HAL Id: hal-03167240
https://hal.archives-ouvertes.fr/hal-03167240
Submitted on 16 Mar 2021
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Distributed under a Creative Commons Attribution| 4.0 International License
A collaborative resource platform for non-human
primate neuroimaging
Adam Messinger, Nikoloz Sirmpilatze, Katja Heuer, Kep Kee Loh, Rogier
Mars, Julien Sein, Ting Xu, Daniel Glen, Benjamin Jung, Jakob Seidlitz, et al.
To cite this version:
Adam Messinger, Nikoloz Sirmpilatze, Katja Heuer, Kep Kee Loh, Rogier Mars, et al.. A collaborative
resource platform for non-human primate neuroimaging. NeuroImage, Elsevier, 2021, 226, pp.117519.
�10.1016/j.neuroimage.2020.117519�. �hal-03167240�
ContentslistsavailableatScienceDirect
NeuroImage
journalhomepage:www.elsevier.com/locate/neuroimage
A
collaborative
resource
platform
for
non-human
primate
neuroimaging
Adam
Messinger
a,
Nikoloz
Sirmpilatze
b,c,
Katja
Heuer
d,e,
Kep
Kee
Loh
f,g,
Rogier
B.
Mars
h,i,
Julien
Sein
f,
Ting
Xu
j,
Daniel
Glen
k,
Benjamin
Jung
a,l,
Jakob
Seidlitz
m,n,
Paul
Taylor
k,
Roberto
Toro
e,o,
Eduardo
A.
Garza-Villarreal
p,
Caleb
Sponheim
q,
Xindi
Wang
r,
R.
Austin
Benn
s,
Bastien
Cagna
f,
Rakshit
Dadarwal
b,c,
Henry
C.
Evrard
t,u,v,w,
Pamela
Garcia-Saldivar
p,
Steven
Giavasis
j,
Renée
Hartig
t,u,x,
Claude
Lepage
r,
Cirong
Liu
y,
Piotr
Majka
z,aa,bb,
Hugo
Merchant
p,
Michael
P.
Milham
j,v,
Marcello
G.P.
Rosa
aa,bb,
Jordy
Tasserie
cc,dd,ee,
Lynn
Uhrig
cc,dd,
Daniel
S.
Margulies
ff,
P.
Christiaan
Klink
gg,∗a Laboratory of Brain and Cognition, National Institute of Mental Health, Bethesda, USA
b German Primate Center – Leibniz Institute for Primate Research, Kellnerweg 4, 37077 Göttingen, Germany c Georg-August-University Göttingen, 37073 Göttingen, Germany
d Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany
e Center for Research and Interdisciplinarity (CRI), INSERM U1284, Université de Paris, Paris, France
f Institut de Neurosciences de la Timone (INT), Aix-Marseille Université, CNRS, UMR 7289, 13005 Marseille, France g Institute for Language, Communication, and the Brain, Aix-Marseille University, Marseille, France
h Wellcome Centre for Integrative Neuroimaging, Nuffield Department of Clinical Neurosciences, John Radcliffe Hospital, University of Oxford, Oxford OX3 9DU, UK i Donders Institute for Brain, Cognition, and Behaviour, Radboud University Nijmegen, Montessorilaan 3, 6525 HR Nijmegen, The Netherlands
j Child Mind Institute, 101 E 56th St, New York, NY 10022, USA
k Scientific and Statistical Computing Core, National Institute of Mental Health, Bethesda, USA l Department of Neuroscience, Brown University, Providence RI USA
m Department of Child and Adolescent Psychiatry and Behavioral Science, Children’s Hospital of Philadelphia, Philadelphia PA USA n Department of Psychiatry, University of Pennsylvania, Philadelphia PA USA
o Department of Neuroscience, Institut Pasteur, UMR 3571 CNRS, Université de Paris, Paris, France p Instituto de Neurobiologia, Universidad Nacional Autónoma de México campus Juriquilla, Queretaro, Mexico q Department of Organismal Biology and Anatomy, University of Chicago, Chicago IL USA
r McGill Centre for Integrative Neuroscience, Montreal Neurological Institute (MNI), Quebec, Canada s Centro Nacional de Investigaciones Cardiovasculares Carlos III (CNIC), Madrid, Spain
t Centre for Integrative Neurosciences, University of Tübingen, Tübingen, Germany u Max Planck Institute for Biological Cybernetics, Tübingen, Germany v Nathan S. Kline Institute for Psychiatric Research, Orangeburg, New York, USA
w International Center for Primate Brain Research, Chinese Academy of Science, Shanghai, PRC x Focus Program Translational Neurosciences, University Medical Center, Mainz, Germany y Department of Neurobiology, University of Pittsburgh Brain Institute, Pittsburgh PA, USA
z Laboratory of Neuroinformatics, Nencki Institute of Experimental Biology of the Polish Academy of Sciences, 02-093 Warsaw, Poland aa Australian Research Council, Centre of Excellence for Integrative Brain Function, Monash University Node, Clayton, VIC 3800, Australia bb Biomedicine Discovery Institute and Department of Physiology, Monash University, Clayton, VIC 3800, Australia
cc Commissariat à l’Énergie Atomique et aux Énergies Alternatives, Direction de la Recherche Fondamentale, NeuroSpin Center, Gif-sur-Yvette, France dd Cognitive Neuroimaging Unit, Institut National de la Santé et de la Recherche Médicale U992, Gif-sur-Yvette, France
ee Université Paris-Saclay, France
ffIntegrative Neuroscience and Cognition Center, Centre National de la Recherche Scientifique (CNRS) UMR 8002, Paris, France gg Department of Vision & Cognition, Netherlands Institute for Neuroscience, Amsterdam, The Netherlands
a
r
t
i
c
l
e
i
n
f
o
Keywords: Open science Resource sharing Toolbox Pipeline Structural Functional Diffusiona
b
s
t
r
a
c
t
Neuroimagingnon-humanprimates(NHPs)isagrowing,yethighlyspecializedfieldofneuroscience.Resources thatwereprimarilydevelopedforhumanneuroimagingoftenneedtobesignificantlyadaptedforusewithNHPs orotheranimals,whichhasledtoanabundanceofcustom,in-housesolutions.Inrecentyears,theglobalNHP neuroimagingcommunityhasmadesignificanteffortstotransformthefieldtowardsmoreopenandcollaborative practices.HerewepresentthePRIMatEResourceExchange(PRIME-RE),anewcollaborativeonlineplatformfor NHPneuroimaging.PRIME-REisadynamiccommunity-drivenhubfortheexchangeofpracticalknowledge, specializedanalyticaltools,andopendatarepositories,specificallyrelatedtoNHPneuroimaging.PRIME-RE caterstobothresearchersanddeveloperswhoareeithernewtothefield,lookingtostayabreastofthelatest developments,orseekingtocollaborativelyadvancethefield.
https://doi.org/10.1016/j.neuroimage.2020.117519
Received31July2020;Receivedinrevisedform15October2020;Accepted24October2020 Availableonline20November2020
1. Introduction
Whennavigatinganunfamiliarcity,peopleorientthemselvesbased onafewlandmarks,useamapfordetailednavigation,andtakeadvice fromlocalsandpreviousvisitorsaboutwheretogoandwhichareas toavoid.Navigatingahighlyspecializedresearch field,suchas non-humanprimate(NHP)neuroimaging,isconceptuallyverysimilar.In or-dertoorient,bothaccesstobasicinformationandsupportfrom knowl-edgeablepeersareinvaluable.HerewepresentthePRIMatEResource Exchange(PRIME-RE, https://prime-re.github.io),anew community-drivenplatformthatpromotescommunicationamongpeersand facil-itatestheexchangeofspecializedknowledgeandresourcesrelatedto NHPneuroimaging.
Translationalneuroimaginginanimalsbridgesthegapbetween non-invasiveimagingstudiesinhumansandinvasiveneuralrecordingsthat areonlycarriedoutinanimalmodels(andoccasionallyinneurosurgical patients).Theapproachispivotalforourunderstandingoftheneuronal basisofneuroimagingsignals(Logothetis,2003;Logothetisetal.,2001;
LogothetisandWandell,2004),guidestheselectionofrecordingtargets forinvasivestudies,andrevealsthebroaderbrainnetworksinvolved inparticularcognitivefunctionsorbrainprocesses.Cross-species com-parisonofneuroimaging resultscan furthermoreprovideinsightinto theevolutionof thebrainanditscapabilities(Friedrichetal., 2020;
Heueretal.,2019).Finally,combiningneuroimaginginanimalswith invasivemethodsthatinfluenceneuronalactivitymakesitpossibleto drawcausalinferencesaboutbrainfunctionandtestpotentialhuman therapies(Klinketal.,2020).
TheNHPbrainissimilartothatofhumansinmanyrespects.Due totheevolutionaryproximityofNHPstohumans,theirabilityto per-formcomplexcognitivetasks,andthevastbodyofexistingneuronal knowledgefrominvasiveNHPstudies,NHPneuroimagingisacrucial branchoftranslationalimaging(Chenetal.,2012; Farivarand Van-duffel,2014; Logothetisetal.,1999).However,thestandardhardware andsoftware used in humanneuroimaging is generally not directly suitableforNHPneuroimaging,whichrequireshighlyspecialized ap-proaches.Asaresult,mostresearchgroupshavedevelopedtheirown in-housesolutionsthat oftenrequiresubstantialfurtherdevelopment forcompatibilitywithothersites.Thesecustomsolutionsoftenremain in-houseandlackextensivedocumentation,whichisfarfromidealfor peerreview,collaboration,bestpractices,andreproduciblescience.In recentyears,theinternationalNHPneuroimagingcommunityhasbegun embracingacultureofdata-sharing,openscience,andcollaboration. TheseeffortsledtothePRIMatEneuroimaging DataExchange initia-tive Milhametal.(2018),aglobalworkshoptodiscussstrategiesfor thefutureofthefield(Milhametal.,2020),andthisspecialissueof NeuroImageonNHPneuroimaging.
OnceNHPneuroimagingdataarecollectedorshared,anewsetof challengespresentsitself.Mostneuroimagingsoftwarepackageshave been developed for use in humans and cannot be easily applied to NHPdatadue tobuilt-inassumptions, suchasalarger field-of-view, standardizedvoxelsize,orahighercontrast betweengrayandwhite matter. Thelarge number ofchoices involvedin neuroimaging data analysis is a hotly-debatedissue in human neuroimaging (Botvinik– Nezeretal.,2020).ForNHPneuroimaging,theseissuesmaybeeven morepressing duetothecustom-solutions thatareoftenrequiredto make existing software compatiblewith NHP data. The fieldof hu-manneuroimaginghasstartedtoaddresstheseissueswithinitiatives for standardized,reproducible analysis pipelines (BIDS,Nipype, etc) (Gorgolewskietal.,2011,2016),datasharing(OpenfMRI,Zenodo, Neu-rovault,etc)(Gorgolewskietal.,2015;Poldracketal.,2013),and collab-orativeandopencoding(GitHub,GitLab,etc).NHPneuroimaginghas beenlaggingbehindinadoptingsuchcollaborativeopenscience
initia-∗Correspondingauthor.
E-mailaddress:C.Klink@nin.knaw.nl(P.C.Klink).
tives,butpeople’swillingnesstoshareandbeopenabouttheresource solutionstheydevelopedissteadilyincreasing(Balbastreetal.,2017;
Tasserieetal.,2020)andnewdevelopmentsininformationtechnology promotetheseinitiatives,whilecreatingavenuestoassignexplicitcredit todevelopers.TheglobalNHPneuroimagingconsortium(Milhametal., 2020,2018)aimstofacilitatethisprogresswiththeintroductionand cu-rationofthePRIMatEResourceExchange(PRIME-RE, https://prime-re.github.io),acommunity-drivenonlineplatformfortheexchangeof knowledgeandresourcesconcerningtheacquisition,analysis,and visu-alizationofNHPneuroimagingdata.
PRIME-REactsasanopenhubwhereresearchersfromallcountries andcareerstagescan findandcontributeinformationandresources related tothechallengesandadvantages of NHP neuroimaging. Dy-namiccollectionsofsoftwaretoolsaregroupedbyanalysis-categoryto provideanoverviewofexistingsolutionstocommonchallenges.Links tocommunicationoutletsallowresearcherstomeetinternational col-leagues,discusschallengesandsolutions,andstartnewcollaborations. Resourcesaremaintainedbytheirrespectivedevelopers,with PRIME-REservingasanevolvingaccesspointwithdescriptionsofthetools, their softwarerequirements,a listofauthors, relevantcitations, and linkstothetoolsthemselves.
2. Methods
ThisMethodssectionwillofferabriefdescriptionofthemain struc-tureofthePRIME-REplatform,includingitsmaincomponents,methods forpeopletocontributeandinteract,andpossibilitiesforfuturegrowth. ThelightweightPRIME-REwebsite(https://prime-re.github.io/)is in-tendedtobedynamicandcontinuouslyevolvebasedoninputfromthe researchcommunity.Itcanservebothasastartingpointforresearchers thatarenewtothefieldofNHPneuroimagingandasameetingground formoreexperiencedresearchersinthefield.Thisinitiativeemerged fromtheGlobalCollaborativeWorkshop2019ofthePRIME-DE con-sortiumandsubsequentBrainHack Milhametal.(2020)andhassince attractedacriticalmassofactivecontributorsdedicatedtoopenand col-laborativeNHPneuroimaging.TheplatformishostedonGitHub,which greatlyfacilitatescommunity-drivencontributionswhilealso maintain-ingacompleteversionhistoryofitscontentandevolution.
Content-wise,PRIME-REdoesnotaimtohost,curateandmaintain all available toolsand documentationregardingNHP neuroimaging. Instead,itchoosesanagileapproachwithstandardizedlistingsof re-sources,categorizedbythetypeofchallengetheyaddress.Theselistings includeabriefdescription,developercontactinformation,andalink tothedeveloper-maintainedresource.Thismodelfurthermoreensures thatthemostrecentversionofaresourcewillbefoundatitssourceand thatdevelopersreceivecreditfortheirefforts.Thestandardizedlistings alsomakeiteasyfordeveloperstosubmittheirresourceforinclusionon PRIME-REthroughapre-formattedsubmissionforminwhichthey an-swerafewbasicquestionsabouttheirresource(Table1).Theseanswers formthebasisofaresource’slistingonPRIME-RE.Ifthedevelopersare alreadyhostingtheirresourceonline,asubmissiontoPRIME-REcanbe completedinafewminutes.
Submissions to PRIME-RE are approved for inclusion through community-driven evaluation.Once asubmission form is submitted, a member of the community with access to PRIME-RE’s backend can opt toinclude it on the website.It is up tothe community to evaluate a resource’s usefulness. A simple community support rat-ing system has been implemented toallow anyone to express sup-port for a resource by ‘liking’ it. This system could help the users of PRIME-RE to navigate the collection of resources and find tools thathaveaccumulatedalotofcommunitysupport.Ifconcernsarise, they can be discussed through any of the offered communication channels.Directcommunitydiscussionsareavailablethrougha
ded-Table1
StandardizedinformationtableforthesubmissionofaresourceforinclusiononPRIME-RE.Whendevelopersclicka‘Contribute’buttonthey aretakentoapre-formattedformwheretheycanprovidethisinformation.Anymemberofthecommunitywithaccesstothewebsite’sbackend canthenincludethenewresourceunderthecorrectwebsitecategory.Inadditiontothistable,thesubmissionformalsohascheckboxeswith whichthesubmittercanconfirmthatresourcedevelopmentoccurredinaccordancewithallapplicabledirectivesandguidelines.
Name Name of the resource
Authors Names of the developers or authors
Description A brief description of the resource. What challenge was it developed for? What does it do? Documentation Provide a link to more extensive (external) documentation, e.g. on the developers’ website. Link Where can the resource be found or downloaded?
Language What kind of programming languages are used and/or required to use this tool? Publication If there is a publication associated with this resource you can mention it here. Communication How can (potential) users contact developers?
Restrictions Are there any restrictions for using this resource? Does it only work on certain systems? Are users obliged to cite something? Category What would be the most suitable place on PRIME-RE to list this resource?
icated “prime-re” chat-channel on the Brainhack Mattermost server (https://mattermost.brainhack.org/brainhack/channels/prime-re),and moreconventionalmailing-listorforumstylediscussionisavailable us-ingtheNeurostarsforum(https://neurostars.org)withthetag "prime-re".TheNeurostarsforumis hostedbytheInternational Neuroinfor-maticsCoordinatingFacility(INCF)andalreadyhasabroaduser-base ofneuroscienceresearchers.Itisalsopossibletosubmitacontactform withcomments,questions,orsuggestionsdirectlytoPRIME-RE’s back-end, and often also to individual developers of a resource through theirprovidedcommunicationinformation.Tofacilitatecollaboration amongitsusers,PRIME-REalsopartnerswiththeBrainWebinitiative (https://brain-web.github.io).
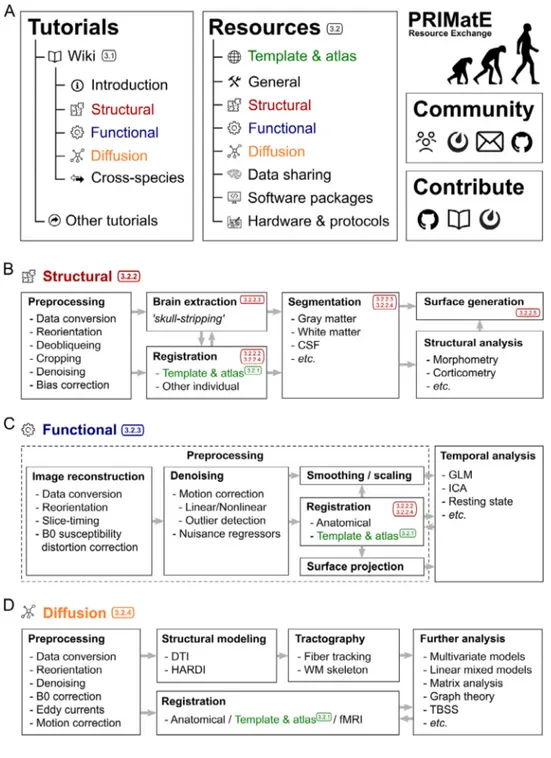
Atthemoment,PRIME-REhastwomaincomponents(Fig.1A),1) theprimarywebsitewithcategorizedresourcelistings,and2)an evolv-ingwikitodocumentthechallengesandbestpracticesofNHP imag-ing.TheresourcecategoriesthatarecurrentlylistedonthePRIME-RE websiteare:1)Templatesandatlases;2)Generalanalysis;3)Structural analysis;4)Functionalanalysis;5)Diffusionanalysis;6)Datasharing; and7)Softwarepackages.Theresultssectionofthispaperwillpresent anoverviewofsomeofthecustomtoolslistedunderthesecategories atthetimeofwriting.TheSoftwarePackagessectioncontainsalistof commonlyusedneuroimagingpackagesthatwerenotspecifically de-velopedforNHPneuroimaging.Manyofthespecializedtoolsintegrate withsomeofthesepackagesoremploysomeoftheirsub-functions.The mainwebsitealsoprovideslinkstotheabove-mentioned communica-tionchannelsandtothewiki.ThewikicontainsaprimeronNHP neu-roimagingthatdocumentscommonchallengesandpotentialsolutions. Ithasitsownversioncontrolledback-endandiscurrentlyopenfor con-tributionsforanyone withaGitHubaccount.Whilethecurrent wiki alreadyconstitutesateameffort,thedocumentispresentedasa start-ingpointthatcouldpotentiallyevolveintoa‘bestpractices’guidewith dynamicinvolvementofthebroaderNHPneuroimagingcommunity.
MostofthetoolsthatarecurrentlylistedonPRIME-REwere devel-opedformacaqueneuroimaging,buttheplatformaimstobean inclu-siveplacewhereresources forallnon-humanprimatespeciescanbe found.Severaltoolsformarmosetneuroimagingarealreadylisted,but thespeciesdiversityislikelytofurtherincreaseasPRIME-REevolves.
3. Results
ThefirstversionofthePRIME-REplatformwentliveinNovember 2019.Itcurrentlydescribesover30customresourcesfromcontributors allovertheworldandhousesanextensiveprimeronNHPneuroimaging intheformofawikiwithreferencestotheresourceslistedon PRIME-RE.Italsoprovidesacomprehensiveoverviewofneuroimagingtools thatarenotnecessarilytailoredtowardsNHPneuroimaging(although someofthesepackagesdoofferbuilt-insolutionstofacilitateworking withnon-humandata).Here,wewillpresentasynopsisofthewiki docu-ment(https://github.com/PRIME-RE/prime-re.github.io/wiki)andan
overviewofsomeofthespecializedresourcesthatarepartof PRIME-REatthistime.
3.1. Thewiki:aprimeronNHP-MRI
Thewiki-pagesstartwithsectionsthataddressthevarious motiva-tionsforNHPneuroimagingandsomeofthecommonchallenges en-counteredindatacollection(FarivarandVanduffel,2014).These sec-tions includestudy preparationissues,such as obtainingethical ap-provalforyourstudy,responsibleanimalhousing,handling,surgery, andtransportation.Thechoiceofcoiltypeis discussedandthe con-sequencesofusingcustomcoilsareclearlyexplained.Otherdiscussed challengesindataacquisitionincludenon-standardbodyorientations, subjectmotion,theuseofanesthesiaorcontrastagents,variable fields-of-view,andareducedsignal-to-noise-ratio(SNR)asaresultofhaving toscansmallNHPbrainsathigherresolutionthanhumanbrainsatthe samemagneticfieldstrength.
The nextsectionbrieflyaddresses thetopicof dataorganization. StandardizationeffortsinhumanneuroimaginghaveyieldedtheBrain ImagingDataStructure(BIDS)standard,awidelyadoptedfile-naming anddata-structureconventionthatfacilitatesdataandresourcesharing. WhereasarangeoftoolsonPRIME-REunderstanddatainBIDSformat, thecompatibilityofNHP datawiththeBIDSstructureisnotperfect, againduetosomeuniquechallengesinNHPneuroimagingthatarenot presentinhumanneuroimaging.Thereare,however,ongoingeffortsto eitherexpandtheBIDSformatsothatitcanbetterincorporatethe id-iosyncrasiesofNHPdata,orcreateaderivativeNHPversionthatfulfills specificrequirementstothisresearchfield.
Theremainderofthewikilargelyfollowsthecategoriesthatarealso used inPRIME-RE’s resourcelistings.Thereareseparatesectionsfor structural,functional,anddiffusiondataprocessing(Fig.1A).The sec-tiononstructuraldataprocessingincludesdataprocessingsteps,such asorientationcorrection,de-obliqueing,cropping,denoisingand aver-agingmultiplevolumesorimages.Itlistsseveraloptionstohandle bias-correction,brainextraction(skull-stripping),andsegmentation.Italso linkstoseveralpipelinesavailablefromPRIME-REthatoffermoreor lesscompletesolutionstoarangeoftheseissues.
Thesectiononfunctionaldataprocessingpointsoutthatthisstepis infactnotthatdifferentfromhumanneuroimagingsincethesametypes ofstatisticalmodelscanbeappliedtodifferentspecies.Onemajor differ-encethatdoesexististheextenttowhichdataneedtobepre-processed. NHPdata,especiallydatafromawakeNHPneuroimagingstudies,tend torequirealotmoremotionanddistortioncorrectionthanthetypical humandataset.Thesectionondiffusion-weightedimagingdiscussesthe differencesbetweeninvivoandpost-mortem(exvivo)diffusionMRIand theirconsequencesfordataanalysis.Severalcaveatsandsolutionsfor tractographyinNHPsarealsolaidout.
The wikiends withasection on cross-speciescomparison.While challenging,this is alsoone ofthemost powerful uses ofNHP neu-roimaging sinceit hasthe potentialto yieldimportant translational
Fig. 1. Organization of PRIME-RE and commonNHP-neuroimaginganalysis work-flows.A)Themaincomponentsofthe PRIME-REwebsite(https://prime-re.github.io)arethe TutorialsandResourcessections.TheTutorials sectionhasacommunity-drivenWiki explain-ingthebasicsofNHPneuroimagingaswellas linkstoexternaltutorials.TheResources sec-tionprovidesdescriptionsofthevarioustools, withlinks tothe relevant files, documenta-tion,andmanuscripts.Boththewikiandthe Resourcessection areorganizedthematically (e.g.,structural,functional,diffusion),shown hereindifferentcolors.Thewebsitealso in-cludesaCommunitysection,whereNHP neu-roimagingresearchersanddeveloperscan con-nectandfindrelevantliterature,anda Con-tributesectionthatexplainshowpeoplecanget involvedin the PRIME-REinitiativethrough GitHub,theWiki,ortheMattermostchannel.
B-D) Exampleworkflows for theanalysis of structural(B),functional(C),anddiffusion(D) NHPneuroimaging data,indicatingcommon analysissteps.Thecolored numbersrefer to section numbers in this paper that describe toolsandpipelinesdevelopedtoaddressthese analysissteps.Notethatthereisnota standard-izedworkflowforthesetypesofanalysesand thatindividualresearchersmayincludeoromit certainstepsandproceedinanorderthatis fit-tingfortheirownneedsandcircumstances.
orevolutionaryinsights.Thesection explainstherecentlydeveloped commonfeaturespaceapproach(Marsetal.,2018a,2018b),aswell ascomparisonsbasedonactivitydynamics(Mantinietal.,2012),and brainmatchingbasedonhomologoussulci(Auziasetal.,2011). Sev-eralresources,suitablefortheprocessingofNHPdata,aresuggestedto implementtheseapproaches.
ThePRIME-REwikiisnotmeantasadefinitiveguidetoNHP neu-roimaging,butratherasacommunitycuratedanddynamicallyupdated collectionof‘best’(or‘optimal’)practices.Improvementsandadditions fromthecommunityarehighlyencouraged.
3.2. PRIME-REresources
TheresourcesthatarecurrentlylistedonPRIME-REaredividedinto anumberofcategoriesbasedontheirgeneralpurpose(Fig.1).Belowwe highlightthesecategoriesandaselectionoftoolsfromeach.Please con-sultthewebsiteforacompleteandup-to-dateoverviewoftheavailable
resources(https://prime-re.github.io/resources).Allresourceslistedon PRIME-RE,includingthosedescribedhere,weredevelopedandassessed withdatasetscollectedinaccordancewithlocallyapprovedguidesand legislationconcerninganimalwellbeing,includingtheNIHGuidefor CareandUseofLaboratoryAnimals,theU.K.Animals(Scientific Proce-dures)Act,1986,EuropeanDirective2010/63/EU,andtheAustralian CodefortheCareandUseofAnimalsforScientificPurposes.
3.2.1. Templates&atlases
Alignment of an individual’s(f)MRI data toa standard template spaceprovidesseveraladvantages.Firstly,itprovidesadetailed anatom-ical image for evaluating and presenting functional results or other maps.Secondly,itpresentsastandardorientationandcoordinate sys-temforreportingfindings,comparingresultsacrosssubjectsandlabs, andaligningdataforgroupanalysis.Thirdly,templatesfrequently in-cludeancillarydatasetssuchasbrainmasks,tissuesegmentationmasks, anatomicalatlases,surfaces,morphometry,anddatafromother
modal-itiesthatputavarietyof analysistools attheuser’sdisposal.These resourcescaneliminatetime-consumingmanualprocessingand encour-agetheuseofconsistentandreproducibleprocessingstreams.For ex-ample,abrainmaskcanbeusedfortemplate-basedbrainextractionin thesubject’sspaceoranatlascanbeusedforaregionofinterest(ROI) analysisinthetemplateorsubjectspace.
Thechoiceandadoptionofastandardtemplatespaceisbecoming especiallyrelevantnowthatdatasharinginitiativesmakeitmuchmore feasibletoobtainlargersamplesizes.Forhuman(f)MRI,theMNI152 template(Fonovetal.,2011)providesawidelyadoptedstandard vol-umetricspace,whilethefsaverage(Freesurferaverage)(Fischl,2012) templateisapopularstandardspaceforsurface-basedanalysis.For non-humanprimate(NHP)MRI,thereareavarietyoftemplates.PRIME-RE currentlylistsseveralmacaqueandmarmosettemplates, aswellasa mouselemurtemplate.Inadditiontospecies,templatesdifferwith re-gardtoimagingmodality,invivoversusex vivoscanning,single sub-jectversuspopulationaverage,fieldstrength,resolution,etc.Forthe rhesusmacaque,theNIMHMacaqueTemplate(NMT)hasbeenwidely adoptedandhasbeen incorporatedinanumberof theresources on PRIME-RE(Seidlitzetal.,2018).Anupdatedversionofthisinvivo popu-lationtemplate(NMTv2)ispresentedinthisspecialissueanddescribed below(Jungetal.,2020).TheNMTv2anditsassociatedaverage sur-face(seeSection3.2.2.5.1),togetherwiththeirtissuesegmentationsand atlases,constituteacompletesetofcomplementaryvolumetricand sur-facespacesforrepresentingmacaquedata.
Twonovelmacaqueatlasesarepresentedinthisspecialissueand detailedbelow.BotharedefinedontheNMTv2andmanuallyrefined tothistemplate.Thefirstisanatlasofthecortex(CHARM,Jungetal., 2020)andthesecondisanatlasofthesubcortex,coveringtheforebrain, midbrain,andhindbrain(Hartigetal., 2020).These complementary atlasesarearrangedhierarchically,describingregionsatvarious lev-elsofgranularity.ThoughtheseatlaseshavebeentailoredtotheNMT v2,theycanbealignedtopreviousmacaquetemplatesalreadyusedby somelabs,andatlasesinthosetemplatescansimilarlybealignedtothe NMTv2.TheRheMAPresource,whichisdescribedinthesubsequent sectiononStructuralMRITools(see Section3.2.2.2.2),stores nonlin-earalignmentwarpsbetweenvariousmacaqueanatomicaltemplatesto facilitatesuchconversionsbetweentemplatespaces(Klinkand Sirmpi-latze,2020),SirmpilatzeandKlink(2020).
TwomarmosetresourcesarealsoincludedinPRIME-RE.The Mar-mosetBrainMappingProjectisamultimodalhigh-resolutiontemplate andatlasofthemarmosetbrainthatincludeswhitemattermapsbased ondiffusionMRIdata.Themostrecentversionisapopulationtemplate basedoninvivoscansthatincludessurfaces(Liuetal.,2020).In ad-dition,theMarmosetBrainConnectivityAtlas(Majkaetal.,thisissue) providesalargecollectionofanatomicaltracerdataandhistological ma-terialinaninteractiveplatform,equippedwithvariousanalysistools.
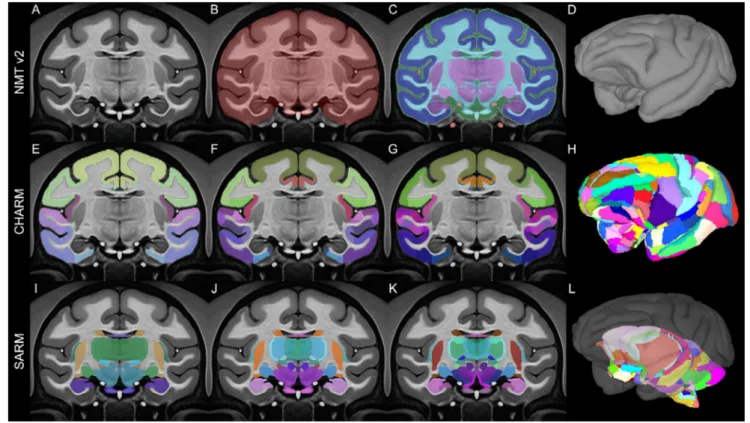
3.2.1.1. NIMHmacaquetemplate(NMTv2)andhierarchicalatlasofthe cortex(CHARM). Amacaquetemplateinstereotaxiccoordinateswith a multi-scalecorticalatlas.
The National Institute of Mental Health (NIMH) Macaque Tem-plate (NMT) is an anatomical MRI template of the macaque brain thatservesasastandardizedspaceformacaqueneuroimaginganalysis (Seidlitzetal.,2018).TherecentlyreleasedNMTversion2(Jungetal., 2020)provides a complete overhaul of the NMT,including a fully-symmetrictemplateinstereotaxicorientation(i.e.,intheHorsley-Clarke plane; HorsleyandClarke,1908).Coordinatesinthisstereotaxicspace aremeasuredfromearbarzero(i.e.,theintersectionofthemidsagittal planeandalinethroughtheinterauralmeatus).Theadoptionof stereo-taxicorientationandcoordinateswillassistuserswithsurgicalplanning andfacilitatesreportingofcoordinatescommensuratewiththoseused withothertechniques(e.g.electrophysiology,intracerebralinjection).
TheNMTwascreatedbyiterativelyregisteringT1-weightedscans of31 adultrhesusmacaquebrainstoaworkingtemplate,averaging thenonlinearlyregistered scans,andthenapplyingtheinverse
trans-formationstobringtheworkingtemplateclosertothegroupaverage (Seidlitzetal.,2018).ThesymmetricNMTwasgeneratedthroughthe same processexceptthat eachsubject’s anatomicalwasinputtwice, onceinitstrueorientationandoncemirroredaboutthemidline. Modi-ficationstothescanaveragingandpostprocessinghaveimproved tem-platecontrastandspatialresolution.Brainmasks,segmentations,and variousothertissue masksareprovided(Fig.2A-C). Theavailability ofsymmetricandasymmetricvariantsoftheNMTv2allowsusersto choosetheversionthatbestsuitstheiranalysis.Othertemplate vari-antsincludeanexpandedfullheadfield-of-viewandalower resolu-tionversionforfasterprocessing(0.5mmisotropic,down-sampledfrom 0.25 mm isotropic). Surfacesfor NMT v2, generated using thenew CIVET-Macaqueplatform(Lepageetal.,2020),areprovidedforeasy datavisualization.Forsurface-basedgroupanalysis,theNMTaverage surface,whichcomeswithanatomicallabelsfromtheD99andCHARM atlases,canserveasasurface-basedregistrationtargetforrepresenting corticalsurfacesinacommonframeworkregardlessoftheprocessing pipeline.
AtlasesareanimportantaspectofgroupandROI-basedanalyses.The NMTv2packagecomeswithmultipleanatomicalatlasesthathavebeen manuallytailoredtothetemplate’smorphology.Theseatlasesinclude theD99atlas(Reveleyetal.,2017) andthenewCorticalHierarchy AtlasoftheRhesusMacaque(CHARM;Jungetal.,2020).Thelatterisa novelanatomicalparcellationofthemacaquecerebralcortex,wherethe corticalsheetissubdividedintosix-levelsofincreasinglyfine-grained parcellation(Fig.2D-F).Thebroadestlevelconsistsofthefourcortical lobesandthefinestlevelisbasedontheD99atlas,withmodifications thatmaketheregionsmorerobustwhenappliedtolowresolution(e.g. fMRI)data.Differentscalesof CHARMcan becombinedso that,for example,atracerinjectionortheseedregionforarestingstateanalysis can bedescribed usingafinescale,while theregion’s anatomicalor functionalconnectivitycanbesuccinctlydescribedonabroaderscale. Thisway,wholebraindatacanbecharacterizedonaspatialscalethat ismost suitabletoastudy’sfindings.UserscanalsoselectaCHARM levelaprioribasedonhowmanyregionsitcontains,andthuscontrol therequireddegreeofmultiplecomparisoncorrectionfortheiranalysis. While the NMT v2 works with any neuroimaging platform that accepts NifTI/GifTIformat, thetemplate has been designedtohave additional functionalitywithintheAFNI ecosystem(Cox, 1996). En-hancementsincludesupportfortheNMT2standardizedspace, recog-nitionoftheNMTv2inthe@animal_warperalignmentpipeline(see
Section 3.2.2.4.2) and the functional processing pipeline generator afni_proc.py (see Section3.2.3.1),anddownloadable demosshowing howAFNIcanperformstructuralandfunctionalanalyses(task-basedor restingstate)usingtheNMTv2.SeetheaccompanyingarticlebyJung etal.(Jungetal.,2020)forfurtherinformation.
3.2.1.2. Subcorticalatlasoftherhesusmacaque(SARM). AcompleteMRI atlasofthemacaquesubcortexsuitedforneuroimaging.
TheSubcorticalAtlasoftheRhesusMacaque(SARM)isan anatomi-calparcellationoftheentiresubcortextailoredformagneticresonance imaging(MRI)(Hartigetal.,2020).Theregions-of-interest(ROIs)are hierarchicallyorganized,withgroupinglevelssuitedforbothfine struc-turalandspatiallybroaderfunctionalanalyses.SARMaimstofacilitate theidentification,localization,andstudyofneuralinteractions involv-ingsubcorticalregionsofthebrain.
Ahigh-resolutionstructuralMRIofanexvivorhesusmonkeybrain wassegmentedinto180anatomicalROIsbymatchingdistinctly con-trastedstructureswiththeirhistologicalcounterparts.ROIsfollowedthe recentlyupdatednomenclatureanddelineationsoftheRhesusMonkey BraininStereotaxicCoordinatesatlas(Paxinosetal.,2020).Theexvivo
MRIwasnonlinearlyregisteredtothesymmetricpopulation-level tem-plateoftherhesusmacaqueNMTv2(seeSection3.2.1.1andJungetal., 2020). TheSARM was warped using AdvancedNormalization Tools (ANTs; Avantsetal.,2008)andrefinedusingAFNI(Cox,1996).ROIs weredelimitedusingthetissuesegmentationmasksfortheNMTv2,
Fig.2. MacaqueTemplatesandAtlases.ThesymmetricvariantoftheNIMHMacaqueTemplatev2(NMTv2),withtissuesegmentation(toprow)andanatomical regions(middleandbottomrows)depictedincoloronacoronalslice(11mmanteriortoearbarzero)andthepialsurface(rightcolumn).TheNMTv2includesA) thepopulationaveragevolume;B)amanuallyrefinedbrainmaskthatindicateswhichvoxelscontainbraintissue;C)a5-classtissuesegmentationthatdifferentiates cerebrospinalfluid(green),graymatter(darkblue),whitematter(lightblue),subcortex(purple),andvasculature(red);andD)renderingofsurfaces.Themiddle rowshowstheCorticalHierarchyAtlasoftheRhesusMacaque(CHARM)andthebottomrowshowstheSubcorticalAtlasoftheRhesusMacaque(SARM).Theatlases provideanatomicallabelingofcorticalandsubcorticalregions,respectively,atsixprogressivelyfinerspatialscales.ParcellationsforE,I)Level2,F,J)Level4,and
G,K)Level6areshown.H,L)Surfacerepresentationofthefinest(Level6)parcellations.
smoothedbycomputingthemodeoverneighboringvoxels,and indi-viduallyrefinedbyhandtomatchtheaveragestructuraltemplate.The SARMisorganizedinto6hierarchicallevels.Thefinestlevelpresents eachindividualROI(Fig.2G),whereasthebroadestlevelcorresponds to developmental subdivisions (i.e., tel-, di-,mes-, met-, and myel-encephalon).TheSARMwasvalidatedin3monkeyswithafunctional MRI(fMRI)paradigmknowntoactivatethelateralgeniculatenucleus (LGN)(Logothetisetal.,1999).
TheSARMoffersneuroimagingresearchersacompletesubcortical segmentationof therhesusmacaquebrain.Theatlascan beusedfor arangeofstructuralandfunctionalanalysesandhasbeenspecifically tailoredtoworkwiththeCHARMandNMTv2(Jungetal.,2020).The SARMisalsoavailabletodownloadviaZenodo,andhasconveniently beenincorporatedwiththeatlasesavailableinAFNI.Additional infor-mationisavailableintheaccompanyingarticle(Hartigetal.,2020).
3.2.1.3. Marmosetbrainmapping project(MBM). MRI-basedmarmoset brainatlasesandtoolsforneuroimagingandconnectomestudies
The Marmoset Brain Mapping Project (www.marmosetbrainmapping.org) includes three atlases and tem-plates:
1) Version1focusesoncorticalparcellations(Liuetal.,2018).A3D digitalgraymatteratlaswasconstructedfromhigh-resolution(150
𝜇misotropic) exvivo MRIimages,includingmagnetization trans-ferratio(aT1-likecontrast),T2wimages,andmulti-shelldiffusion MRI(dMRI).Basedonthemulti-modalMRIcontrasts,54cortical areasand16subcorticalregionsweremanuallydelineatedonone hemisphereofthebrainusingadata-drivenapproachthatwas devel-opedtominimizemanualdrawingerrors.The54corticalareaswere
thenmergedinto13largercorticalregionsaccordingtotheir loca-tionstoyieldacoarseversionoftheatlas,andalsosubdividedinto 106regionsusingadMRIconnectivity-basedparcellationmethod.A Paxinos-stylecorticalatlas(Majkaetal.,2016)wasfusedandrefined intothehigh-resolutionMRItemplatetoprovideinteroperabilityto othermarmosetdatabases.Theatlassetprovidesareadilyusable multi-modaltemplatespacewithmulti-levelanatomicallabelsthat canfacilitatevariousneuroimagingstudiesofmarmosets.
2) Version2focusesonfine-detailedwhitematterpathways(Liuetal., 2020).Inthisversion,ex-vivoMRIdataofthemarmosetbrainwas collectedwiththehighestspatialresolution availabletodate, in-cluding80𝜇mand64𝜇mmulti-shelldMRI,80𝜇mMTR,and50𝜇m
T2wandT2∗wimages.Thedataallowedbuildingafine-grained3D
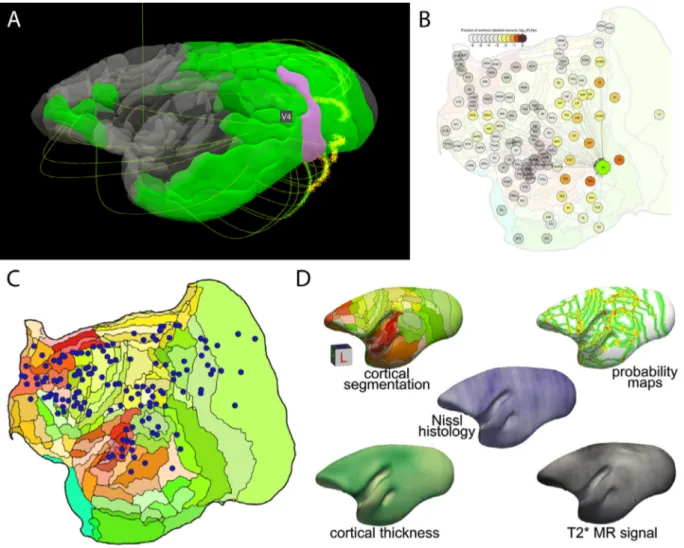
whitematteratlas,whichdepictsmany fiberpathwaysthat were eitheromittedorincorrectlydescribedinpreviousMRIdatasetsor atlasesoftheprimatebrain.BycombiningdMRItractographyand neuronaltracing data(Majkaetal., 2020),adetailedfiber path-waymappingofcorticalconnectionsisprovided. Thewhite mat-teratlas,fiberpathwaysmapsanddMRIdata,includingbothraw andprocesseddata,arepubliclyavailableontheproject’swebsite (Fig.3A,B).
3) Version3focusesonpopulationstandardtemplatesandcortical sur-faces(Liuetal.,2020).While versions1and2werebasedona fewexvivobrainsamples,andlackedessentialfunctionalitiesforin vivostudiesoflarge animalcohorts,version3isbasedon invivo
populationdata.Standardtemplatesarederivedfrommulti-modal dataof 27 marmosets,includingmultiple typesof T1wandT2w contrastimages,DTIcontrasts,andlargefield-of-viewMRIandCT images.Multi-atlaslabelingofanatomicalstructureswasperformed onthenewtemplatesandhighlyaccuratetissue-typesegmentation
Fig.3. Marmosetatlases.A-B)ExamplevisualizationsavailableonthewebsiteoftheMarmosetBrainMappingProject.A)3D-viewerthatdemonstratesthe connectivityprofileofanareaselectedbyclicking(hereV4).B)Comparisonwiththeweightedanddirectedconnectivitygraphbasedontheresultsofmonosynaptic retrogradefluorescenttracerinjectionsfromtheMarmosetBrainConnectivityAtlas(https://analytics.marmosetbrain.org/graph.html).Thegraphviewhighlights thespatialrelationsbetweenconnectedareas.Clickinganoderevealsfurtherinformation,includinganaverageconnectivityprofile,interactivevisualizationsof thedataforeachinjection,andmetadata.C-D)MarmosetConnectivityAtlas.C)Locationsof143tracerinjectionsregisteredtoastereotaxictemplateillustratedin a2-dimensionalflat-mapofthemarmosetcortex(C).D)Themaindatalayersofthehistology-basedaveragemorphologyoftheadultmarmosetbrainshowcasing theinteroperabilityofthisdatasetwithneuroimagingresearch(T2∗MRsignal).
mapswereconstructedtofacilitate volumetric studies.Fully fea-turedbrainsurfacesandcorticalflatmapsfacilitate 3D visualiza-tionandsurface-basedanalyseswithmostsurfaceanalyzingtools. Thepopulation-basedtemplatewillsignificantlyaidawiderange ofneuroimagingandconnectomestudiesthatinvolveacross-subject analysis.
3.2.1.4. Marmosetbrainconnectivityatlas. Marmosetanatomical tracer dataandconnectivityanalysistoolsintegratedwithanatlasandhistological material.
TheMarmosetBrainConnectivityAtlas(Majkaetal.,2020)allows explorationofagrowingcollectionofdatafromretrogradetracer injec-tionsinthemarmosetneocortex.Atpresent,itincludesdatafromover 140 experiments coveringalmost50% of currentlyrecognized areas ofmarmoset cortex,encompassingsubdivisionsof prefrontal, premo-tor,superiortemporal,parietal,retrosplenialandoccipitalcomplexes. Datafromdifferentanimalsarepresentedagainsthigh-resolution im-agesoftheunderlyinghistology,andregisteredtoatemplatebasedon thePaxinosetal.(Paxinosetal.,2012) stereotaxicatlasusingan al-gorithmguidedbyanexpert delineationof histologicalborders.The portal(http://www.marmosetbrain.org) implementsbest practicesin termsofsharing theconnectivitydata. Theresourceprovidesaccess
toprimaryexperimentalresults,aswellasconnectivitypatternsthat arequantifiedaccordingtothecurrentlyacceptedparcellationof the marmosetcortex.Toenablegraph-basednetworkanalyses,theportal incorporatestoolsfordataexplorationrelativetocytoarchitectural ar-eas(http://analysis.marmosetbrain.org),includingstatisticalproperties suchasthefractionoflabeledneurons,thepercentageofsupragranular neurons,andgeodesicdistancesbetweenareasacrossthewhitematter. Importantly,italsoprovidesthecellularconnectivitydatainapurely spatial(parcellation-free)format,includingthestereotaxiccoordinates of ~2millionneuronslabeledby thetracerinjections.These results can bedownloadedin3Dvolumeformatin thetemplate space (Ma-jkaetal.,thisissue),sotheycanbecomparedwithMRI-based topolo-gies.Theportalalsomakesdataavailableinamachine-mineableway (http://analytics.marmosetbrain.org/wiki/api) tofacilitate large-scale modelsandsimulations.
Apart from the tracer-based connectivity data (Fig. 3C), the portal hosts a collection of other unique datasets typically inac-cessible in non-invasive imaging, such as high-resolution images of cytoarchitectural characteristics for each of the marmoset corti-cal areas, stained with a neuron-specific antibody (Atapour et al., 2019) (http://www.marmosetbrain.org/cell_density). Moreover,
a unique histology-based average morphology of the adult mar-moset brain provides the basis for probabilistic registration of digital datasets to cytoarchitectural areas (Majka et al. this issue;
http://www.marmosetbrain.org/nencki_monash_template). Spatial transformations to other marmoset brain templates are included to enablepromptintegrationwithmagneticresonanceimaging(MRI)and tracer-basedconnectivitystudies(Fig.3D).
3.2.2. StructuralMRItools
3.2.2.1. Overview. Pre-processingofstructuralandfunctionalMRI in-volvesalignmentofvolumetricdataacross scanningsessionsorwith aspecies-specific anatomicaltemplate/atlasin astandardizedspace; segmentation(i.e.,differentiation)ofthebrainfromskull,muscle,and othertissues(a.k.a.“skull-stripping”);segmentationofdifferenttissue classeswithinthebrain(e.g.grayandwhitematter).Additionally, vol-umetriccorticaldataaresometimesrepresentedonaflatmaporsurface mapbycreatingindividualizedsurfacesorprojectingdatatoacommon surface.HerewebrieflydescribesomePRIME-REresourcesforhandling volumetricdata(Fig.1B).Typically,anatomicalscansareanalyzedin detailandtheresultsarethenalsoappliedtoanyfunctionaldata col-lectedinthesamescanningsession.
Some of the structural tools described perform a single pre-processingstep,allowingthemtobeflexiblyusedwithothertools. Ex-amplesofdedicatedalignmenttoolsareReorient,whichdoesrigidbody alignmentandcroppingthebordersofascan,andRheMAP,which pro-videsthenon-linearwarpsbetweenvariousmacaquetemplates. Ded-icatedsegmentationtools includetheinteractiveThresholdmannand themachine-learningbasedUNet,bothofwhichgeneratebinarybrain masks,andBrainBox,whichiswellsuitedforthecollaborative devel-opmentofbinaryormulti-valuedmasks.
Otherresourcesperformmultiplestepswithinasinglesoftware en-vironmentorbycallingonvariousothertools.Forexample,Macapype usesseveraltoolsinterchangeablytoperformalignment,normalization, andsegmentationoperations.Similarly,AFNI’s@animal_warper com-putesandappliesanonlinearalignmentbetweenasubjectanda tem-plateandcanwarpthetemplate’smaskstocreateabrainmask, tis-suesegmentation,andatlasparcellationfortheindividual.Forother tools,alignmentandsegmentationareprecursors toadditionalsteps suchassurfacegeneration(e.g.,CIVET-Macaque,PREEMACS)or func-tionalanalysis(e.g.,C-PAC).
PRIME-REoffersfourtoolsforgeneratingsurfacesfromvolumetric monkeydata.TheNHP-Freesurfer,PREEMACS,andPrecon_alltoolsall utilizetheFreesurfersoftwarepackageinconjunctionwithseveralother tools.Incontrast,CIVET-macaqueisanoveladaptationofCIVETforuse withmonkeydata.Itcanbeusedtocharacterizesurfacemorphology suchascorticalthickness(Lepageetal.,2020).Allfourprogramsrequire aT1-weightedanatomicalscan.BothCIVET-macaqueandPREEMACS cantakeanoptionalT2-weightedscanaswell,andbothprovidequality controltools.
3.2.2.2. Alignmentandregistrationtools. HavingproperlyorientedMRI datasetsisimportantformanyneuroimagingworkflowsandoften cru-cialforfurthersegmentationorregistrationacrossfunctional, anatomi-calandtemplateimages.Fornon-humanscans(bothinvivoandexvivo), orientations andfield-of-viewscan vary tremendously, which means thatacorrectionthroughreorientationandcroppingisoftenrequired beforeanystandardtoolscan beused.Groupanalysisof(f)MRIdata iscommonlyperformedinastandardtemplatespacetowhich individ-ualdataareregistered.Herewedescribetoolsforaligningdatasetstoa templateortoeachotheraswellasprecalculatedregistrationsbetween templates(RheMAP).ThesetoolsincludeReorient,whichaidsinvisual rigidbodyalignment,andnonlinearalignmenttoolssuchasmacapype and@animal_warper(see Section3.2.2.4).
3.2.2.2.1. Reorient. Anintuitivewebtoolforreorientingandcropping MRIdata.
Reorient (https://neuroanatomy.github.io/reorient) is an open sourcewebapplicationfortheintuitivemanualalignmentandcropping ofMRINifTIvolumes.TheMRIscanisdraggedontoawebinterfaceand visualizedinaninteractivestereotaxicviewer.Userscanthentranslate androtatethebrainbysimplydragginginsidethethreeviewplanes. Anadjustableselectionboxdeterminesthecroppingofanimage. Re-sultingaffinematrices,selectionboxes,aswellasthereorientedand croppedvolumecanbesaved.Theaffinematrixandselectionboxcan thenbeusedinscriptedworkflowstomakethesestepsreproducible. Existingrotationmatricescanalsobeloadedorappendedintheweb interface.Reorientcomplementsexistingtoolsbyprovidinganintuitive approachformanualimagereorientationandallcomponentsforafully reproducibleworkflow.Ithasbeenusedextensivelytoreorientscans frommanydifferentvertebratespecies,including60differentprimate species.
3.2.2.2.2. RheMAP. Precomputednonlinear warpsbetweencommon rhesusmacaquebraintemplates.
Thegenerationofaccuratenonlinearregistrationsbetweendifferent brainimagesisatime-consumingandcomputationallyexpensive opera-tion.TheRheMAPprojectwascreatedtogenerateasetofpre-computed nonlinearregistrationwarpsthatallowthedirectremappingof(f)MRI dataacrossdifferentcommonmacaquetemplatebrains.Thenon-linear warpsweregeneratedusingANTs(Avantsetal.,2008)andthePython codethatwasusedinRheMAPtocomputethewarpsisavailableon GitHub(https://github.com/PRIME-RE/RheMAP)togetherwith docu-mentation andexample codethatexplains how tousethe warpsto remap data between template spacesor registerasingle anatomical scantooneormoretemplatespaces(KlinkandSirmpilatze,2020).The datasetofcomputedwarpsisfreelyavailablefordownloadfrom Zen-odo(SirmpilatzeandKlink,2020).Registrationqualitycanbevisually assessedwiththehelpofimagesthataredistributedwiththecodeand thedataset.RheMAPalreadyincludesacomprehensivesetoftemplates — NMT(Jungetal.,thisissue;Seidlitzetal.,2018),D99(Reveleyetal., 2017),INIA19(Rohlfingetal.,2012),MNImacaque(Freyetal.,2011) andYerkes19(Donahueetal.,2016)— buttheprovidedcodeallowsfor easyinclusionofadditionaltemplatebrainsaswell.Moreover,the gen-eralwarpcomputationworkflowcanbeeasilyadaptedforotheranimal species,sincetheunderlyingANTsregistrationfunctionsdonotrelyon priorknowledgeaboutbrainsizeandshape.
3.2.2.3. Segmentationtools. Obtainingappropriatetissuemasks(brain vs.surroundingtissue,ordifferenttissuetypeswithinthebrainitself) canbeparticularlydifficultinnon-humanbrainimaging,asstandard au-tomaticsegmentationtoolsstrugglewithsurroundingmuscletissue,the skull,andthestrongintensitygradientsthatareoftenpresent. PRIME-RElistsseveraltoolsthatcanfacilitatesegmentationofNHPbrainscans.
3.2.2.3.1. Thresholdmann. Awebtoolforinteractivelycreating adap-tivethresholdstosegmentNifTIimages.
A simple intensity threshold can often provide a good initial guess for tissue segmentation, but in the presence of strong inten-sity gradients, a threshold that works well for one brain region can easily fail elsewhere in the same brain image. Thresholdmann (https://neuroanatomy.github.io/thresholdmann) is an open source webtoolfortheinteractiveapplicationofspace-varyingthresholdsto NifTIvolumes.Itdoesnotrequireanysoftwaredownloadsor installa-tionandallprocessingisdoneontheuser’scomputer.NifTIvolumesare draggedontoawebinterfacewheretheybecomeavailableforvisual ex-plorationinastereotaxicviewer(Fig.4A).Aspace-varyingthresholdis thencreatedbysettingcontrolpoints,eachwiththeirownlocal thresh-oldvalue.Theviewerisinitializedwithonecontrolpointatthecenter ofthebrain.Theadditionoffurthercontrolpointsproducesa space-varyingthresholdobtainedthroughradialbasisfunctioninterpolation. Eachlocalthresholdcanbeadjustedinrealtimeusingsliders.Finally, thethresholdedmask,thespacevaryingthresholdandthelistof con-trolpointscanbesavedforlateruseinscriptedworkflows. Threshold-manncomplementsavarietyofexistingbrainsegmentationtoolswith
Fig.4. Segmentationtools.A)Thresholdmannisabrainextractiontoolthatcreatesamaskfromthosevoxelsthatarebrighterthanaspatiallyvaryingthreshold. Thethresholdisconstrainedbycontrolpoints(bluedotsflaggedwithnumberarrows)thatareaddedbyclickingatlocationsinthewebviewer(left).Adjustable sliderstotherightcontrolthethresholdinthevicinityofeachcontrolpoint.Intheexamplesshowntotheright,themaskinitially(top)excludesventralbrainareas butisfilledout(bottom)bytheadditionofventralcontrolpoints.B)Brainboxisaweb-basedtoolforcollaborativebrainsegmentation.Intheinterface,eachMRI (left)hasapagewhereinformationonallprojectsutilizingthisdatasetiscentralized.Underprojectsettings(right),userscanassembledatasetsintoprojects,add collaborators,manageaccessrights,andaddannotationlayers.C)TheUNettoolusesaneuralnetworkmodelapproachforbrainextraction.Localaccuracyateach voxelisestimatedbytheregionalDicecoefficient(top).BottompanelsshowresultsforthreeexamplesubjectsfromthePRIME-DErepository,withregionsassessed tobebraintissueshowninred.
aneasyinterfacetomanuallycontrolthesegmentationonalocalscale. Theresultingmaskscanserveasstartingpointsformoredetailed man-ualeditingusingtoolssuchasBrainBox(https://brainbox.pasteur.fr)or ITKSnap(http://www.itksnap.org).Theinteractiveapproachis espe-ciallyvaluablefornon-humanbrainimagingdata,whereithasbeen successfullyusedtocreateinitialbrainmasksforavarietyofvertebrate brains– includingmanyNHPdatasets(Heueretal.,2019)– aswellas developmentaldata.
3.2.2.3.2. BrainBox. Awebapplicationforrealtime,collaborative vi-sualization,annotation&segmentation
Automaticbrainsegmentationsoftenneedtobemanuallyadjusted andinsomecasesacompletemanualsegmentationisrequired.Manual segmentationcanhoweverbedifficult,timeconsuming,anditisoften performedbyresearchersworkingin isolation,evenfor shared pub-licdata.BrainBox(https://brainbox.pasteur.fr)isanopensourcetool forthevisualization,collaborativecurationandanalysisof neuroimag-ingdata.BrainBoxmakesitpossibleforseveralresearchersto simul-taneouslyworkonthesamedatasetinasharedvirtualworkingspace. BrainBoxcanbeusedtovisualizeandprovidealayerofhuman annota-tiontoanydatasetavailableontheWeb,forexample,todatastoredin Zenodo,FigShare,GoogleDrive,Dropbox,AmazonS3,GitHub,orthe OpenScienceFramework.AfterprovidingtheURLofadataset, Brain-Boxpresentsaninteractiveviewer,andaseriesofeditingtools,e.g.for segmentingbrainregions,erasing,orbuilding3Dmodels(Fig.4B). Sev-eraldatasetscanbecombinedinto“projects” andprojectcreatorscan invitecollaborators.Accesstothedatacanrangefromcompletely pri-vatetocompletelyopenprojects.Finally,BrainBoximplementsa
REST-fulAPI(ApplicationProgramInterface)whichallowsprogrammatic ac-cesstoallitsdata.Forexample,annotationsandsegmentationscanbe queriedbyalocalpythonscript,orevenasharednotebookrunningon GoogleColab.
BrainBox adds a layer of collaborative curation and analysis to sharedneuroimagingdatasetsusingonlyawebbrowser,allowingusers toincrementallyimproveeachother’swork.Thisincreasesscientific ef-ficiency,improvespublicdataquality,andreducesredundanteffort.All PRIME-DEdatasets(Milhametal.,2018)arealreadyindexedin Brain-Boxprojects,andreadyforcollaboration.
3.2.2.3.3. UNetNHPbrainextractiontool. Aconvolutionalneural net-workmodelforskull-strippingofNHPimages.
TheUNetbrainextractiontoolprovidesaflexiblebrainlabeling so-lutiontofacilitatethepreprocessingofNHP neuroimagingdata, par-ticularlyformulti-sitedatasetswithdifferentscanacquisition,quality, andsurgicalsituations(e.g.,withhead-holderimplants)(Fig.4C).The UNetmodelwasinitiallytrainedonalargesampleofhuman neuroimag-ingdataandtransferredtotheNHPwithadditionalmacaquetraining samplesfromthePRIME-DEdatarepository(Milhametal.,2018).In ordertorunthetoolwiththedefaultmodel,auseronlyneedsto spec-ifytheinputT1-weightedimage.Thefullprocessrequires~20sona GPU(NVIDIAGTX1070TI,700MBgraphicscardmemory)and~2– 15 minon a CPU, which meansitcan easily be run on a personal computer.Thecurrentdefaultmodelhasbeensuccessfullyappliedto 136macaquemonkeyscansfrom19sitesonPRIME-DEwithgood per-formance (https://github.com/HumanBrainED/NHP-BrainExtraction). ThetoolalsoprovidesamodulefortheusertoupgradetheUNetmodel
foraspecificdataset(e.g.,rawT1wfromahighfieldscan,MP2RAGE, etc.).Unlikethetraditionalneuroimagingtools,nostrongimaging back-groundisrequiredfortheusertoadjustprocessingparametersfora spe-cificdataset.Instead,thetoolonlyrequiresasmallmacaqueimage-set trainingsample(n=1–2)toupgradethemodelandimproveperformance accuracyforthatspecificdataset.
3.2.2.4. Alignmentandsegmentationpipelines. Inadditiontotoolsthat specializeinasinglestepofstructuralanalysis,therearealsoseveral pipelineslistedonPRIME-REthatcantakecareofmultiplestructural analysissteps.
3.2.2.4.1. Macapype. An open multi-software framework for non-humanprimateanatomicalMRIprocessing
Macapype (https://github.com/Macatools/macapype) provides an open-sourceframework tocreate customizedNHP-specific MRI data processingpipelinesbasedonNipype(Gorgolewskietal.,2011).Nipype isa widely-adoptedPythonframeworkforhumanMRIdata process-ing, which provides wrappers of various commands and functions from well-known neuroimaging software (e.g., AFNI, FSL, SPM12, ANTs). InMacapype,customscripts specifictoNHP dataprocessing arealsowrapped. These includea brainextraction tool(AtlasBREX;
Lohmeieretal.,2019)andascriptforcomputingregistrationsbetween asubjectbrainandtheNMTmacaquetemplate(NMT_subject_align.csh;
Seidlitz et al., 2018). Users can thus flexibly construct customized pipelinesbyputtingtogethervariousprocessingmodules(and parame-ters)thatareoptimalfortheirdataset.
Macapype consists of several configurable modules for: 1) Data preparationsteps,suchasimagereorientation,deoblique-ing,cropping, andtheregistrationandaveragingofmultipleimages;2)Anatomical preprocessingstepslike bias-correctionwithANTsN4BiasCorrection; (Tustisonetal.,2010)and/orT1wxT2wbiasfieldcorrection(Rilling et al., 2012), denoising using the adaptive non local means filter (Manjónetal.,2010),brainextraction(e.g.withAtlasBREX),andbrain segmentation(e.g.usingAntsAtroposN4orSPMOldSegment).
Twoexamplesofmodularanatomicalpipelinesarealready imple-mentedin thedistributedversionof Macapype.Theyarecustomized forthepreprocessing,brainextractionandsegmentationof macaque anatomicaldata,andhaveperformedrobustsegmentationson differ-ent datasets. The first pipeline creates a high quality segmentation in nativespaceforsurfacereconstruction.Briefly,thepipelinetakes theT1wandT2w imagesasinputs,andfirstapplies cropping, bias-correction(viaT1wxT2wandN4intensitybiascorrection),and de-noising(adaptivenon-localmeansfiltering)toimproveimagequality. Next,brainextractionisperformedusingAtlasBREX.Forsegmentation, transformationsbetweenthesubjectskull-strippedimageandtheNMT macaquetemplatearefirstcomputedusingtheNMT_subject_align.csh script(Seidlitzetal.,2018),andthenappliedtoregistertheNMTtissue segmentationstothesubjectimage.Finally,segmentationisperformed usingAntsAtroposN4.shtosegmentthesubjectimageinnativespace, withthetemplatesegmentationaspriors.Thesecondpipelinereaches thesamegoalwithdifferentpackages.Thispipelineprovidesan itera-tivesequencefornormalization(sourcetotemplatespace)ofT1wand T2w,andprovidessegmentationwithbothSPM12(OldSegment)and FSLFAST.
Adockerfileisincludedinthepackagetoallowuserstogetafully embeddedversionofthepipelinesworkingonanycomputerwithout previous installationof MRI processing software.Both pipelines are compatiblewithBIDS(Gorgolewskietal., 2016) formatteddatasets. The two pipelines have been demonstrated toachieve robust skull-stripping and segmentation on anatomical data from different pri-matespecies:Pipeline1wastestedonbothmacaque(Milhametal., 2018) and marmoset datasets, while Pipeline 2 was tested on both human andmacaque datasets. Documentation and an extensive de-scription of the segmentation results from the macaque and mar-moset brain extraction and segmentation are available on Github (https://macatools.github.io/macapype).
3.2.2.4.2. @animal_warper. AFNIprogramthatalignsstructuraldata toatemplate.
@animal_warperisabidirectionalalignmentprogrammadefor an-imalneuroimaging.Whilehumanneuroimaginghastypicallyrequired astandardtemplate,manyanimalresearchersmayprefertokeeptheir datainthenativeacquisitionspaceandmoveatlasregionsandtissue segmentationsintothenativespaceofthesubject.Alternatively,data can be transformedtoastandard templatespacetoallowvoxelwise groupanalysisandhavetheadvantageofastandardizedcoordinate re-portingsystem.
The @animal_warper program proceeds in a series of alignment steps.First,thecenteroftheinputdatasetismovedtomatchthecenter ofthetemplate.Then,anaffinealignmentusesa12-parameter transfor-mationtomatchtranslation,rotation,shearandscale.Finally,a non-linearwarpiscomputedtoalignstructuraldetailstothetemplate.The inversewarpsandinverseaffinetransformationsarecomputedand ap-pliedforthereversedirectionoftemplatetonativespace."Follower" datasets,likeROIsdrawninthenativespace,canalsobetransformed intoatargetspace.DatasetsandROIstypicallyfollowfromnativeto templatespace;templates,atlasesandsegmentationsfollowfrom tem-platetonativespace.
Thewidely varyingkindsof datausedinanimalimagingrequire differentcostfunctionsforalignmenttoflexiblydealwiththevoxel res-olutionandwiththekindsoftissuecontrastineachimagingmodality. Severalfeaturesof@animal_warperaddresssomeoftheidiosyncrasies of animal alignment.First, theusercan specifya“featuresize” that controlsblurringandcostfunctions.FormanymacaqueMRIdatasets,a valueof0.5mm,whichroughlymatchestheapparentvoxelresolution, isausefulfeaturesize.Otherspecies,suchasmouse,marmoset,andrat, mayrequireafeaturesizethataccommodatesthefinervoxelresolution typicallyusedwiththeseanimals.Animalbrainsizescanvary dramati-callyfromeachotherandfromanyparticulartemplate,soa"supersize" optionallowsforuptoa50%differenceinsize.Theprogramappliesa kindofspatialregularizationforROIandatlasregionsthatgoesbeyond thetypicalnearestneighborinterpolation;amodalsmoothingisapplied toalltransformedROIs.Thiskindofsmoothingassignstoeachvoxel themostcommonvoxelvalueinauser-specifiedradiusaroundit.
@animal_warper is developed within the larger AFNI software ecosystem. The computed transformations serve as input to a gen-eral fMRI processing pipeline implemented with afni_proc.py (see
Section3.2.3.1).Outputdatasets innativespaceareviewablein the AFNIGUIandmarkedwiththeappropriatespace.Datathathavebeen transformedtoastandardspacehaveallthefunctionalityofthe "where-ami"atlascommandlineandGUI.WhileNifTIdatasetsaremarkedas beinginascanner,Talairach,MNIorotherspace,@animal_warperadds anidentifyingspacetothedata.Atlasregionsaregeneratedinthe na-tivespacebothasvolumes(Fig.5A)andasindividualsurfacesinGifTI format.Everyregioncanbe shownbyitselforwithanyorall other atlasregionsalongwithasimplesurfacerenditionofthetemplatein thenativespaceofthesubject.Qualitycontrolreportsaregeneratedas simplepngimages.
Theprogramhasbeentested withawidevarietyof themacaque data availableon thePRIME-DE website(Milhametal., 2018). The AFNIwebsitehoststheD99,NMTv1(Seidlitzetal.,2018),the stereo-taxicNMTv2templatesaswellastheD99atlasandtheCHARM(see
Section3.2.1.1andJungetal.,2020)andSARM(seeSection3.2.1.2and
Hartigetal.,2020)hierarchicalatlases.
3.2.2.5. Surfacegenerationpipelines. Surfacevisualizationsofferarich viewofcorticalfMRIactivity.Thegenerationofsurfacevisualizations fornon-humanbrainscanhowever beratherinvolved.Thepipelines belowwerespecificallydesignedtofacilitatethisprocess.
3.2.2.5.1. CIVET-macaque. Apipelineforgeneratingsurfacesand cor-ticalthicknessmapsfromanatomicalMRIs
TheCIVETpipelineforautomatedreconstructionofcorticalsurfaces from invivo humanMRI hasbeenextended forprocessing macaque
Fig.5. AnatomicalMRIpipelines.A)SixsagittalslicesillustratingtheD99atlastransformedtothenativespaceofamacaquesubjectusing@animal_warper.B) CIVET-macaqueresultsfortheD99subject.Toprow(lefttoright):anatomicalscan,brainmask(red)frombrainextraction,tissueclassification(CSFindarkblue, corticalGMingreen,portionsofnon-corticalGMinwhite,WMandothernon-corticalGMinyellow)fromsegmentation,andgeneratedcorticalsurfacesoverlaidon theanatomicalscan(WMsurfaceinred,pialsurfaceingreen).Bottomrow(lefttoright):3D-renderingsofthewhiteandpialsurfaces;corticalthicknessindication frommorphometricanalysis(1.0mminlightblueto3.5mminred)andD99surfaceparcellation(atlas)viewedonthepialsurface.C)PREEMACSresults.Toprow: brainmask(red)createdbyaDeepLearningconvolutionalneuralnetworkmodel(left)andvolumetrictissuesegmentation(right).Middlerow:whitematterand pialsurfaceestimation(left)andrenderedwhitemattersurface(right).Bottomrowleft:surfaceregistrationtothePREEMACSRhesusparameterizationtemplateto obtainvertexcorrespondence(i.e.,surfacecoregistration)betweensubjects.Bottomrowright:individualcorticalthicknessestimationfrommorphometricanalysis. TheresultingsurfacescanbeanalyzedinthegeometricspaceofthePREEMACSaveragesurface.
brains.CIVET-macaquerequiresonlyaT1-weightedMRIimageto gen-eratehighresolutioncorticalsurfaces,capableofintersubject surface-basedcoregistrationwithper-vertexcorrespondencebetweengrayand whitemattersurfaces.Preprocessingandsurfacereconstructionwere customizedformacaquestoaddressspeciesrelatedissuessuchas vari-ationsinheadsizeandbrainmorphology(Lepageetal.,2020).
ThepipelinefeaturesregistrationtotheNMTv1.2population tem-plate(Seidlitzetal.,2018),brain-masking,tissueclassification,cortical surfacereconstruction,andcalculationofcorticalmorphometrics (thick-ness,surfacearea,andcurvature).Surface-basedregistrationtotheNMT averagesurfaceallowsforgroupanalysisandregionalanalysisbasedon eithertheD99orCHARManatomicalparcellations(Jungetal.,2020).
Fig.5Billustratesatypical CIVETrun withthepreprocessingstages (toprow:registration,brain-masking,tissueclassification)andthe gen-erationofthecorticalsurfaces(bottomrow).Thepreprocessingstages, whichcanbegeneratedwithoutcorticalsurfaces,canserveasthe foun-dationfortheanalysisofotherinvivomodalities(fMRI,qMRI,PET,EEG, MEG,sEEG,etc.)aswellaspost-mortemhistologicalandgeneticdata. Imagesoftheoutputsfromvariousviewpointsareprovidedforquality assessment.
Thefullpipelinewassuccessfullyrunontheinvivoscansfromthe 31subjectsthatwereusedtomaketheNMTandontheexvivoscanof theD99subject.Thepreprocessingstagesofthepipelinewererunon 95PRIME-DEsubjectsfrom15sitesthatprovidedscansinthesphinx positionwithcompletesexandageinformation(Milhametal.,2018). EffortshavealsobeguntofurtherextendCIVETtothemarmosetand, usingarchiveddata,thechimpanzee.Thepipelinethushasthepotential tomakeitpossibletostudysurface-basedmorphometryacrossarange ofprimatespecies.
3.2.2.5.2. NHP-Freesurfer. CreatingFreesurfer surfaces of macaque brainscans
Freesurfer(Fischl, 2012)is oneofthemost widelyusedsoftware packagesforthecreationofsurfacesfromvolumetricMRIdatain hu-mans.Whileitcanalsobeusedtoprocessnon-humanbrains,its con-venientrecon-allpipelinethatautomatesmostoftheprocessingsteps cannotbereadilyappliedtomacaquebrainsduetoitsdifferencesin sizeandcontrast.NHP-Freesurferoffersapipelinethatdoesworkfor macaque brains.Itisbased onFreesurfertoolsandanumberof ad-ditionalworkarounds.Allthesestepsareexplainedandpre-codedin annotatedJupyterNotebooksbasedonthebash-kernelthatallowthe usertorunthecodedirectlyfromthenotebook.Unlikerecon-all,which ismostlyautomated,creatinghighqualitysurfacesofmacaquebrains requiressome manualadjustmentsthatcan bedifficulttofigureout fornoviceusers.NHP-Freesurferprovidesafullguidestartingwithone ormore T1-weightedanatomicalscansandendingwithsurfaces and flatmaps.Tofacilitate this process,NHP-Freesurfer usesfreely avail-abletools fromtheNMT templatebrain(Seidlitzetal.,2018),ANTs (Avantsetal., 2008),andAFNI(Cox,1996).Inadditiontothecore Freesurferfunctionality,NHP-Freesurferalsoexplains howtoproject volumetricstatisticalresultstotheFreesurfersurfaceswithFreesurfer tools,butthisworkflowhasbeensupersededbytheNHP-pycortex pack-age(see3.2.3.5)thatusesanadaptedversionofPyCortex(Gaoetal., 2015) toobtain similar results witha much more flexibleapproach basedonthePythonprogramminglanguage.
3.2.2.5.3. Precon_all. An automated Freesurfer andHCP compliant surfacepipelineforpreclinicalanimalmodels
Precon_all is a surface reconstruction pipeline inspired by Freesurfer’s recon-all and adapted for use with non-human brains (Fischl, 2012). The great success of Freesurfer’s recon-all is in part due totheeaseofusefor theend useraswellastheautomationof surface generation from volumetric data. Precon_all aims to export this ease of use andautomation totheanimal imaging community.
Inorder toachieve theflexibilitythatmakes theprecon_allpipeline workwithalargerangeofspecies,asetof5easilydrawnmasksare requiredas input. Guidelines for drawingthese masks are provided in the precon_all documentation.Once they aremade, they can be used for an individual subject or added tothe precon_all standards directoryandusedtoautomatesurfacereconstructionforanewspecies. Currently,precon_allcomespreloadedwithstandardsfortheNMTv1.2 (Seidlitzetal.,2018)andapigtemplate.
Precon_allmakesuseofcommonlyusedsoftwarepackages, includ-ingFSL,ANTs,Freesurfer,andConnectomeWorkbench(Avantsetal., 2008;Fischl,2012;Glasseretal.,2013;Jenkinsonetal.,2002).Written asasetofshellscripts,precon_alliscalledfromthecommandlineand, likeFreesurfer’srecon-all,itcanberuninstagesformanualeditingand correctionof surfaces.Thesestagesincludebrainextraction, segmen-tationviaFASTorANTs,cuttingandfilling,surfacegeneration,and optionalsurfacedown-sampling.Outputsarecontainedwithina sub-jectdirectorysimilartothatofrecon-all,allowingFreesurferusersto easilytransitionfromhumantoanimalimaging.Finaloutputsinclude FreesurferandHumanConnectomeProjectcompatiblecorticalsurface models,aswellasallrequiredNifTIimagesandtransformscreatedin thesurfacereconstructionprocess.Asetofscriptsisprovidedtoeasily generategroupaveragesurfacesandregistrationtemplateswhichcan thenbeusedtocreateacustomFSaverageforindividualspecies.
3.2.2.5.4. PREEMACS. Brainsurfacesandcorticalthicknessfromraw structuraldata
PREEMACS (pipeline for PREprocessing and Extraction of the MACaquebrainSurface)isasetofcommontools,customizedforRhesus monkeybrainsurfaceextractionandcorticalthicknessanalysis.Some oftheadvantagesofusingPREEMACSare:1)itavoidsmanual correc-tion,2)thepipelinewasdevelopedinthestandardMNImonkeyspace, 3)itprovidesvisualreportsineachmoduleasqualitycontrol,and4) itshighlyaccuratesurfaceextractionwithvertexcorrespondence be-tweensubjects.PREEMACShasamodulardesign,withthreemodules runningindependently. These modules perform the canonical work-flowforMRIpreprocessing(Alfaro–Almagroetal.,2018;Estebanetal., 2019;Glasseretal.,2013)usingdifferentpreviously-availablefunctions from FSL(Jenkinson etal., 2002), ANTs(Avants et al., 2008), MR-trix(Tournieretal., 2012),MRIqc (Esteban etal.,2017),DeepBrain (https://github.com/iitzco/deepbrain), andFreesurfer (Fischl, 2012). Inputstothepipelineshouldbeone(orpreferablymore)T1-and T2-weightedvolumesperanimalfromthesamesession.Module1prepares therawvolumesfor initialpreprocessingin six steps:volume orien-tation,conformation,imagecropping,intensitynon-uniformity correc-tion,imageaveraging,resamplingandskull-stripping.Module2isthe qualitycontrolmodule.ItwasadaptedfromtheMRIQualityControl tool(MRIqc)(Estebanetal.,2017)forhumanstoobtainimage qual-itymetricsandprovideavisualreportfromtheresultsofModule1. Module2usesthesequalitycontrolmetricstoprovideaclassification ofimage qualitythatallowsPREEMACStoestimate whetherthe in-putwillyieldanappropriatesurfacereconstruction.Finally,Module3 obtainscorticalthicknessmeasuresbasedonthebrainsurfaces using anNHP-customizedversionofFreesurferv6.Toevaluatethe generaliz-abilityofthisprocedure,PREEMACSwastestedontwodifferentNHP datasets:PRIME-DE(Milhametal.,2018)(57subjects)andINB-UNAM (5subjects).Results(Fig.5C)showedaccurateandrobustautomatic brainsurfaceextractionforbothdatasets.PREEMACSthusoffersa ro-bustandefficientpipelinefortheautomaticNHP-MRIsurfaceanalysis.
3.2.3. FunctionalMRItools
FunctionalMRI(fMRI)usestime-seriesdatainanattempttomodel thefluctuationsintheimagingsignalwithsometypeofeventstructure (Fig.1C).Theseeventscanbeexternalstimuli(i.e.,task-basedfMRI),or thetimeseriesofactivityinanotherbrainarea(i.e.,functional connec-tivity).Conceptually,theanalysisoffMRIdataisnotradicallydifferent acrossspecies,butthechecksrequiredateachanalysisstepdodiffer. Forexample,animalscannersetupsmayproducedifferentkindsofB0
inhomogeneity anddistortions,due tocustomizedcoilsortheuseof otherspecializedequipmentduringacquisition.Boththepotentialuse of anesthesiaandcontrastagentscan furthermoreimpact processing choicessuchashemodynamicresponsemodelingandnoisefiltering. Importantly,pre-processingstepslikemotioncorrectionandalignment toahighresolutionanatomicalscanortemplatebrainoftenrequire dif-ferentoptionsandparameterscomparedtowhatonewouldchoosefor humandata.Forawakeanimals,bodymotioncancausedistortionsin themagneticfieldresultinginslice-specificnonlineardeformationsthat canbeespeciallydifficulttocorrect.Properhead-fixationandextensive trainingwillkeepsuchproblemstoaminimum,butEPIdistortionand brightnessartifactscanstilloccur,requiringpost-acquisition ameliora-tion.TheresourcesbelowcontainseveralNHPspecificsolutionstodeal withpre-processingandalignmentcorrectly.
3.2.3.1. afni_proc.py. AFNIprogramthatgeneratesacompletefMRI pro-cessingscriptfromalistofdatafilesandthedesiredprocessingstepsand options.
AFNI’safni_proc.pyprogram(Cox,1996)allowsaresearcherto cre-ateacompletefMRIprocessingpipelineforindividualsubjects,from rawinputs,throughalignmenttostandardspace,andregression mod-eling. Touse it,onespecifiesinputdatasets (e.g.,EPIs,anatomicals, tissuemasks),majorprocessingblocks(motioncorrection,warpingto standardspace,blurring,regression,etc.),anddetailedchoicesforeach block(e.g.,whatkindofalignment,theblurradius,orderofthe polyno-mialforbaselinemodelingandtypesofmotionregressors).Thus,one canplantheprocessinghierarchically,forconceptualclarityand orga-nization.Thecommandgeneratesafull,commentedprocessingscript, whichisthenexecutedtocarryoutsinglesubjectprocessing.In prac-tice,atypicalafni_proc.pycommandcontains20–25options,whichis averycompactwaytospecifyafullpipeline.
Forconvenienceandassuredmathematicalcorrectness,afni_proc.py automaticallytakescareofseveralaspectsoftheprocessing.For exam-ple,allwarps(motioncorrection,B0distortion,alignmenttoanatomical andtostandardspace)areconcatenatedbeforebeingappliedtotheEPI datasettominimizesmoothingduetoregridding.Thisallowstheuserto focusontheparameterchoicesinprocessing,ratherthanonthe techni-calprogrammingaspects,whichgreatlyreducesthenumberofbugsin ananalysisstream,particularlyifoneupdatesortweaksexistingcode. Becausetheprocessingscriptitselfisexplicitlycreatedandsaved, re-searcherscancheckexactlywhatstepsareoccurringintheiranalysis, andthewholeprocessisdocumented,whichincreasesreproducibility. Whileafni_proc.pywasmainlydevelopedinconjunctionwith hu-manbrainresearchers,itisfullycompatiblewithanimaldataandhas beenappliedinanimalstudies.Forexample,itintegratesdirectlywith AFNI’s@animal_warpercommand(see3.2.2.4.2)anditslistofpossible hemodynamicresponsefunctionsincludesastimulusresponseshapefor MION(monocrystallineironoxidenanoparticle),acontrast-agentthat is commonlyusedinanimalneuroimaging studies.Whenperforming EPI-anatomicalalignment,onecanfurthermoresettheminimum “fea-turesize” ofstructurestomatchtoavaluethatisrelevantforsmaller animalbrains.
TherearecurrentlytwodemosavailableinAFNIformacaquefMRI processing withafni_proc.py:onefor task-baseddata(visualstimuli, MIONcontrast),andtheotherwithrestingstatedatafromPRIME-DE (Milhametal.,2018).EachdatasetusesthestereotaxicNMTv2asits standardspaceanddemonstratesnonlinearwarpestimationwith @an-imal_warper (see Sections 3.2.1.1, 3.2.2.4.2, and Junget al., 2020). AFNIisfreelyavailable,andmostprogramsarewritteninC(aswell asPython,Randshell),forgeneralityandcomputationalefficiency.
3.2.3.2. C-PAC. Aflexiblepipelineforperformingpreprocessingand con-nectivityanalyseswithvarioustools.
TheConfigurablePipelinefortheAnalysisofConnectomes(C-PAC) isanopen-sourceplatformthatallowsuserstoconfiguretheirown anal-ysispipelineforstructuralandfunctionalMRIdata.Itisdesignedand