for this group of proteins because it accumulates to similar levels in wild-type and albino leaves (average nSpC values are as follows: wild type, 1.35; wtS, 1.16;
ppi2, 1.22; Toc159cs, 1.46) (see Supplemental Data Set 1 online).
We fused PGL35 to green fluorescent protein (PLG35:GFP) (Vidi et al., 2006) and bombarded wild-type and white Toc159cs leaves with the fusion construct attached to gold particles (Figure 9). We observed the accumulation of PLG35:GFP in several wild-type plastids and in smaller plastids of white Toc159cs leaves (Figure 9A). By contrast, the precursor protein of the small subunit of ribulose-1,5-bis-phosphate carboxylase/oxygenase fused to GFP only accumulated in wild-type plastids (Figure 9B). These data confirm our proteomics strategy for the quantification and selection of Toc159-independent proteins.
To extract information about sequence features that might govern import specificity, we aligned the sequences of the 100 first amino acids of the putative dependent and Toc159-independent substrates (see Supplemental Figure 5 online). The enriched amino acids comprise mostly Ser residues, and the pattern did not reveal any specific sequence or domain structure that would allow distinguishing proteins in the two data sets. Other attempts to define relevant information in the amino acid sequences by multiple sequence alignment using ClustalW were not successful, and none of the approaches allowed us to distinguish the proteins in the two data sets by sequence information.
DISCUSSION
The Chloroplast Proteome in the Absence of Toc159 We used quantitative proteomics and transcriptomics to inves-tigate the contribution of the import receptor Toc159 to the assembly of the chloroplast proteome. In addition to the loss-of-function mutant ppi2, we used a Toc159 cosuppression line that allowed us to study the chloroplast proteome composition without exogenous sugar supply (Figure 1). We expected that the natural distribution of source/sink relationships in the
Toc159cs plants better reflects the in vivo situation compared
with the growth of ppi2 on Suc. Interestingly, the qualitative and quantitative accumulation of plastid proteins in leaves was very similar in Toc159cs and ppi2, and also in the wild type and wtS, suggesting that exogenous Suc supply has only a minor effect on the assembly of the plastid proteome.
Based on the above data, ppi2 and white leaves of Toc159cs are equivalent with respect to plastid proteome assembly in the absence of Toc159. Both genotypes accumulate photosynthetic proteins (Figures 2, 4, and 6), and proteins from several other metabolic pathways in the plastids of Toc159-deficient leaves are reduced (Figure 2). Thus, the substrate specificity of Toc159 is not restricted to photosynthetic proteins. Most of the pro-teins that had a lower abundance in ppi2 and Toc159cs were also downregulated at the gene expression level, suggesting that their decreased accumulation is a result of two different
Figure 7. Comparison of N-acetylation Sites Identified in Isolated Plas-tids and Leaves.
(A) Similar distribution patterns of N-acetylation sites identified in isolated the wild-type and ppi2 plastids indicate that plastid proteins were imported and processed correctly. The number of proteins for each blot (the wild type and ppi2) is 68 and 42.
(B) Similar distribution patterns of N-acetylation sites between isolated wild-type chloroplasts and wild-type leaves. The number of proteins for each blot (leaves and plastids) is 42 and 68.
(c) Comparison of the N-acetylation sites between isolated ppi2 chloro-plasts and ppi2 leaves. A major difference (*) is observed at positions 1-10 suggesting the accumulation outside plastids of unprocessed precursor proteins when import is partially impaired in ppi2. The number of proteins for each blot (leaves and plastids) is 33 and 42.
responses, the transcriptional response as well as the import defect (Figure 3). This is especially true for photosynthetic pro-teins. It could be argued that import deficiency directly affects gene expression, but such a scenario would require separate signaling circuits that control the expression of individual nuclear genes encoding plastid proteins, which seems unlikely. A more likely explanation is that nuclear-encoded plastid proteins are regulated in modules, which is supported by the proposed master switch model (Richly et al., 2003; Biehl et al., 2005).
Many plastid proteins accumulate in Toc159-deficient plants, some of them to similar or even above wild-type levels. Appar-ently, the absence of Toc159 is partially compensated by one or more of its homologs: Toc132, Toc120, or Toc90. We could not detect these receptors by MS, either in leaves or in isolated plastids, but transcripts of Toc132 and Toc120 were slightly elevated in ppi2 compared with the wild type (see Supplemental Figure 1 and Supplemental Data Set 4 online). A recent study showed that the overexpression of Toc90 can partially restore
the accumulation of Toc159 client proteins in ppi2 (Infanger et al., 2011). However, the low abundance of the Toc90 protein and the low expression level of its transcript in ppi2 make it unlikely that the residual protein import observed here is exclusively based on Toc90 activity (Bauer et al., 2000; Infanger et al., 2011). It is more plausible that Toc132 and Toc120 sustain protein import in Toc159-deficient plants because their expression is induced in
ppi2. Elevated receptor abundance was previously shown to
restore an albino phenotype to a pale-green phenotype, sug-gesting that receptor abundance is limiting for efficient protein import (Hiltbrunner et al., 2004; Ivanova et al., 2004; Kubis et al., 2004). Together with these observations, our data call for a model in which the import capacity is controlled not only by substrate specificity but also by the abundance of the different receptors and their ability to associate into functional translocon complexes.
A recent report suggested that the A-domain of the Toc159 receptor family mediates precursor selectivity (Inoue et al.,
Figure 8. Transit Peptide Visualization and Extracted Ion Chromatogram Quantification for Phosphoglycerate Kinase1 Peptides Identified in Leaves and Isolated Plastids.
(A) Visualization of peptide detection along the protein. Several spectra matched to the transit peptide region (indicated as TP; 75 amino acids predicted length) in Toc159-deficient (toc159) leaves. By contrast, we could not detect any spectra matching the predicted TP region in wild-type leaves or in isolated ppi2 or wild-type plastids, although the coverage of Phosphoglycerate Kinase1 (PGK1) detection is similar between the different samples (cf. peptide color scheme and peptide localization between the different samples; the darker the shading, the more spectra matching to the indicated peptide were observed).
(B) Extracted exact mass ion chromatogram for the spectra matching to the transit peptide region from leaves (top panel; marked with an asterisk) and corresponding samples from isolated plastids. In three samples from isolated plastids, an ion matching the exact mass of the PGK1 transit peptide
identification (asterisk) was detected at a shifted retention time of;8 min (bottom panels). MS/MS information on these ions revealed that they are
clearly different from the peptide eluting at 48 min (top panel, precursor peptide). The upper peptide represents the peptide matching to the PGK1 transit peptide, the three ions below produced a poor-quality MS/MS spectrum that could not be assigned to any peptide (it was assigned to format dehydrogenase with a score below the threshold). L, leaves; P, plastids.
2010), which explains why overexpression of full-length Toc132 or Toc120 failed to complement ppi2 (Ivanova et al., 2004; Kubis et al., 2004), while constructs containing only the G- and M-domains of Toc132 were able to do so (Agne et al., 2009; Richardson et al., 2009; Inoue et al., 2010). Loss of the A-domain resulted in import receptors with less selective precursor protein recognition and import capacities. It seems that the A-domain confers precursor selectivity to a constitutive import process, whose efficiency depends mainly on the abundance of the different Toc receptors. Initial results suggest that the A-domain may be cleaved upon assembly of the Toc complex (Agne et al., 2010). The conclusion that the A-domain mediates import spec-ificity could explain why our data did not identify sequence specificity determinants of Toc159-mediated protein import (see Supplemental Figure 5 online). Although we identified informa-tion-rich regions within the first 20 amino acids up to position 55 as reported for Toc159-mediated RbcS import (Smith et al.,
2004; Lee et al., 2008; Lee et al., 2009a), we could not identify sequence determinants that distinguish Toc159-dependent from Toc159-independent proteins (see Supplemental Figure 5 on-line). Additional components in the cytosol, such as 14-3-3 proteins, might be involved in determining specific transit pep-tide recognition by different TOC receptors, but further research is necessary to establish the exact role of cytosolic guidance complexes in this process (May and Soll, 2000).
N-Terminal Met Excision and Acetylation of Plastid Precursor Proteins in the Cytosol
We found several unprocessed precursor proteins that were
N-acetylated at position 2 most likely in the cytosol of
Toc159-deficient plants (Figure 5). The detection of the unprocessed precursors raises the question of which specific features allow their accumulation in Toc159-deficient plants. The identified proteins are not simply abundant photosynthetic proteins that may saturate the impaired plastid import machinery in ppi2 and
Toc159cs and thus accumulate because of their high
abun-dance. The accumulation of most high-abundance photosyn-thetic proteins seems to be prevented by regulatory signaling circuits that result in reduced expression of their genes (Figures 2 and 3). Interestingly, some of the unprocessed precursors of nonphotosynthetic proteins were also downregulated at the transcriptional level, but their decrease was not as pronounced as that of photosynthetic proteins (Table 1; see Supplemental Data Set 4 online). This suggests that these nonphotosynthetic proteins are integrated in a signaling circuit that is different from the signaling circuit that controls the expression of photo-synthetic proteins. Together, our data support the view that the accumulation of nuclear-encoded plastid precursor proteins triggers an unimported precursor protein response (Kakizaki et al., 2009; Lee et al., 2009b). This signaling response could act through additional unknown signaling pathways that do not depend on the functional state of the plastids.
Our data show that plastid precursor proteins were modified by NME and subsequent N-acetylation outside the plastid. This modification mechanism is used for 60 to 90% of all proteins in eukaryotes (Goetze et al., 2009), and it is conceivable that it also modifies plastid precursor proteins under normal wild-type con-ditions during their cytosolic synthesis and transition. The se-quence context around the identified N-acetylation sites is consistent with described requirements for this type of modifi-cation (i.e., small, uncharged residues like Ala follow the start Met) (Sherman et al., 1985; Meinnel et al., 2006). Our observation may therefore indicate a general modification pathway for plastid proteins in the cytosol. N-acetylation serves as a degradation signal for the ubiquitin ligase-proteasome system in yeast, and it was proposed to be involved in protein folding quality control (Hwang et al., 2010). Support for a function of plastid precursor protein N-acetylation in vivo came from the characterization of a mutant for a cytoplasmic N-acetyltransferase. This mutant is affected in chlorophyll accumulation and photosynthetic capac-ity, suggesting crosstalk between cytosolic N-acetylation and chloroplast function (Pesaresi et al., 2003).
The fact that plastid precursor stability is tightly controlled in the cytosol was recently substantiated by the detection of a
Figure 9. Toc159-Independent Import of 35S:PGL35 in White Toc159cs Leaves.
(A) Epidermal cells of wild-type and white Toc159cs leaves transformed by biolistic transformation. GFP fused to the transit peptide of PGL35 is imported successfully into wild-type and Toc159cs plastids. Maximal projections of confocal images are shown.
(B) Import into plastids in wild-type and white Toc159cs leaves of GFP fused to the transit peptide of ribulose-1,5-bis-phosphate carboxylase/ oxygenase small subunit (pSSU).
ubiquitin-dependent degradation pathway for nonimported Lhcb4 precursors that is mediated by the heat shock cognate protein 70-4 and ubiquitination by an E3 ligase (Lee et al., 2009b). Inhibition of the proteasome resulted in the enhanced accumu-lation of ubiquitinated Lhcb4 precursors in ppi2, thus indicating the existence of a degradation system for plastid precursors. Based on these observations, it is conceivable that cytosolic
N-acetylation may serve as an additional degradation signal for
nonimported plastid proteins to avoid accumulation of unpro-cessed precursor proteins in the cytosol. A short half-life of nonimported N-acetylated plastid precursor proteins could be the reason why these have not been identified in wild-type plants. Future work is needed to establish the role of N-acetylation in the assembly of the plastid proteome and its contribution to cellular function.
In summary, we used quantitative proteomics in combination with transcriptional profiling to establish a definition of Toc159-dependent and Toc159-inToc159-dependent plastid protein import. Based on our results, we could not identify any amino acid sequence specificity determinants that allow distinguishing Toc159-dependent and Toc159-independent proteins. The nu-merous photosynthetic proteins that accumulate as correctly processed mature proteins in ppi2 and Toc159cs and the distinct functions affected in Toc159-deficient plastids furthermore indi-cate an unexpected client protein promiscuity of Toc159 and suggest that the current model for the import of photosynthetic and housekeeping nonphotosynthetic proteins must be revised. The identification of N-acetylated plastid precursor proteins outside the plastid in Toc159-deficient plants could point to a control mechanism for plastid precursor proteins that may prevent precursor accumulation under normal wild-type condi-tions. Altogether, we present evidence for complex regulation of plastid proteome assembly in import-deficient plastids that en-compasses import preference, transcriptional regulation, and po-tentially differential degradation of plastid precursor proteins by
N-acetylation–triggered processes. The quantitative
contribu-tion of each individual pathway must be assessed under different conditions, and more information on their dynamic regulation is necessary. This information in combination with increasingly sensitive analyses of precursor protein accumulation via detec-tion of unprocessed N termini, possibly by combined fracdetec-tional diagonal chromatography (Gevaert et al., 2007), in the future could provide a more comprehensive understanding of chloro-plast proteome assembly.
METHODS
Plant Material
All experiments used Arabidopsis thaliana plants of accession Wassilewskija.
Plants were grown at 208C in photoperiods of 8 h light and 16 h darkness
with a light intensity of 150mmol m22s21. For protein extraction, leaves
(leaf 6 from 5 to 10 plants per sample) were harvested after 35 d, 1 h before the end of the photoperiod and frozen in liquid nitrogen. The wild-type and Toc159cs plants were grown on soil, whereas wtS and ppi2 were grown on half-strength Murashige and Skoog medium supplemented with 100 mM Suc. For each genotype, three independent biological rep-licates were performed (i.e., plants were grown three times under exactly the same conditions), and leaf 6 was harvested as described above. The
genotypic characterization of ppi2 and Toc159cs was described by Bauer et al. (2000) and in Supplemental Figure 1 online, respectively.
Generation of Toc159 Cosuppression Plants
A fragment from Toc159 corresponding to the GTPase and membrane
domains was amplified by PCR using 5
9-CGGGATCCAAAATGGCTCAG-GATCACCACCACCACCACCACGGCACGAAGCTTTTCTCTATGGAT-39
(includes BamHI) and 5
9-GCTCTAGATTAGTACATGCTGTACTTGTCG-TTCGTC-39 (includes XbaI) primers, digested with BamHI and XbaI, and
inserted into the corresponding NcoI and XbaI sites from the pCHF7 binary vector (a gift from C. Fankhauser, University of Lausanne, Switzer-land). Plants were transformed using the floral dip method as described (Clough and Bent, 1998). Transformants were selected on soil by spraying phosphinothricin (30 mg/L BASTA; Duchefa), and DNA prepared from leaf material was used for diagnostic PCR. Analysis of segregation was performed by germinating seeds on half-strength Murashige and Skoog
medium and 30 mg/mL phosphinothricin. Phosphinothricin-sensitive
plants displayed yellow cotyledons and were easily distinguishable from Toc159 cosuppressing plants with white leaves and cotyledons. Lines with a segregation indicative of a single T-DNA insertion and with high incidence of Toc159 cosuppression were selected. T-DNA insertions were mapped in these lines by three rounds of thermal
asymmetric interlaced PCR using pCHF7_LB_a (5
9-CGAACATCGGTCT-CAATGCAAAAG-39), pCHF7_LB_b (59-CTACCTCGGCTCTGCGAAG-39),
pCHF7_LB_c (59-CGGATACTTACGTCACGTCTTGC-39), and random
primers.
Protein Extraction and Immunoblotting
Protein extracts were prepared from leaves by grinding shock-frozen tissue. Subsequently, extraction buffer was added (40 mM Tris, pH 6.8, 40
mM DTT, 4% SDS, and 23 protease inhibitor cocktail [Roche]).
Homo-genates were centrifuged twice at 20,000 rcf for 20 min at room temper-ature. Protein concentration was determined using a BCA protein assay kit (Thermo Scientific) before adding DTT. Twenty micrograms of protein from leaves were subjected to SDS-PAGE on 12% gels. Immunoblotting
using a-Toc159, a-PGL35, and a-CAB antibodies was performed as
described previously (Ivanova et al., 2004). For each immunoblotting experiment, a second gel was prepared in parallel as loading control, which was stained with Coomassie Brilliant Blue according to standard procedures.
Sample Preparation for Large-Scale Proteomics and MS
Protein extraction was performed as described above, and 600mg of
protein from leaves and 200mg from isolated chloroplasts were subjected
to SDS-PAGE on 12% gels. After electrophoresis, gels were stained with Coomassie Brilliant Blue according to standard procedures and cut into 12 sections for leaves and 20 sections for isolated chloroplasts respec-tively. Each gel slice was diced into small pieces. In-gel digestion was performed according to Shevchenko et al. (1996). After digestion, dried peptides were resuspended in 3% acetonitrile and 0.2% trifluoretic acid and desalted using Sepak cartridges (Waters).
Dried peptides were resuspended in 3% acetonitrile and 0.2% formic acid and analyzed with a linear trap quadrupole Fourier transform–ion cyclotron resonance mass spectrometer (ThermoFischer Scientific) cou-pled to an Eksigent nano liquid chromatography system (Eksigent Tech-nologies). Peptide mixtures were loaded onto laboratory-made capillary
columns (BGB Analytik) of 75-mm inner diameter, 8-cm length, packed
with 3 mm, 200 A˚ Magic C18 AQ beads (Michrom BioResources).
Peptides were eluted from the column by an increased acetonitrile gradient from 3% acetonitrile, 0.2% formic acid to 36% acetonitrile, 0.2% formic acid over 55 min, and from 36% acetonitrile, 0.2% formic
acid to 80% acetonitrile, 0.2% formic acid over 5 min, followed by a 10-min wash step with 80% acetonitrile and 0.2% formic acid. Peptide ions were detected in a survey scan from mass-to-charge ratio 300 to 1600 at 100,000 full width half maximum nominal resolution followed by three data-dependent MS/MS scans (isolation width 2 atomic mass units, relative collision energy 35%, dynamic exclusion enabled, repeat count 1, followed by peak exclusion for 2 min).
Data Mining and Protein Quantification
MS/MS spectra were searched either with Sequest and PeptideProphet (Yates et al., 1995; Keller et al., 2002) using the Trans-Proteomic Pipeline (TPP v2.9) or with Mascot version 2.2.04 (Matrix Science) against the
Arabidopsis TAIR9 protein database (downloaded on June 29, 2009) with
a concatenated decoy database supplemented with contaminants (67,079 entries). For Sequest, the search parameters were as follows: requirement for tryptic ends, one missed cleavage allowed, mass
toler-ance =6 5 ppm, variable modification of Met (M, PSI-MOD name:
oxidation, ModAccession: MOD: 00412, monoD = 15.99491) and static
modification of Cys (C, PSI-MOD name: iodoacetamide derivative,
ModAccession: MOD: 00397, monoD = 57.0214). For PeptideProphet,
the cutoff was set to a minimum probability of 0.9. For Mascot, the search parameters were as follows: requirement for semitryptic ends, one
missed cleavage allowed, mass tolerance =65 ppm. Besides
carbami-domethylation of Cys residues as fixed modification, oxidation of Met, N-terminal protein acetylation, and N-terminal acetylation were included as variable modifications. N-terminal acetylated peptides were accepted with a Mascot ion score higher than 24 and a Mascot expectation value smaller than 0.05. All peptide assignments from Sequest and Mascot except those of contaminants were filtered for ambiguity, and the peptides (N-terminal acetylated peptides included) matching to more than one protein were excluded from further analysis. This does not apply to different splice variants of the same protein or to different loci sharing exactly the same sequence. Different N-acetylated peptides matching to the same protein were also excluded from further analysis. Furthermore, spectrum assignments to decoy database peptides were excluded. All remaining spectrum assignments were entered into the pep2pro data-base and are available from the pep2pro website at www.pep2pro.ethz. ch (data set Plastid Proteome Assembly in Absence of Toc159). From the final data, PRIDE 2.1 XML files were created and exported to the PRIDE database (accession numbers from 14834 to 14839) (Vizcaı´no et al., 2010). The spectrum false discovery rate was calculated by dividing the number of decoy database spectrum assignments by the number of spectrum assignments in the final data set. The false positive rate was between 0.34 and 0.74% for all measured biological replicates of leaves and of isolated chloroplasts.
Protein quantification with nSpC was done according to Baerenfaller et al. (2008). Briefly, the expected contribution of each individual protein to the samples total peptide pool was calculated correcting the values with a normalization factor, which balances for the theoretical number of tryptic peptides per protein and sample depth according to the following formula:
nSpCK¼ SpectraK3
TTPK3MS
MP
!2 1
where nSpCKis the normalized spectral count for protein K, TTPKis the
theoretical tryptic peptides of protein K, MS is the total number of measured spectra in the data set, And MP is the total number of theoretical tryptic peptides of the identified proteins in the data set.
For the determination of the number of TTPk, the whole protein database
was digested in silico. If Arg or Lys was followed by Pro (KP/RP site), the site was both cut and not cut (resulting in three tryptic peptides). If several of these sequence pairs followed each other, we only considered cutting one KP/RP site per time. The resulting peptides were labeled as theoretical tryptic peptides of 400 to 6000 D and at least six amino acids.
Statistical Analysis of the Data
In the statistical analysis of the data, only proteins identified with at least 10 spectra were taken into account. Four separate t tests (two-sided, Welch test) were done comparing the wild type and wtS, the wild type and
Toc159cs, wtS and ppi2, and Toc159cs and ppi2. Proteins considered
significantly more abundant in the wild type or the Toc159-deficient
samples had to fulfill the following criteria: P valuewild type_Toc159cs< 0.05,
P valuewtS_ppi< 0.05, P valuewild type_wtS$ 0.05 or not applicable, and
P valueToc159cs_ppi2$ 0.05 or not applicable. In addition to the
signif-icance test, the proteins had to show an at least 1.5-fold change in the comparisons wild-type/Toc159cs and wtS/ppi2. To the proteins fulfilling these criteria, we added those that were not identified at all in wild-type conditions but in at least two of the three replicates both for ppi2 and
Toc159cs (category “Only in Toc159cs/ppi2”), and those that were not
at all identified in Toc159-deficient samples, but in at least two of the three replicates both for wild-type and wtS (category “Only in wild type/wtS”). All analyses were done with R (R Development Core Team, 2010).
Microarray Hybridization and Evaluation
RNA was extracted from 35-d-old plants. The experiment was performed with three biological replicates, and Affymetrix Arabidopsis AGRO-NOMICS1 microarrays were used. Labeling of samples, hybridizations, and measurements were performed as described (Hennig et al., 2004; Rehrauer et al., 2010). Signal values were derived using the robust multiarray analysis algorithm implemented in the statistical language R (R Development Core Team, 2010) using probe sets comprising exonic probes based on the TAIR9 genome annotation. For details of probe set definition and low-level data analysis, see Rehrauer et al. (2010). Differ-entially expressed genes were selected using LIMMA (Smyth, 2004) followed by multiple testing correction according to Storey and Tibshirani (2003). Genes were considered as differentially expressed if P value < 0.05. This resulted in a list of 15,882 genes. Downstream analysis was performed on a subset of the differentially expressed genes that had a fold change of more than 2.Significance of overlaps of gene sets was calculated with a Fisher’s exact test. The microarray data have been submitted to ArrayExpress with the experiment number E-TABM-951.
Chloroplast Localization, Sequence Alignment, and Functional Annotation
Chloroplast localization annotation was based on a chloroplast protein reference table that comprises 1155 proteins (reviewed in Baginsky and Gruissem, 2009). Plastid localization of these 1155 proteins were vali-dated in different proteomics studies by software tools that integrate targeting prediction with coexpression data and several other criteria that were combined and weighted with a naı¨ve Bayesian classifier. Amino acid sequence alignment and visualization were performed with Weblogo (weblogo.berkeley.edu) (Crooks et al., 2004). In silico localization predic-tion was done using TargetP (Emanuelsson et al., 2000). Funcpredic-tional annotation was based on MapMan (mapman.mpimp-golm.mpg.de) (Usadel et al., 2009), and the overrepresentation of functional categories was assessed with Fisher’s exact test.
Chloroplast Isolation
Intact chloroplasts from 35-d-old wild-type or ppi2 plants grown on soil or on half-strength Murashige and Skoog medium supplemented with 100 mM Suc medium were isolated according to Fitzpatrick and Keegstra (2001) with the following modifications. Cellulase Onozuka R-10 and macerozyme R-10 (Serva; catalog numbers 16419 and 28302) were diluted
to 1 and 0.25% (w/v), respectively.L-ascorbate (0.1% [w/v]) and 0.05%