Species boundaries in the Himantura uarnak species complex (Myliobatiformes: Dasyatidae)
Texte intégral
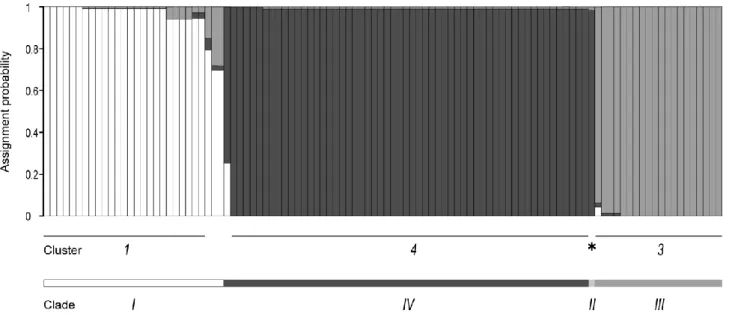
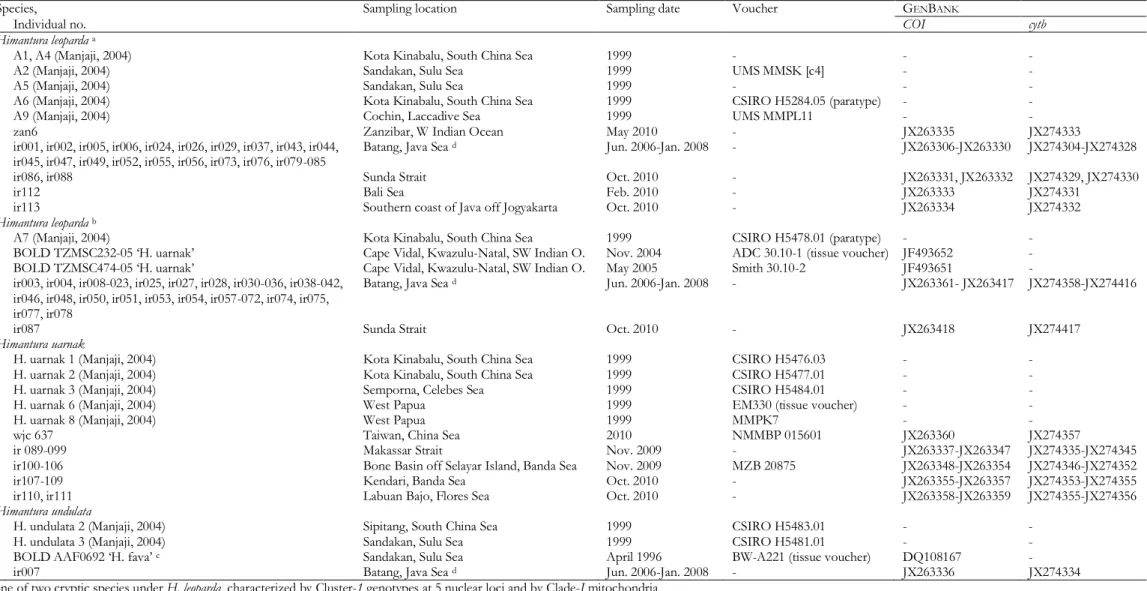
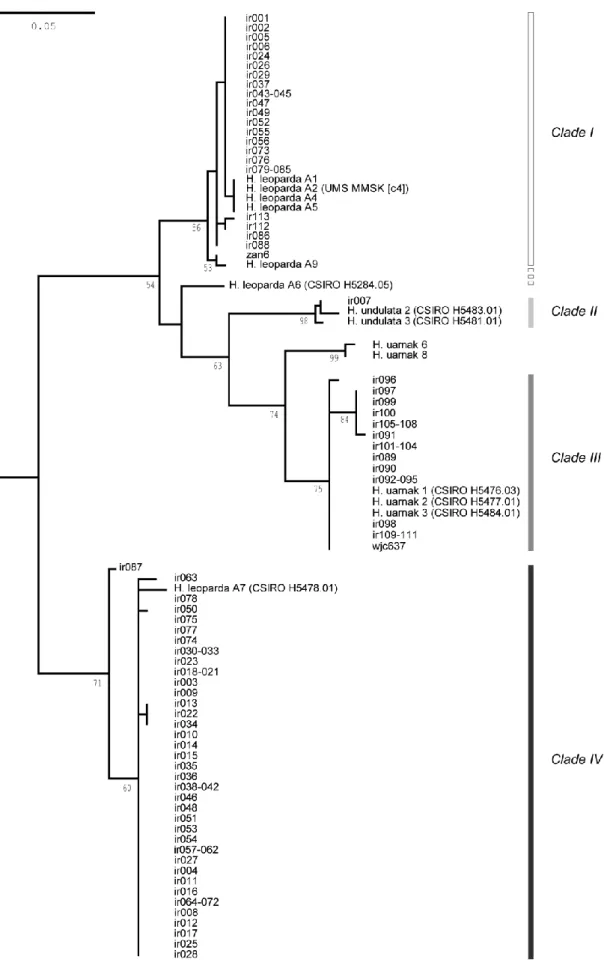
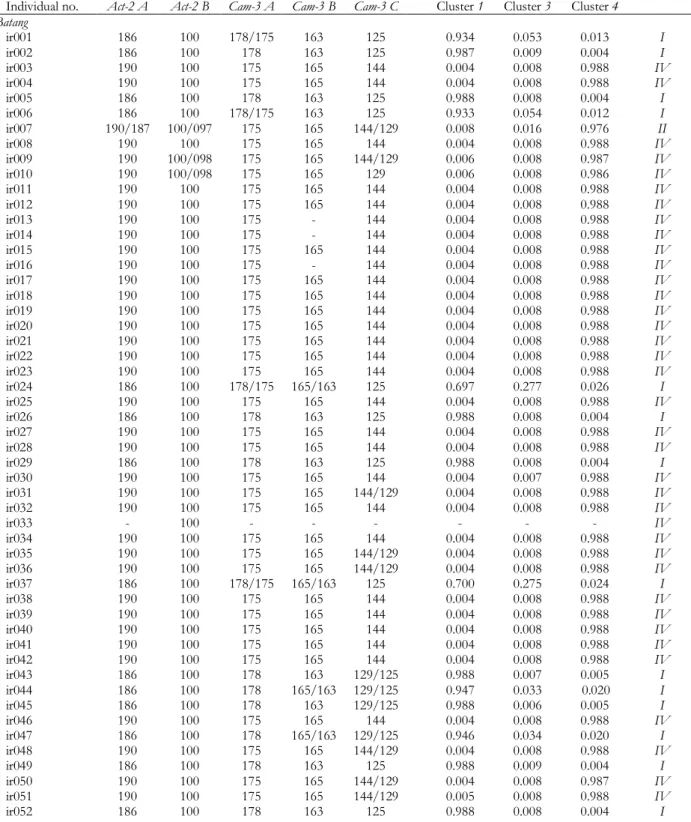
Figure

Documents relatifs
Interestingly, RIB 3 and 8, which are closely related ribotypes based on ribosomal phylogenies but considered as different cryptic species based on CBCs and k-mer analysis,
Table 1: The minor species investigated in this study, their upper limits (3-sigma) by the previous studies, and their theoretical detection limits by NOMAD solar
Chromosomal inversion polymorphism in 2 marginal populations of the endemic Hawaiian species, Drosophila silvestris... Original
Félix Massiot-Granier, Etienne Prévost, Gerald Chaput, Ted Potter, Gordon Smith, Atso Romakkaniemi, Etienne Rivot.. To cite
Supplementary Table S2 Variable nucleotide sites from the cytochrome b gene sequence dataset used to diagnose blue-spotted maskray species formerly under Neotrygon kuhlii (Müller
The blue-spotted maskray from Guadalcanal Island (Solomon archipelago) is distinct by its colour patterns from Neotrygon kuhlii with which it was previously confused, and belongs to
In fact, as might have been expected, it has been previously demonstrated that a specific expression of the extrapolation of the species accumulation curve
A calibration database was created by gathering spectra from various large poaceae species: sorghum, millet, sugarcane, maize, elephant grass, miscanthus from various tropical