HAL Id: inserm-02177298
https://www.hal.inserm.fr/inserm-02177298
Submitted on 8 Jul 2019HAL is a multi-disciplinary open access archive for the deposit and dissemination of sci-entific research documents, whether they are pub-lished or not. The documents may come from teaching and research institutions in France or abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est destinée au dépôt et à la diffusion de documents scientifiques de niveau recherche, publiés ou non, émanant des établissements d’enseignement et de recherche français ou étrangers, des laboratoires publics ou privés.
Cloning and comparative analysis of the human
pre-T-cell receptor a-chain gene
Paola Porto, Ludovica Brunot, Marie-Genevieve Mattei, Harald von Boehmer,
Claude Saint-Ruf
To cite this version:
Paola Porto, Ludovica Brunot, Marie-Genevieve Mattei, Harald von Boehmer, Claude Saint-Ruf. Cloning and comparative analysis of the human pre-T-cell receptor a-chain gene. Proceedings of the National Academy of Sciences of the United States of America , National Academy of Sciences, 1995, 92, pp.12105 - 12109. �10.1073/pnas.92.26.12105�. �inserm-02177298�
Vol. 92, pp. 12105-12109, December 1995
Immunology
Cloning and comparative analysis of the human pre-T-cell
receptor
a-chain gene
PAOLA DEL
PORTO*, LUDOVICA
BRUNOt, MARIE-GENEVIEVE MATrEIt, HARALD
VONBOEHMER*t,
AND
CLAUDE SAINT-RUF*§
*Unite Institut National delaSanteetdelaRechercheMedicale373,Institut Necker,156 rue de Vaugirard, 75730ParisCedex 15, France;tBaselInstitutefur Immunology, 487Grenzacherstrasse, CH-4058 Basel, Switzerland;andtUnit6Institut National de laSanteet de la RechercheMedicale 406,Facultedemedecine, 27Boulevard JeanMoulin,F-13385 Marseille Cedex05,France
Communicatedby Philippa Marrack National Jewish Center, Denver, CO, September 14, 1995
ABSTRACT In immature T cells the T-cell receptor (TCR)
18-chain
gene is rearranged and expressed before the TCR a-chain gene. At this stage TCR18
chain can form disulfide-linked heterodimers with the pre-T-cell receptor a chain(pTa). Using the recently isolated murine pTa cDNA as aprobe, we have isolated the human pTa cDNA. The complete nucleotide sequence predicts a mature protein of 282 aa consistingof an extracellularimmunoglobulin-likedomain, a connecting peptide, a transmembrane region, and a long cytoplasmic tail. Amino acid sequence comparison of human pTa with the mouse pTa molecule reveals high sequence homology in the extracellular as well as thetransmembrane region. In contrast, the cytoplasmic region differs in amino acidcomposition and in length from the murine homologue. The humanpTagene is expressed in immature but not mature Tcells and is located at thep2l.2-pl2 region of the short arm of chromosome 6.Intrathymic T-cell development is accompanied by ordered rearrangement of T-cell receptor (TCR) genes as well as by changes in expression of surface markers (1). At a certain developmental stage, CD4-, CD8-, CD3l0w,CD44+, CD25+ T-cell precursorsdownregulateCD44expression and begin to rearrange theTCR
(3-chain
(TCRI3)
locus. After productiveTCR(3
genes areformed, thymocytes become CD25-,acquireCD4/CD8
coreceptors on the cellsurface, and rearrange the TCRa locus (2-5). At the CD4+ CD8+TCRaf+
stage, thymocytes are subjected to positive and negative selection depending on the quality of TCR-major histocompatibility complex ligand interaction (6,7).
While theTCRao3
controls latethymic development, early developmentalsteps are con-trolledbythepre-TCRcomposedof the variantTCR,3
chain that is disulfide linked to the invariant pre-TCRa (pTa) protein recently described in mice (7-9). TheTCRf3-pTa
heterodimerisassociatedwith CD3 molecules(10) andsignals triggered by the pre-TCR induce expansion and differentiation of immature precursorcells (8, 9,11).
Here wereport on the isolationandcharacterizationof thehuman pTacDNAand its comparison withthe murinehomologue.1
Inaddition weprovide data onthe chromosomal locationaswellas onpTaexpression inthymic subsets.MATERIALS AND
METHODS
cDNA Isolation andSequencing. A3-year-old human thy-muscDNAlibrary constructedinAgtlOvector(1 x
106
clones) (Clontech;HL1127A)wasscreened with aprobe correspond-ing to the full-length murine pTa. Prehybridization and hy-bridization were done at 65°C in a solution containing 1 M NaCl,50 mM TrisHCl
(pH 7.2), 10% dextran sulfate, 1% SDS,Thepublicationcostsof this articleweredefrayedinpartbypagecharge
payment.This article musttherefore beherebymarked"advertisement" in
accordancewith18U.S.C.§1734 solelytoindicate this fact.
and 0.250 mg of salmon sperm DNA per ml. Filters were washed in 2x SSC first at room temperature and then at65°C. The murine cDNA was used as a probe and labeled with
[a-32P]dCTP
by random priming (DNA labeling kit; Boeh-ringer Mannheim). Inserts from positive clones were sub-clonedin the EcoRIsite ofpBluescriptSK(-)vector.Plasmid DNA was purified according to Birnboim andDoly (12) for double strand and according to Sambrook (13) for single strand. Dideoxynucleotide sequencingwas carriedouteither on single-stranded template or on double-stranded template obtained from thepBluescript sub'clone. Reading ambiguities were resolvedbysubstitution of dGTP and 7-deaza-dATPfor dGTPanddATP,respectively, insequencing reac-tion mixtures. Sequenase from United StatesBiochemicaland T7 DNApolymerase from PharmaciaBiotechhavebeenused. DNAsequence analysiswas done with the GCG program (Genetics Computer Group, program manual for the GCG package, version 7, April 1991). Homology searches in the EMBL (release 42.0, March 1995), GenBank (release 88.0, April 1995), and Swiss-Prot (release 29.0, June 1994) data banks were done with the FASTA(14)program. Prediction of thesecondarystructureandhydrophilicitysearchesweredone with thePEPTIDESTRUC=TRE program.Cloning of the 5' and 3' Ends. Poly(A)+ RNAs were extracted from3-month-oldhumanthymus by usingthecitric acid method (15).cDNAsynthesiswasperformedon 1 ,ug of
poly(A)+
RNAwith anoligo(dT)
adaptor 5'-GACTCGAG-TCGACATCGAT17-3' accordingtothesupplier's recommen-dations(cDNA cycle kit;Invitrogen).
Toobtainthe3'end,
aprimer specificfor the humanpTa sequence
(5'-CCTGCCT-TCTGAGGAGCTG-3')
asdeducedfromanalysis of the pos-itiveAgtlO recombinant clone andtheoligo(dT) adaptorwere used for PCR.Thirty cycles of amplification (94°Cfor 40 sec, 50°Cfor 2min, and72°C for3min)
werecarried outfollowed bya 10-min final extension at72°Cin 100ptl
of PCRmixture containing2,ul of cDNA and 50pmolof eachprimer.Asecond round ofamplificationwas performed on amplifiedmaterial after sizepurification.
Theprimers
usedforthe second amplifi-cation were anestedpTa-specific
primer(5'-TGGGGCTGAG-GGTCACAGCA-3')
and theadaptor
(5'-GACrCGAGTCGA-CATCGATT-3').
Theamplified
materialwasmigratedon a1% agarosegel, purified by adsorptiononglass beads,andligatedinto thepCR-Script
SK vector(Stratagene).For the 5' end amplification, the cDNAwas
synthesized
using the pTa-specific primer
5'-GCCTCTCCTGACAGAT-GCAT-3'.The
dG-tailing
wasperformed
inafinal vol of 50,ul oftailing buffer with 15 units of terminaldeoxynucleotidyl-transferase
(GIBCO/BRL).
The firstamplification
ofdG-Abbreviations:TCR, T-cell receptor;TCRf3and -a, TCR13 chain and TCRachain; pTa, pre-TCRa; PKC,proteinkinaseC;C, constant; V, variable.
§Towhomreprint requests should be addressed.
1The sequence reported in this paper has been deposited in the GenBank data base(accessionno.U36759).
12106 Immunology: Del Porto etal.
tailedcDNA was performed in 100 ,ul of PCR bufferusing as
primersananchorpoly(C) primer (16)andahumanpTa-specific primer (5'-GGGAGAGATGGGCCAAGTTG-3'). Amplifica-tionwas done for 25cyclesat94°Cfor 1 min, 55°Cfor 1.5 min, and 72°C for 2.5 min. A second round of amplification was
performed on size-selected amplified material with a nested pTa-specific primer
(5'-TGATTGGTGGGGCCAGAGAAG-3')
and theadaptor primer(16).
ThePCRproductwasclonedasdescribed above.
Southern Blot.A Zoo-Blot from Clontech was hybridized either with the entire humanpTa cDNA,witha 300-bpPst I insert codingfor the human pTa cytoplasmic tail, orwith a
PCR product corresponding to the murine cytoplasmic
se-quence.Prehybridizationandhybridizationweredoneat65°C
asdescribed. Membraneswerewashed in 2x
SSC/0.1%
SDSatroomtemperature andthenat65°C. Stringentwasheswere
performedin 0.5x
SSC/0.
1% SDSat650C when the Zoo-Blotwashybridizedwith the entireprobe.Probeswereremovedin 0.5% SDS at90-1000C.
MonoclonalAntibodies.Thefollowingmonoclonal antibod-ies were used in this study: phycoerythrin-coupled Leu-3a (anti-CD4 antibody; Becton Dickinson), fluorescein isothio-cyanate
(FITC)-labeled
Leu-4 (anti-CD3 antibody; Becton Dickinson), biotinylated Leu-2a (anti-CD8 antibody; BectonDickinson).
Surface StainingandCell Sorting.Cellsweresuspendedin PBS containing2% fetal calfserum(FCS) at 107 cells per ml and incubated with the desired antibodies atoptimal
concen-tration for 10-20 min on ice. After incubation, cells were
washed twice in PBSplus2% FCS and resuspendedeitherin
the same solution
(in
the case ofdirect stainingwith phyco-erythrin-conjugated, FITC-conjugated monoclonalantibod-ies)
or(in
the case of indirect staining with biotinylatedantibodies)
inoptimallydiluted APC(Southern BiotechnologyAssociates)
and incubated foranadditional 10-20 minonice. Cellsuspensions
were filtered and sorted onFACStarPLUs
(Becton Dickinson) instruments. Sorted populations were
reanalyzed to test theirpurity, whichwas alwaysfound tobe >99%.
ReverseTranscription PCR. Cells (5 x 104)were directly sorted into 500 ,tl of RNAzol
(Cinna/Biotecx
Laboratories,Friendswood, TX);
nonsorted cellswere washed in PBS and resuspendedin RNAzol. TotalRNA wasextracted accordingto the manufacturer's protocol. cDNA was prepared with random hexamer primers and reverse transcribed with a
Superscript
kit(GIBCO/BRL).
For PCR, primersusedwereoligonucleotides
recognizingsequencesin the 5' and 3'regions of the pTa(5'-GGCACACCCTTTCCTTCTCTG-3'
and5'-GCAGGTCCTGGCTGTAGAAGC-3')
and actin(5'-ACA-CTGTGCCCATCTACGAGGGG-3' and 5'-ATCATGGAG-TTGAAGGTAGT TTCGTGGAT-3') cDNAs. PCRs were
done in 30
jil
of reaction mixture containing50 mM KCl, 10mM TrisHCl
(pH
8.3), 10 mM mixed dNTP, 10 pM eacholigonucleotide
primer,
and 1 unit ofAmpliTaq DNApoly-merase
(Roche Diagnostics).
DNAwasamplifiedfor 35cyclesat an
annealing
temperature of55°C
with a thermal cycling machine(Perkin-Elmer/Cetus).
A 9-,lI portion ofeacham-plified product
wasseparatedthroughout
a1% agarosegel byelectrophoresis
and stained withethidium bromide.Chromosomal LocalizationbyinSituHybridization.In situ
hybridization
was carried out on chromosome spread prepa-rations obtained fromphytohemagglutinin-stimulated
human lymphocytes cultured for 72 hr. 5-Bromodeoxyuridine wasadded for the final 7 hr of culture
(60
,ug permlofmedium)to ensure a
posthybridization
chromosomalbanding ofgoodquality.
ThepTa
clonecontaining
an insert of 800bp
inpBluescript
was3H
labeledby
nick-translation to a specificactivity
of 1.4 x 10dpm
,ug-1.
The radiolabeled probe washybridized
tometaphase
spreadsat afinal concentration of 25 ng perml ofhybridization
solution asdescribed (17).After coating with nuclear track emulsion (Kodak NTB2), the slides were exposed for 21 days at +4°C and then devel-oped. To avoid any slipping of silver grains during thebanding procedure, chromosome spreads were first stained with buff-ered Giemsa solution and metaphase spreads were photo-graphed. R-banding was then performed by the fluoro-chrome/phytolysis/Giemsa method and metaphase spreads
wererephotographed before analysis. RESULTS
Isolation andAnalysis of Human pTa cDNA. Screening of a human thymus cDNA library led to the isolation of three positive clones, which were subcloned into pBluescriptSK(-) and sequenced.Sequence analysis ofa931-bp insert from the longest clone(F6) revealed the presenceofalong open reading frame. To obtain the complete sequence of the human pTa and to confirm it independently, a rapid amplification of cDNA ends (RACE) (18) was undertaken with primers lo-cated in the immunoglobulin-like domain. Sequencing of 3' RACE products allowed the identification of 104 additional nt with one putativepolyadenylylationsignal 20 bp upstream of thepoly(A) tail. Sequencing of the 5' products identified 60 additional nt in the 5' untranslated region. By the combination of these techniques a sequence of 1097 nt was obtained. These data are in agreement with the 1.1-kb band observed in Northern blots of human thymus mRNA hybridized with mouse orhuman pTa probe (data notshown). The included open reading frame contained 846 nt encoding a 282-aa protein including the N-terminal methionine (Fig. 1). The calculated molecular weight of the protein was 27,793. The overallidentity with the murine pTawas 63%.Positionwise, the human ATG that heads a long open reading frame corresponds to the second in-frame ATG in the murinepTa sequence,suggesting that in the mouse the second ATG in the 5' region represents the initiator codon.
The deduced human amino acid sequence reveals a 5' hydrophobic region of 16 aawith 75% identity at the nucle-otide level and70% identityat theamino acid level with the murine pTaleader sequence. The following 130aashow83% identity withthree cysteines at the conserved positions 31, 91, and 119.Theputative extracellular region presents features of
an immunoglobulin-like structural constant (C) domain as
defined by Williams and Barclay (19). The sequence
encom-passing aa 1-116 shows the characteristic seven ,B-strands, which could fold in two 13-sheets stabilized by an intrachain disulfide bondformed by the cysteinesatpositions31 and 91 andatryptophan in position 46 that is involved inestablishing the tertiarystructure(20). This structural organization is very wellconserved between mouse andhuman. While the murine extracellulardomain exhibits twopotential sites of glycosyla-tion, the human sequence contains only one.The conserved cysteineatposition 119 is assumed to participate in formation ofthe interchain disulfide bond withTCR,B and is included in
a small sequence of 15 aa that form theconnecting peptide, which showsonly 60% identity to the murine sequence. The following20aa share 80% identityto the murine
transmem-brane region. They are mostly hydrophobic and the region contains two polar residues (arginine and lysine) that are
separated by thesame4aaobserved in the murine sequence. This feature is also conserved in human and murine TCRa chains and is assumed to be essential for the assembly and transportof the
TCRaf3-CD3
complex (21).The homology between human and murine pTa ends abruptly 5 aa after the predicted transmembrane region. Furthermore, the length of the human pTa cytoplasmic tail (114 aa)differs from that of themouse(31 aa).Therearethree potential proteinkinase C(PKC)sites in this part of the human pTa as opposed to two in the mouse, which correspond positionwise to the two first PKC sites in the human pTa.
A B C
-16-signalpeptide-1 v
-CHO-m MARTWLLLLLGVRCQALPSGIAGTPFPSLAPPITLLVDGRQHMLVVCLVLDAAPPGLDNPVWFSAGNGSAL h --G---G-P---T-VG---M---K-Q-V---V---S-I---D L F a 56 v -CHO- v m DAFTYGPSLAPDGTWTSLAQLSLPSEELEAWEPLVCHTRPGAGGQNMSTHPLQLSGESSTARSCFPEPLGG h ---T---H---AS---G---E-HS-.--Q-MH----A----T-PQ---R-127 +-M-+ -PKC- -PKC-m TQRQVLWLSLLRLLLFKLLLLDVLLTCSHLRLHVLAGQHLQPPPSRKSLPPTHRIWT* h -PGGA---GV---L---C-CDPAGPLPSPATTTRLRALGSHRLHPATETGGREATSSPRP -PKC- -PKC- -PKC-198 h QPRDRRWGDTPPGRKPGSPVWGEGSYLSSYPTCPAQAWCSRSRLRAPSSSLGAFFRGDLPPPLQAGAA*
FIG. 1. Comparison ofthededuced amino acidsequenceencoding the humanandmousepTacDNA. Humansequence(h) isalignedwith the
previouslydescribed murine pTa sequence(m) (accessionno.U16958).Amino acid residue numbers startwith -16 asthe firstposition of theleader
andproceedfrom+1of theputativematureproteins; locationsofthepotential N-linkedglycosylation site (CHO), the predicted leader (peptide signal), the transmembrane (TM),and thepotentialPKCphosphorylationsitesareindicated. The threecysteinesatpositions31, 91,and 119are
marked(v).Theconservedbasic amino acidsarginine andlysine, located in the transmembraneregion,areindicated(+) in the lower line.The
conservedposition of the,3-strands (A, B, C, D, E, F,andG) deduced bycomputeranalysisof thesecondarystructureof themurineand thehuman
sequenceareshown above thetwosequences.Hyphens replace residuesinthe human pTathatareidenticaltothecorresponding residues inmouse
pTa.
Unlike the murinepTa, the human sequence doesnotcontain
aPPGHR CD2-like motif, which is believedtoplayarole in CD2-mediated T-cell activation (22). Except for a proline tripletatthe 3'end, the human pTa cytoplasmic tail doesnot
contain a proline-rich region that could constitute an SH3 domainbinding region (23), as found in the murinepTa.
Interspecies pTa Hybridization. To determine whether nucleotide sequence homologies could be detected by
cross-hybridization at low stringency, we performed Southern blot analysisofEcoRI-digestedDNA fromseveralspecies. When theentire human pTa cDNAwasusedas aprobeatleastone
hybridizing band was detected in all species analyzed. The
same result wasobtained with aprobe corresponding to the human extracellular immunoglobulin-like domain only. In
contrast,withprobescorrespondingtothecytoplasmictails of either humanormurinepTa,hybridizationwasobtainedonly with human and monkey DNAs or murine and rat DNAs, respectively(Fig.2). Thus, among differentspecies,the
extra-cellular immunoglobulin-like domain appears much better conserved than thecytoplasmic tail,aresult consistent with the sequence data ofhuman and murine pTa.
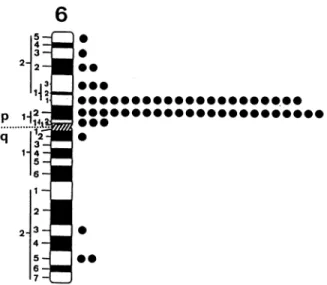
Human pTa Chromosomal Location. In 100 metaphase cells examined afterin situhybridization,therewere214silver grains associated with chromosomes and 58 of these (27.1%)
werelocatedonchromosome6; theirdistributionon
chromo-some 6 was notrandom: 44 of the 58 silvergrains(76%)were
localized on the p2l.2-p12 region of the short arm of
chro-A
mosome 6 (Fig. 3). Thus, the human pTa gene maps to the 6p21.2-6pl2 region of the human genome.
Recently, the murine pTa genewas localized in the D.E1 region of chromosome 17 (24), which is homologous to the short arm of human chromosome 6.Thus, in both human and mouse the pTa gene is localized in the area of the major histocompatibility complex.
Intrathymic Expression of Human pTa. HumanpTa RNA
was detected by PCR analysis using the primers described. Human thymocytes were separated into immature CD4-CD8-,
CD3I0W,
CD4+ CD8-,CD3I0w,
CD4+ CD8+, CD3+ aswell as matureCD4+CD8-CD33+, CD4-CD8+CD3++,and CD4- CD8-CD3++ thymocytes (25, 33).Asshown in Fig. 4, pTa RNA is detected in the immature but not the mature subsets, and its expression is strongest in the CD4+ CD8-CD3l0w fraction of cells thatare on their wayto differentiate into CD4+ CD8+ thymocytes.
DISCUSSION
Inthis paper,wereportthenucleotide sequence of the human pTa cDNA. Analysis of the human and murine pTa cDNA shows the existence of conserved and nonconservedregionsin theproteinthat maybe related to its function.Alignmentof theextracellular regions of the human andmousepTa shows that they are identical in length. The highest degree of homology is detected in thepeptidicstretchcorrespondingto
theimmunoglobulin-likeC domain(20),where83% ofidentity
B C 1 2 3 4 5 6 kb 23,7-9.4 -6.6 -4.3 -kb 23,7-9.4 -6.6 -4.3 -1 2 3 4 5 6 kb 23,7-9.4 -6.6 -4.3 -1 2 3 4 5 6
FIG.2. InterspeciesSouthern blotanalysis.EcoRI-digestedDNAfrom human(lanes 1), monkey(lanes2),rat(lanes 3),mouse(lanes4), dog (lanes 5),andcow(lanes 6)wasanalyzedby Southern blotting.The blotwassuccessivelyhybridizedwithprobescorrespondingtotheentirehuman
pTacDNA(A)and part of the cDNA encodingthecytoplasmictails of human(B)and mouse(C) pTa.
12108 Immunology: Del Porto etal.
6
5 -4 -3 -2- 2 -1. p 1 i2-... ...1 q 32-1- 4 - 5-6-
1-21
2-34-I
U
0 00*
0*0 1000 0 0 00FIG. 3. Idiogram of the humanG-banded chromosome6 illustrat-ingdistribution of the labeled sites with the human pTa probe.
is observed. This score is greater than the identity found between the human and murine Cregions of TCRa
("60%).
This remarkable conservation in the extracellular region of pTa couldreflect the constraints imposed by the interaction withaconservedthymic(or extrathymic) ligand,which could promote expansion and differentiation ofthe T-cell lineage throughsignaltransductionby the pre-TCR.Alternatively, the conservationcould be duetopairingwith theTCR,B protein. Some
of.the
conservation could also beimposed because,asin pre-B cells, an additional variable (V) domain in pre-Tcells might benoncovalently associated with the immunoglobulin-like domainofpTa. Thiscouldbecompatiblewith the notion that theTCRl3-pTaheterodimer isinefficiently transportedtothe cell surface ina matureT-cell line(9).Onthe otherhand, there are several pieces of evidence that suggest that the
structure ofTCRf3 differs from that of the immunoglobulin heavy chain.
Vp
andC, are notstructurallyautonomousunits (26)unlike the V and the Cregionsofimmunoglobulin heavy chain(27).
This is confirmed by analysis of the three-dimensionalstructureofTCRI3
(28), showingthat theV, and C, domainsareinintimatecontactinthecrystal
structure.TheVp
is folded on theC,3
domain in a rigid conformation, indicatingthat theinteractionbetween the Vregionsof theaand ,3 chains could be reduced. Because of the stability of
TCRJ3
byitself,there may benorequirementforanassociated Vpre-T-cellregioninthepre-TCR. Furthermore,it has been shown that expression of a mutilated V,3 gene permits the intrathymic double-negative to double-positive transition to occur(29).
This could suggest that the Vdomain of TCR13is not anessential part of thepre-TCR complextoachievesignal transduction and, therefore, that interaction of a V pTaI + + + + C t00 C0 t co C0-0 00 0 00 0in00 0 0 0 0 0 0 0 0 0 0 0 0 000 00 0 7 7: 7 7 z c: c) 7
domain with the
Vp
domain, analogous to the binding of V pre-B cell toV,1
in pre-B cells, is not needed to promote differentiation. These considerations are at least consistent withthe idea that thepTa-TCRI3
complexcould be sufficient to drive thymocytedevelopment without the association ofaV-like domain.
Thepeptide connecting the pTa transmembrane region with theimmunoglobulin-likedomain shows lesshomology(60%) butcontains aconservedcysteineinposition 119, expected to participate inthe disulfidebond withTCRf.Ahigh degreeof conservation is observedinthetransmembraneregion of pTa, whichcontains thebasic amino acidsarginine andlysinethat arealso found in TCRa andTCR8 and appear necessaryfor interaction with the CD3molecules(21).These data indicate thatthehuman pTa molecule shares with its mouse counter-partall the features necessarytoform acomplexwithTCR,B and the CD3 complex. It has beenreported (10)that a weak association exists between the pre-TCR
ap
and the ;chains of the CD3complex.Ahighly conservedphenylalanine at posi-tion 195 of the TCRa Cregionhas been shown to be essential fora high avidity interaction with the ;chains (30, 31). The absence of this phenylalanine residue in the corresponding part of the extracellular regions of human and murine pTa couldaccountfor the poor association ofpre-TCR aj3chain with the rchain.The homology betweenhum-an and mousepTa breaks off abruptlyatthe end of thetransmembrane region.Infact,the 127aathatarepredictedtocomposethe humancytoplasmic region do not show any significant identity with the 30 aa
residues found in the mouse pTa cytoplasmic tail. In this regionofthe human molecule,inadditiontothe presenceof potential PKC sites present in the two molecules, we could identifyneitheraCD2-like motif(PPGHR) (22)nor proline-richregions potentially involved in SH3binding(23).
Theseresultsargueagainst the previously discussed hypoth-esis (9)that thecytoplasmicpartofpTa couldplayarole in signal transduction of the pre-TCR through binding with proteinsbearing SH3 domainslike
p56Ick.
Indeed, the fact that pre-TCR isassociatedwith thesignaltransduction CD3 mol-ecules and that thephosphorylationpatternofT-cell activation is very similar afterpre-TCRorTCRaBcrosslinking (unpub-lished observations) could suggest that the cytoplasmic pTa tail hasno specialrole in signaltransduction.Thelackofidentitybetween the human andmurine cyto-plasmic tails couldreflect a divergence in gene structure. In
contrast to the human sequence, the murine pTa sequence containsaB2repetitive elementinthe 3' untranslatedregion thatprovidesthepolyadenylylationsite. This type ofelement is knowntogeneraterecombinationortranslocationeventsby insertionalmutagenesis. Moreover,adifference between the gene structure of the murine pTa gene and the TCRa and
TCR6
genes is that the pTa transmembrane region, the cytoplasmic tail, andthe 3' untranslatedregionareencodedbyasingleexon (24),whereas in TCRa and
TCR8
genes the3'+ + + + + C0 00 cs000O) C00 0 000 000 a 000cc C)OC)C )C)L )C)OuO I 11 -I 0 0 0 0 00. _, r, 0 0 0 0 0 0 0 00 00 0
o7
or
8 8 oi: pTa actinFIG. 4. Semiquantitative analysis of pTaexpressiononhumanthymocyte subsets.PCRamplifications of pTa- andactin-specific sequenceswere
performed onserial dilutionsof cDNAs. Amplifiedmaterialwasseparated byagarosegelelectrophoresis and stained withethidium bromide. Phenotypesof thethymic populations areindicated.
untranslated region is encoded by a separate exon. These differences could suggest that the 3' part of the pTa molecule has been subjected to variation. Analysis of the genomic organization of the human pTa should help to define whether differential splicing sites exist between human and murine
pTa.
The Zoo-Blotanalysis indicates again that a portion of the extracellular part of pTa is conserved in all the species analyzed and is compatible with the idea that some type of selective pressure may have conserved the extracellular do-main of the pTa but notthecytoplasmic region.
Overall, the human and murine pTa sequences for both the extracellular andcytoplasmic regions shownosimilarities with anypublished sequences in the data banks. We did not find any evidencefor the existence of a pTapseudogene when screen-ingthe library or in Southern blot analysis. Thus, unlike the A5 gene(32), the pTa gene maynotbelongto aclusterof similar genes.
The chromosomal location of pTa on the short arm of human chromosome 6 wasexpected after itwasshown that the murine pTa gene is located in theD/E1region of chromosome 17. In fact, these regions are syntenic and harbor the major histocompatibilitycomplexlocus.
Finally, theexpression pattern of pTa in the human thymus correspondstothatpreviouslyobserved in the murinethymus. Taking into consideration the differences in thymocyte surface markers in the two species, our data indicate that in both human and mouse pTa expression is developmentally regu-lated.
We thank M. Reth(Freiburg)forhelpful advice concerninganalysis of thepTasequence.TheBasel Institute forImmunologyissupported
by F. Hoffmann-La RocheLtd,Basel. This work was alsosupported
byInstitut National de la Santeetde la RechercheScientifique,Unite 373. H.v.B. is supported bythe Human Frontier Science Program
Organization.P.D.P. isaEuropean MolecularBiologyOrganization postdoctoralfellow.
1. Kisielow, P. & von Boehmer, H. (1995) Adv. Immunol. 58, 87-209.
2. Raulet,D.H., Garman, R. D.,Saito,H. &Tonegawa,S. (1985)
Nature (London)314, 103-107.
3. Pearse, M., Gallagher, P.F., Wilson, A., Wu, L., Fisicaro, N.,
Miller,J. F.A.P.,Scollay,R.&Shortman,K.(1988)Proc.Natl. Acad. Sci. USA 85, 6082-6086.
4. Snodgrass,H.R.,Dembic, Z.,Steinmetz,M. &vonBoehmer,H.
(1985)Nature (London)315,232-233.
5. Godfrey,D.I. &Zlotnik,A.(1993) Immunol. Today 14,547-553.
6. vonBoehmer,H.(1994) Cell76, 219-228.
7. Nossal, G. J. V. (1994)Cell 76, 229-239.
8. Groettrup, M.& vonBoehmer, H.(1993)Eur. J. Immunol. 23, 1393-1396.
9. Saint-Ruf, C.,Ungewiss, K., Groettrup, M., Bruno, L., Fehling, J. & vonBoehmer, H. (1994) Science 266, 1208-1212. 10. Groettrup, M., Baron, A., Griffiths, G., Palacios, R. & von
Boehmer, H. (1992) EMBO J. 11, 2735-2745.
11. Fehling, H. J., Krotkova, A., Saint-Ruf, C. & von Boehmer, H.
(1995)Nature (London)375,795-798.
12. Birnboim, H. C. & Doly, J. (1979) Nucleic Acids Res. 7, 1513-1525.
13. Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular
Cloning:A LaboratoryManual(Cold SpringHarbor Lab. Press, Plainview, NY), 2nd Ed.
14. Pearson, W. M. R. & Lipman, D. J. (1988)Proc.Natl.Acad. Sci. USA 85, 2444-2448.
15. Schibler, U., Tosi, M., Pittet, A. C., Fabiani, L. & Willauer, P. K.
(1980)J. Mol. Biol. 142, 93-116.
16. Loh, E. Y., Elliot, J. F., Cwirla, S.,Lanier, L. L. & Davis, M. M.
(1989)Science 243, 217-220.
17. Mattei, M. G., Philip, N., Passage, E., Moisan, J. P., Mandel, J. L. & Mattei,J. F. (1985)Hum. Genet. 69, 268-271.
18. Frohman, M. A.,Dush,M. K. &Martin, G. R.(1988)Proc.Natl. Acad. Sci. USA 85,8998-9002.
19. Williams,A. F.&Barclay,A. N.(1988)Annu.Rev. Immunol. 6, 381-405.
20. Hunkapiller, T.&Hood, L. (1989)Adv. Immunol. 44, 1-63. 21. Cosson, P., Lankford, S. P., Bonifacino, J. S. &Klausner, R. D.
(1991)Nature (London) 351,414-416.
22. Chang,H.C.,Moingeon, P., Pedersen, R.,Lucich,J.,Stebbins, C. & Reinherz, E. L.(1990)J.Exp. Med. 172, 351-355.
23. Yu, H., Chen, J. K., Feng, S., Dalgarno, D., Brauer, A. W. &
Schreiber, S. L.(1994) Cell76, 933-945.
24. Fehling,H.J.,Laplace, C., Mattei, M.G., Saint-Ruf, C. &von
Boehmer,H.(1995)Immunogenetics 42, 275-281.
25. Galy, A., Verma,S., Barcena, A. & Spits, H.(1993)J.Exp. Med. 178, 391-401.
26. Casorati, G., Traunecker, A. & Karjalainen, K. (1993) Eur. J.
Immunol. 23,586-589.
27. Simon,S. &Rajewsky,K. (1990)EMBO J. 9, 1051-1056. 28. Bentley, G. A., Boulot, G., Karjalainen, K. & Mariuzza, R. A.
(1995)Science267, 1984-1987.
29. Ossendorp,F.,Jacobs,H.,vanderHorst,G.,deVries,E., Berns, A. & Borst, J. (1992) J. Immunol. 148, 3714-3722.
30. Caspar-Bauguil,S.,Arnaud, J.,Huchenq,A.,Hein, W. R., Geis-ler,C. & Rubin,B. (1994)Scand. J.Immunol. 40, 323-336. 31. Geisler,C.,Rubin,B.,Caspar, B. S.,Champagne,E.,Vangsted,
A.,Hou, X. &Gajhede,M.(1992)J.Immunol. 148, 3469-3477. 32. Melchers, F., Karasuyama, H., Haasner, D., Bauer, S.,Kudo,A.,
Sakaguchi, N., Jameson,B. &Rolink,A.(1993) Immunol. Today
14, 60-68.
33. Kraft,D.L.,Weissman,I.L. &Waller,E. K.(1993)J.Exp. Med. 178, 265-277.