A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome
Texte intégral
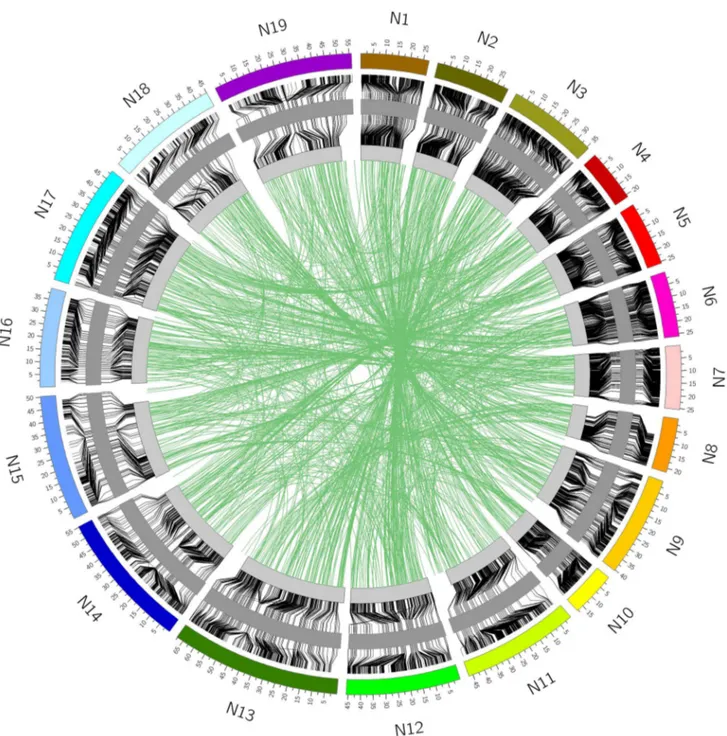
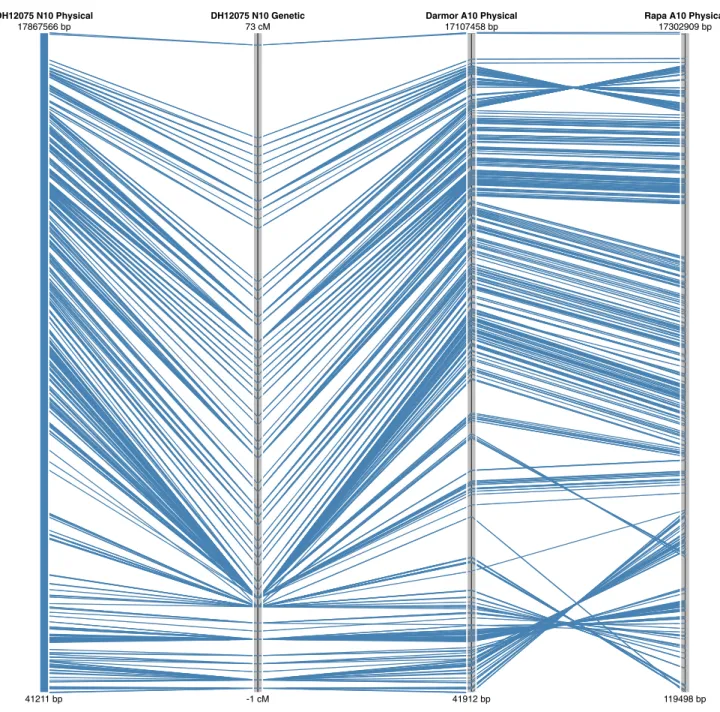
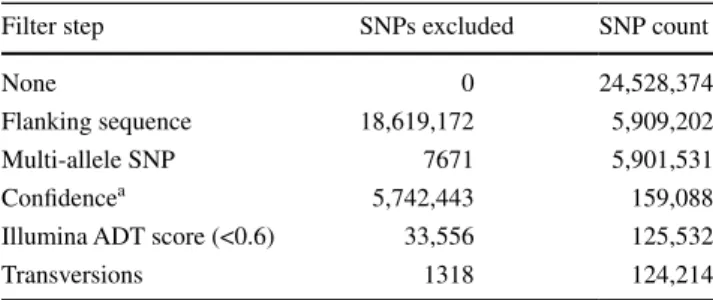
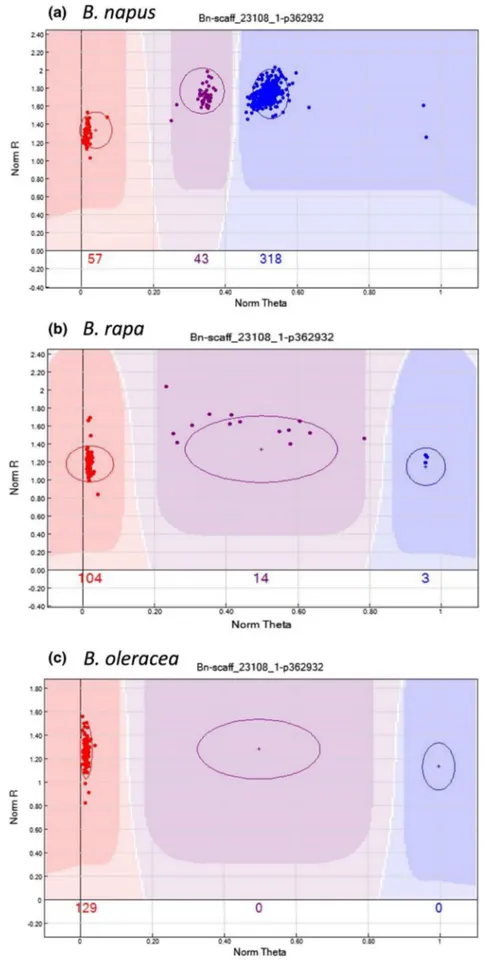
Figure
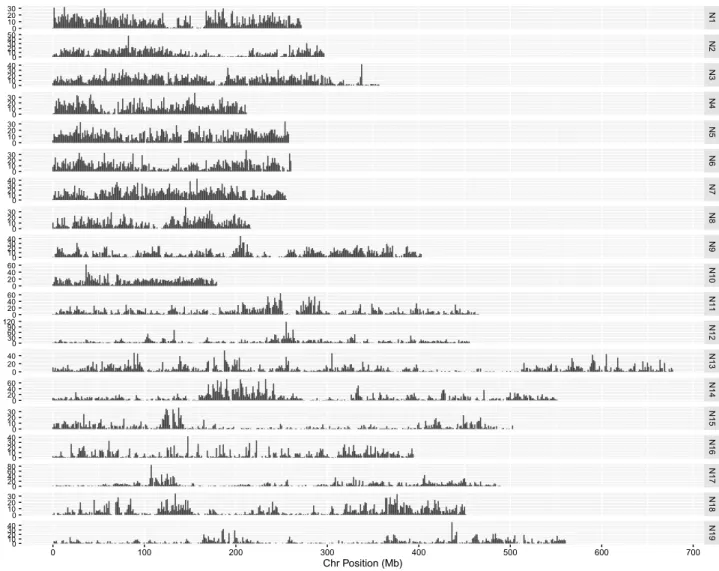
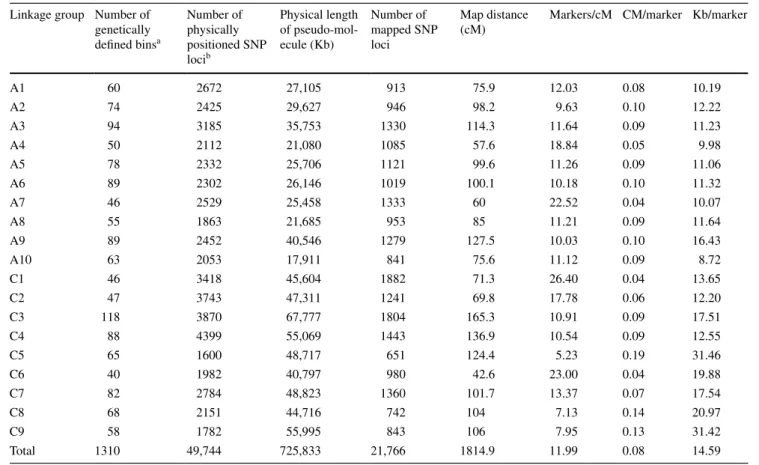
Documents relatifs
The experimental sideband intensities in the 31 P spectrum of Figure 4a, obtained after spinning a sam- ple at 400 Hz for 14 hours, are well described by a powder distribution of
In our framework, we focus on the antecedents of the fit between intended and realized brand personality that play an important role in terms of conceptual (derived from
The objectives of the work presented here are (1) to use SNPs previously identified in maize to develop a first reliable and standardized large scale SNP genotyping array; (2)
L’évaluation faite sur la prise en charge de l’anémie chez 130 patients en insuffisance rénale chronique terminale au niveau des services de néphrologie et d’hémodialyse de Bouira
Par contre si le scénario d’accords agricoles alternatifs a augmenté les prix aux producteurs et l’équité, il a aussi augmenté les dépenses publiques pour le financement
1) Postural recovery of a three link human model standing in gravity was simulated. 2) The simulations implicated that the proposed model can implement the
Figure 3: Single Linkage (left), Average Linkage (mid), Ward method (right) are used for clustering the simulated time series; Dashed lines represent the ratio of correct
Quantitative Trait Loci Mapping for Sugar-Related Traits in Sweet Sorghum based on High-Density SNP Markersd. Room: Grand