Molecular Ecology (2005) 14, 1151–1162 doi: 10.1111/j.1365-294X.2005.02476.x
Blackwell Publishing, Ltd.
Phylogeographical footprints of the Strait of Gibraltar
and Quaternary climatic fluctuations in the western
Mediterranean: a case study with the greater white-toothed
shrew,
Crocidura russula
(Mammalia: Soricidae)
J E A N - F R A N Ç O I S C O S S O N ,* R A I N E R H U T T E R E R ,† R O L A N D L I B O I S ,‡ M A U R I Z I O S A R À ,§
P I E R R E T A B E R L E T¶ and P E T E R V O G E L**
*Centre de Biologie et Gestion des Populations, INRA UMR 1062, Campus International de Baillarguet, CS 30016, 34988 Montferrier/ Lez cedex, France, †Zoologisches Forschungsinstitut und Museum A. Koenig, D-53113 Bonn, Germany, ‡Unité de Recherches Zoogéographiques, Institut de Zoologie, B-4020 Liège, Belgium, §Dipartimento di Biologia Animale, Università de Palermo, I-Palermo, Italy, ¶Laboratoire d’Ecologie Alpine (LECA), CNRS UMR 5553, Université Joseph Fourier, BP 53, F-38041 Grenoble cedex 9, France, **Department of Ecology and Evolution, University of Lausanne, 1015 Lausanne, Switzerland
Abstract
We used mitochondrial cyt b sequences to investigate the phylogenetic relationships of
Crocidura russula (sensu lato) populations across the Strait of Gibraltar, western Europe, Maghreb, and the Mediterranean and Atlantic islands. This revealed very low genetic divergence between European and Moroccan populations. The application of a molecular clock previously calibrated for shrews suggested that the separation of European from Moroccan lineages occurred less than 60 000 BP, which is at least 5 million years (Myr) after the reopening of the Strait of Gibraltar. This means that an overwater dispersal event was responsible for the observed phylogeographical structure. In contrast, genetic analyses revealed that Moroccan populations were highly distinct from Tunisian ones. According to the molecular clock, these populations separated about 2.2 million years ago (Ma), a time marked by sharp alternations of dry and humid climates in the Maghreb. The populations of the Mediterranean islands Ibiza, Pantelleria, and Sardinia were founded from Tunisian populations by overwater dispersal. In conclusion, overwater dispersal across the Strait of Gibraltar, probably assisted by humans, is possible for small terrestrial vertebrates. Moreover, as in Europe, Quaternary climatic fluctuations had a major effect on the phylogeographical structure of the Maghreb biota.
Keywords: Europe, human-assisted dispersal, mtDNA, North Africa, phylogeography, Quatern-ary history, speciation
Received 13 September 2004; revision received 1 January 2005; accepted 1 January 2005
Introduction
The relationships between the species currently found in Europe and Maghreb (the northern parts of Africa, i.e. Morocco, Algeria, and Tunisia) are thought to have been influenced mainly by the Strait of Gibraltar which acted as a geographical barrier for gene flow (Beerli et al. 1996; De Jong 1998; Prüser & Mossakowski 1998; Castella et al. 2000;
Palmer & Cambefort 2000; Gantenbein & Largiadèr 2003). During the Messinian period in the late Miocene, 5–6 Ma, the Mediterranean basin largely dried out because of the closing of the Strait of Gibraltar (Hsü et al. 1977; Krijgsman 2002). The resulting land corridors allowed biotic inter-changes between southern Europe and the Maghreb. The strait reopened at the start of the Pliocene, about 5 Ma, causing the refilling of the Mediterranean and the clos-ing of the terrestrial connection between Europe and the Maghreb. This event probably accounts for the vicariance observed in many Mediterranean lineages (Cheylan 1990; Zardoya & Doadrio 1998; Blondel & Aronson 1999; Dobson Correspondence: Jean-François Cosson, Fax: 04 99 62 33 45; E-mail:
1152 J . - F . C O S S O N E T A L .
& Wright 2000; Sanmartín 2003) and for the genetic differentiation between some Iberian and Maghreb biota (Castella et al. 2000; De Jong 1998; Harris & Sá-Sousa 2002; Gantenbein & Largiadèr 2003). Later on, particularly since 2.4 Ma, the Quaternary climatic oscillations profoundly affected the phylogeographical structure of Mediterranean biota (Hewitt 1996; Taberlet et al. 1998; Hewitt 1999). The ranges of most European species underwent several con-secutive contractions and expansions, with regular extinc-tions of the northern populaextinc-tions during ice ages, followed by subsequent northward expansions from southern Mediterranean peninsulas during interglacial periods. The isolation of populations in separate Mediterranean regions during the ice ages led to allopatric differentiation and strong phylogeographical structures (Hewitt 2001, 2004; Michaux et al. 2001). Comparatively little is known about the Quaternary evolutionary history of the fauna and flora of North Africa (Hewitt 2000). North Africa was regularly affected by alternating humid and hyperarid phases dur-ing the mid-Pliocene to Pleistocene (Street & Gasse 1981; Quezel & Barbero 1993) and a few recent studies suggest that these changing climatic conditions led to conspicuous phylogeographical footprints for some terrestrial animals (Brown et al. 2002; Guiller et al. 2001).
The greater white-toothed shrew (Crocidura russula) is
a good example of a mammalian species that has under-gone drastic changes in its taxonomic definition. The high
level of morphological variability within Crocidura species
resulted in numerous redescriptions of the same species
under different names since its first description as Sorex
russulus Hermann, 1780. The morphological similarity of populations of different species resulted in many errone-ous species-level assignments (Ellerman & Morrison-Scott 1966; Corbet 1978). The extensive use of karyological
tech-niques elucidated the distribution of C. russula, showing
that it is limited to the Maghreb and continental western Europe, from Portugal through Spain and France, Switzer-land, Germany and Austria (Reumer & Meylan 1986;
Genoud & Hutterer 1990; Vogel et al. 1990). Morphological
investigations covering the entire range of C. russula
re-vealed substantial differences between the populations of eastern and western Maghreb (Sarà & Vogel 1996; Contoli
& Aloise 2001). A molecular study (Vogel et al. 2003) using
the cytochrome b gene confirmed a deep split within
C. russula, with a western clade formed by samples from Spain and Morocco, and an eastern clade represented by a sample from Tunisia. Finally, a study based on mitochondrial
simple sequence repeats and 12S rRNA gene confirmed the
existence of two different clades (Lo Brutto et al. 2004).
The evolutionary history of C. russula is relatively well
documented by palaeontological records (Rzebik-Kowalska 1988). However, as in taxonomy studies, the difficulty of precise species identification using mandibles and skulls can lead to misinterpretation of fossil records. Based on the
compilation of the literature about fossil reports, Dobson (1998) and Dobson & Wright (2000) concluded that C. russula was native to Iberia and reached the Maghreb only recently. In contrast, based on chronological analyses
of some fossil assemblages, Poitevin et al. (1986) stated
that C. russula arrived in the south of France only after the
last glacial age. Along with Catzeflis (1984) and Vogel &
Maddalena (1987), Poitevin et al. proposed that the species
originated in the Maghreb.
The aim of our study was to determine the influence of the major climatic events that have occurred in the western Mediterranean since the reopening of the Strait of Gibraltar at the beginning of the Pliocene, on the evolutionary his-tory of the greater white-toothed shrew. We used a phylo-geographical approach to trace the history of the species in the region and to estimate the chronology of diversification events within the species. The phylogenetic relationships among populations were determined by analysis of
cyto-chrome b in numerous specimens collected from the entire
range of C. russula. The evolutionary history of the species
was profoundly influenced by Quaternary fluctuations in the Maghreb and by its ability to disperse across different stretches of water to reach continental Europe and the Atlantic and Mediterranean islands. We show that small terrestrial vertebrates dispersed across the Strait of Gibraltar and that, like in Europe, Quaternary climatic fluctuations profoundly influenced the phylogeographical structure of some Maghrebian biota.
Materials and methods
Forty-five Crocidura russula samples were used in this
study (Fig. 1). Specimens, origins, and identification codes
are given in Table 1. We included Crocidura russula osorio
from Gran Canaria in the data set, as this taxon was
recently shown to be conspecific with C. russula (Molina
et al. 2003; Vogel et al. 2003). Shrews were brought to the laboratory alive or preserved in liquid nitrogen in the field,
then stored at −70 °C, and finally preserved in 80% ethanol
until DNA extraction. In some cases, we extracted DNA directly from bones collected in owl pellets, as was the case from several localities in France and Belgium.
Total genomic DNA was extracted by digestion with
proteinase K for 4–8 h at 37 °C. The resulting DNA was
purified by extracting twice with phenol–chloroform and once with chloroform. The sample was then desalted and concentrated by ethanol precipitation. DNA was extracted from owl pellets as described by Taberlet & Fumagalli (1996). Double-stranded DNA was amplified with the primers L14841: TCAAACATCTCATCATGATGAAA, H15149:
CCT-CAGAATGATATTTGTCCTCA and H15915:
TCATCT-CCGGTTTACAAGAC. Polymerase chain reaction (PCR) conditions consisted of 30–35 cycles of 30–60 s denaturation
P H Y L O G E O G R A P H Y I N T H E W E S T E R N M E D I T E R R A N E A N 1153
at 72 °C (depending on the length of the sequence amplified).
PCR products were then subjected to electrophoresis on a 1% agarose gel and stained with ethidium bromide staining to verify product size. Bands of the predicted sizes were excised then purified using the QIAquick PCR Purification Kit (QIAGEN), according to the manufacturer’s instructions. The PCR products were sequenced using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit
(Perkin-Elmer) in a 20-µL volume containing 15–80 ng of
purified DNA (depending of the length of the PCR prod-uct) and 3.2 pmol of primer, following the manufacturer’s specifications. The sequencing conditions were 25 cycles of
30 s at 96 °C, 30 s at 50–58 °C (depending on the primer),
and 4 min at 60 °C on a PE 9600 thermocycler. Excess dye
terminators were removed by spin-column purification. Sequencing reactions were subjected to electrophoresis for 4–7 h on an ABI PRISM 377 DNA sequencer (Perkin-Elmer) on a 5% Long Ranger gel (FMC).
Most of the cytochrome b gene (1074 pb) was amplified
and sequenced (one direction) with the primers L14841
and H15915 for 15 selected individuals. Additional short sequences were amplified for 30 more samples with the
primers L14841 and H15149. Two long sequences
belong-ing to other Palaearctic Crocidura species (Crocidura
suaveo-lens and Crocidura leucodon) were included for comparison
and phylogenetic reconstruction (Vogel et al. 2003). Table 1
gives the lengths of the sequenced fragment for each
sam-ple. The long cytochrome b sequences (998 bp) were added
to the GenBank/EMBL databases. Nucleotide diversity, π,
and other statistics concerning the 273-bp DNA sequences
were estimated using the dnasp 3.99 program (Rozas &
Rozas 1999).
Nucleotide sequences were aligned by eye. As is
common for cytochrome b sequences from closely related
species, no insertions or deletions were observed. Three different phylogenetic analysis methods (distance, maxi-mum parsimony, and maximaxi-mum likelihood) were carried
out using paup* version 4.0b8 PPC (Swofford 1998). As no
evidence of saturation for the cytochrome b gene within
European Crocidura species was previously observed
(Vogel et al. 2003), all substitutions were taken into account
for phylogenetic analyses. A network of 998-bp long haplo-types was also constructed using the statistical parsimony
method with the tcs software (Clément et al. 2000).
For maximum-likelihood (ML) analyses, we selected a mutation model according to the protocol of Posada
& Crandall (1998) using modeltest 3.6 witch test for 56
models of evolution. This is a TrN + I + G model in which the proportion of invariable sites is estimated from the data (0.568), with an unequal distribution of rates at variable Fig. 1 Geographical distribution of the sample sites of the greater white-toothed shrew (Crocidura russula) and haplotypes identified in this study. Actual locations are indicated in Table 1.
1154 J . - F . C O S S O N E T A L .
sites (gamma shape parameter = 1.419) and three different substitution types [rate (A-G) = 12.12; rate (C-T) = 17.28; and other rates = 1.00]. ML analyses were performed assum-ing this model and usassum-ing the full heuristic search option
with a stepwise addition of sequences. As the sequence of taxon entry in phylogenetic reconstruction can bias the position in the tree (Maddison 1991), we systematically used 10 repeated randomized input orders for ML analyses. Table 1 Species sequenced, geographical origin of the samples, voucher codes (IZEA, Department of Ecology and Evolution, University of Lausanne; JFC, Centre de Biologie et Gestion des Populations, Montferrier), length of the cytochrome b gene sequenced and haplotype designations for short and long sequences
Sample location Identification codes
Cyt b sequences
Length (bp) Haplotype designation
Crocidura russula russula Breskens, the Netherlands IZEA1084 998 Ru1 Ru1.2
Crocidura russula russula Bonn, Germany IZEA752 998 Ru1 Ru1.3
Crocidura russula russula Liège, Belgium JFC-R10 273 Ru1
Crocidura russula russula La Chapelle d’Huin, France JFC-R13 273 Ru1
Crocidura russula russula Bréhat, France JFC-R9 273 Ru1
Crocidura russula russula Roscoff, France IZEA5541/842 273 Ru1
Crocidura russula russula Molène, France JFC-R8 273 Ru1
Crocidura russula russula Sein Island, France 9/JFC-R7 998 Ru1 Ru1.4
Crocidura russula russula Sein Island, France 19/JFC-R6 998 Ru1 Ru1.4
Crocidura russula russula Glénan (St Nicolas), France JFC-R5 273 Ru1
Crocidura russula russula Groix, France JFC-R4 273 Ru1
Crocidura russula russula Touchay, Cher, France JFC-R11 273 Ru1
Crocidura russula russula Morges, Vaux, Switzerland IZEA812 273 Ru1
Crocidura russula russula Laubertie, France JFC-R2 273 Ru1
Crocidura russula russula Commarque, France JFC-R3 273 Ru1
Crocidura russula russula Montpellier, France JFC-R12 273 Ru1
Crocidura russula russula Nice, France JFC-R13 273 Ru1
Crocidura russula russula Banyuls, France JFC-R1 273 Ru1
Crocidura russula russula Candelario, Salamanque, Spain IZEA5936 998 Ru1 Ru1.1
Crocidura russula russula Albaracin, Teruel, Spain IZEA650 273 Ru1
Crocidura russula russula Albaracin, Teruel, Spain IZEA652 273 Ru1
Crocidura russula russula Peniche, Madure, Portugal IZEA5897 273 Ru1
Crocidura russula russula Murillo, Logroño, Spain IZEA5788 998 Ru1 Ru1.2
Crocidura russula yebalensis Skhirat, Rabat, Morocco IZEA2641 998 Ru2 Ru2.1
Crocidura russula yebalensis Moulay Bou Selham, Morocco IZEA2642 998 Ru2 Ru2.2
Crocidura russula yebalensis Oukaimeden, Asni, Morocco IZEA1239 273 Ru2
Crocidura russula yebalensis Imlil, Asni, Morocco IZEA2631 273 Ru2
Crocidura russula yebalensis Imlil, Asni, Morocco IZEA2633 273 Ru2
Crocidura russula yebalensis Marrakech, Morocco IZEA5058 273 Ru2
Crocidura russula yebalensis Marrakech, Morocco IZEA5060 273 Ru2
Crocidura russula yebalensis Casablanca, Morocco IZEA5073 273 Ru2
Crocidura russula yebalensis Oukaimeden, Asni, Morocco IZEA1572 998 Ru3 Ru3.1
Crocidura russula yebalensis Aguelmane Aziga, Khenifra, Morocco IZEA1237 998 Ru4 Ru4.1
Crocidura russula yebalensis Aguelmane Aziga, Khenifra, Morocco IZEA1582 273 Ru4
Crocidura russula yebalensis Moulay Bou Selham, Morocco IZEA2644 273 Ru5
Crocidura russula yebalensis Skhirat, Rabat, Morocco IZEA2639 273 Ru6
Crocidura russula agilis Aïn Draham, Tunisia IZEA4011 998 Ru7 Ru7.1
Crocidura russula agilis Tunisia IZEA4126 273 Ru8
Crocidura russula ichnusae Gallura, Sardinia, Italy IZEA649 273 Ru9
Crocidura russula ichnusae Gallura, Sardinia, Italy IZEA5680 998 Ru9 Ru9.1
Crocidura russula ibicensis Ibiza, Balearic Archipelago, Spain Libois-I36 273 Ru10
Crocidura russula ibicensis Ibiza, Balearic Archipelago, Spain Libois-I42 273 Ru10
Crocidura russula cossyrensis Montagna Grande, Pantelleria, Italy IZEA3895 998 Ru11 Ru11.1
Crocidura russula cossyrensis Siba, Pantelleria, Italy IZEA4184 998 Ru11 Ru11.2
Crocidura osorio Gran Canaria, Canary Islands, Spain IZEA3707 998 Os1 Os1.1
Crocidura suaveolens Candelario, Salamanca, Spain IZEA1203 998 Su1 Su1.1
P H Y L O G E O G R A P H Y I N T H E W E S T E R N M E D I T E R R A N E A N 1155 Neighbour-joining (NJ) phylogenetic trees were constructed
using the previously selected ML genetic distance using heuristic search with tree-bisection–reconnection (TBR) branch-swapping. Parsimony analyses were performed using the branch-and-bound search option; all characters having the equal weight.
Maximum likelihood, distance and parsimony results were compared for congruence of tree topologies. To test the robustness of nodes, we conducted a thorough boot-strap procedure using ML reconstructions with the TrN + I + G model, TBR branch-swapping algorithm, and 10 random input orders with stepwise-addition of sequences (500 pseudoreplicates for each). Support for nodes was also evaluated with the bootstrapping method using 500 pseudo-replicate data sets for parsimony and distance analyses.
The molecular clock hypothesis was tested using long sequences and according to Posada & Crandall (1998), by calculating the log-likelihood score with molecular clock enforced and comparing it with the log likelihood pre-viously obtained without enforcing the molecular clock. In our case, the degree of freedom for the likelihood ratio test was 13 (number of OTUs = 2). Relative-rate tests were also done between the different lineages observed within C. russula. Tests were conducted with each lineage against the remaining lineages on the proportions of synonymous (Ks) and nonsynonymous (Ka) substitutions using rrtree version 1.0 (Robinson et al. 1998). The NJ tree rooted with C. suaveolens and C. leucodon sequences was chosen as the reference.
Divergence time was estimated from the molecular data according to the calibration developed for Soricidae by Fumagalli et al. (1999). This calibration is based on an estimate of 20 Myr for the split between Crocidurinae and Soricinae shrews and was developed considering cytochrome b sequence divergences based on third-position
transver-sions (tv). The estimate of divergence rate at third-position tv in shrews is 1.36% per Myr.
Results
The sequencing of the PCR fragments revealed 12 different haplotypes among the 998-bp sequences, and 11 different haplotypes among the 273-bp sequences. Within the long sequences, 247 sites were variable, of which 121 (49%) were parsimony-informative (including outgroups).
Pairwise sequence divergences (Kimura 2-parameter distances was chosen for comparison with other studies) varied from 0.001 to 0.091 across the different lineages of Crocidura russula (Table 2). Most of these values are within the range of variation observed between mammalian taxa at the intraspecies level, although the maximal values (up to 0.091) are particularly high (Avise et al. 1998; Johns & Avise 1998; Bradley & Baker 2001).
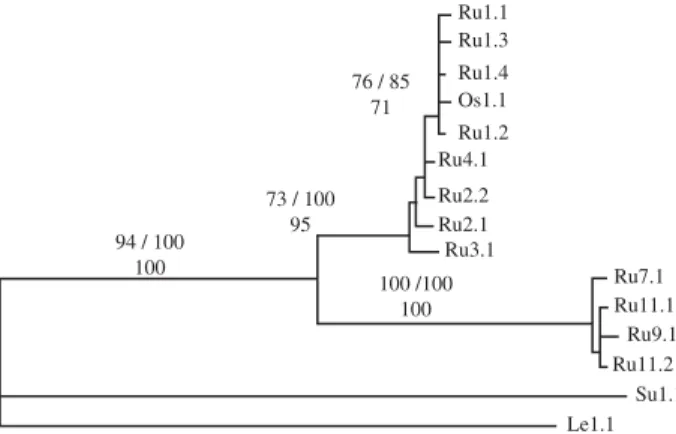
Phylogenetic relationships between haplotypes are given in Figs 2 and 3. As all methods, either using short or long sequences, gave similar topologies, only the tree resulting from maximum likelihood is shown, completed for long sequences with the bootstrap values from the NJ and MP analyses. The ML method gave one tree of score –ln L = 2771.20. The distance method gave one tree of minimum evolution score equal to 0.596. The parsimony analysis provided 32 most parsimonious trees (CI = 0.81 excluding uninformative characters) with a tree length of 318 steps. The majority-rule consensus of the 32 MP trees had similar topology as the ML tree shown in Fig. 2.
The three phylogenetic methods strongly support the integrity of the clade grouping all sequences from the spe-cimens referred to as C. russula, apart from those of the two outgroups, Crocidura suaveolens and Crocidura leucodon. Within the C. russula lineages, the only differences between
Table 2 Nucleotide divergence values for the 998-bp sequences calculated with the Kimura 2-parameter method
Ru1.1 Ru1.2 Ru1.4 Ru1.3 Ru1.5 Os1.1 Ru2.1 Ru2.2 Ru3.1 Ru4.1 Ru7.1 Ru11.1 Ru11.2
Ru1.1 — Ru1.2 0.00301 — Ru1.4 0.00503 0.00201 — Ru1.3 0.00503 0.00201 0.00402 — Ru1.5 0.00807 0.00503 0.00301 0.00705 — Os1.1 0.00705 0.00402 0.00604 0.00604 0.00705 — Ru2.1 0.00705 0.00402 0.00604 0.00604 0.00503 0.00604 — Ru2.2 0.00604 0.00301 0.00503 0.00503 0.00402 0.00503 0.00100 — Ru3.1 0.01419 0.01215 0.01215 0.01419 0.01113 0.01418 0.01012 0.00910 — Ru4.1 0.00705 0.00402 0.00604 0.00604 0.00503 0.00604 0.00302 0.00201 0.01115 — Ru7.1 0.08951 0.08723 0.08723 0.08951 0.08496 0.08827 0.08503 0.08620 0.08973 0.08620 — Ru11.1 0.08951 0.08723 0.08723 0.08951 0.08496 0.08827 0.08503 0.08620 0.08973 0.08620 0.00909 — Ru11.2 0.08958 0.08730 0.08730 0.08958 0.08503 0.08834 0.08510 0.08627 0.08981 0.08627 0.00706 0.00201 — Ru9.1 0.09062 0.08834 0.08834 0.09062 0.08606 0.08938 0.08613 0.08730 0.09084 0.08730 0.01010 0.00503 0.00503
1156 J . - F . C O S S O N E T A L .
the different phylogenetic methods were the relative posi-tions of the sequences into three well-defined clades. The first clade groups all European specimens, referred to as subspecies Crocidura russula russula, and specimens of Crocidura russula osorio from Gran Canaria. This grouping
is supported by high bootstrap values (76% for ML, 71% for MP, and 85% for NJ). The second clade groups sequences from the first clade with sequences from Moroccan speci-mens with high bootstrap values (73% for ML, 100% for MP, and 95% for NJ). Finally, the third and independent clade, groups sequences from Tunisia with those from the Mediterranean islands, is also supported by very high bootstrap values (100% for ML, 100% for MP, and 100% for NJ).
The statistical parsimony network estimated that the parsimony connection limit was 13 mutations in our data set (i.e. meaning that the maximum number of differences among haplotypes that are not the result of multiple substitutions at a single site is 13 with a 95% statistical confidence). As a result of their strong differentiation, east-ern (Tunisia and Mediterranean islands) and westeast-ern (Morocco & Europe) lineages are not connected by the network reconstruction (Fig. 4). The number of steps (i.e. mutations) separating the two clades was estimated to be 80. Within the western clade, haplotypes from Europe and Gran Canaria were derived from one ancestral haplotype not sampled in our data set. Within the eastern clade, haplo-types from the Mediterranean islands were derived from a common ancestral haplotype sampled in Pantelleria (as shown also by the phylogenetic reconstruction in Fig. 3).
Haplotype and nucleotide diversities within the 273-bp sequences varied greatly according to geographical origin, with North African ones showing high diversities and European ones showing no diversity at all. Among the North African haplotypes, those from Tunisia and Medi-terranean islands were slightly more diverse than those from Morocco (Table 3).
The likelihood ratio test led us to accept the molecular clock hypothesis for the long cytochrome b sequence data
(χ2 = 12.93, d.f. = 13, P = 0.55). The relative rate tests
re-vealed no significant heterogeneity between the different lineages for synonymous or nonsynonymous substitutions (P > 0.20 for each test).
According to Fumagalli et al. (1999) the divergence rate at third-position tv in shrews is 1.36% per Myr. This calibration helped us to estimate the divergence times between the main lineages within C. russula, although it Fig. 2 Phylogeny of the 998-bp cytochrome b sequences analysed
with the maximum-likelihood method using the TrN + I + G model of substitution and TBR branch swapping. The values at the branches are bootstrap values for maximum likelihood/distance analyses (percentage of 500 replications for each of 10 random orders for stepwise addition of sequences). Bootstrap values for the parsimony analysis using the branch-and-bound option are also indicated (percentage of 500 replications). Codes are as in Table 1.
Fig. 3 Phylogeny of the 273-bp cytochrome b sequences analysed with maximum-likelihood method using the TrN + I + G model.
Table 3 Number of samples and haplotypes, number of polymorphic sites, haplotype diversities and nucleotide diversities for the 273-bp sequences from different geographical regions
Europe North Africa Morocco Tunisia & islands
No. of examined samples 23 21 13 8
No. of different haplotypes 1 10 5 5
Polymorphic sites 0 13 7 6
Haplotype diversity 0.000 ± 0.000 0.848 ± 0.069 0.681 ± 0.132 0.893 ± 0.086
P H Y L O G E O G R A P H Y I N T H E W E S T E R N M E D I T E R R A N E A N 1157
could not be used directly because of the low numbers of third-position tv observed. We thus estimated the cor-relation between the third-position tv and the maximum-likelihood distances inferred from an ML tree constrained to clock-like evolution after adding published sequences of the main lineages of Crocidura species in Europe (Vogel et al. 2003). A fairly good correlation was observed (r = 0.973, P < 0.001, Fig. 5) giving a rate of divergence of 0.051 increase in ML distance per Myr, with a 95% confidence interval of 0.044–0.070 per Myr. The ML distances between main lineages were then estimated using the formula in Edwards (1997) which corrects for ancestral mtDNA poly-morphism within lineages. The rate of ML distance
increas-ing with time (0.051 per Myr) was then applied to the dif-ferent dichotomies within C. russula, suggesting that the European and Moroccan populations separated from the Tunisian and Mediterranean island populations 2.21 (1.61– 2.57) Ma; and that the European populations separated from the Moroccan populations 0.06 (0.05–0.08) Ma. These divergence dates are only approximate because of the low precision in dating fossil records and stochastic events occurring during lineage evolution within species (Edwards & Beerli 2000; Nichols 2001). However, they give useful information about the timing of the major splits that occurred within C. russula over its present range.
Discussion
Basic remarks about the phylogeography of the Crocidura
russula group
Our investigation was based on a large sample covering the entire distribution of the Crocidura russula group. Our results confirm the monophyly of the shrews sharing a karyotype of 2N = 42 chromosomes, as already described by Vogel et al. (2003). Shrews from Gran Canaria, Ibiza, Sardinia, and Pantelleria were clearly included in the C. russula clade. Haplotypes from North Africa are far more diverse than those collected in Europe. This supports the hypothesis that the C. russula group basically evolved in North Africa (Vogel & Maddalena 1987). We observed a deep split between western haplotypes (Morocco) and eastern ones (Tunisia) in the Maghreb. Lineages from Europe and Gran Canaria were closely related to those from Morocco, whereas haplotypes from Mediterranean islands were closely related to Tunisian lineages.
Fig. 5 Correlation between the number of third-position trans-versions and maximum-likelihood distances inferred with the TrN + I + G model of substitution for published cytochrome b sequences of the main lineages of the Crocidura species in Europe.
Fig. 4 Relationships between the 998-bp cytochrome b sequences reconstructed with the maximum-parsimony method and tcs. Small, white circles represent hypothetical haplotypes not found in the sample.
1158 J . - F . C O S S O N E T A L .
The colonization of Europe across the Strait of Gibraltar
The divergence between European and Moroccan popula-tions was unexpectedly low, suggesting overwater dispersal across the 10- to14-km wide Strait of Gibraltar, rather than a vicariance event dating back to the reopening of the strait. This very low diversity (Table 3) and the star-like topology of the network of European haplotypes (Fig. 4) suggest a strong bottleneck event, consistent with the recent invasion of the area by a fewer number of indi-viduals. The relationship between European and Moroccan haplotypes suggests that the source population evolved in Morocco. However, the lack of samples from the south of the Iberian Peninsula does not make it possible to rule out the possibility of divergence of populations from the south of Spain followed by a recent expansion to the northern part of Europe, presumably following the last glacial age. Further studies are required to resolve this question.
The Strait of Gibraltar is deep and steep-sided. Its width did not decrease much when sea levels were low, and the strait did not dry out during the Quaternary. However, bathymetric maps of the western part of the strait indicate the occurrence of substantial shoals that became large islands during ice ages when sea levels were low. The length of the strait itself was increased to the west where some islands reduced the passage towards the Atlantic Ocean, forming a somewhat small interior and safe sea (Fig. 4 in Nehren 1992; Fig. 1 in Collina-Girard 2001). Islands probably formed visible land masses covered by vegetation, completely changing the appearance of the strait from either shore, and providing a stepping stone for humans to cross from Africa to Europe (Flemming et al. 2003). When sea levels were low, the maximum distance between two land masses from Morocco to Spain was only about five kilometres.
Crocidura russula is clearly unable to swim the distance between Africa and Europe, and even between either of the two continents and the Mediterranean islands. However, rafting on a natural support may potentially have occurred even though biogeographical data concerning the western Mediterranean (Dobson 1998) and other parts of the world (Heaney 1986) suggest that such events are extremely rare. The alternative means of crossing the Strait of Gibraltar are even less plausible. The connection of Moroccan and Iberian populations through a terrestrial route along the southern Mediterranean shore, via corridors along the Mediterranean coasts of the Sahara and the Levant, up to eastern Mediterranean, then back to the western Mediter-ranean along the northern shores, is hard to believe based on the existing palaeontological literature for the period and the area (Kowalski 1991; Geraads 1995; Tchernov 1996) and given that the species is currently absent from eastern parts of Europe and North Africa. Likewise, a crossing through the Sicily channel, which separates Tunisia from
Sicily (Italy), is not really compatible with the current ranges of the two C. russula lineages, as Tunisian lineages are well differentiated from Iberian and Moroccan ones.
Given the number and variety of small mammal species introduced to the Mediterranean islands (Vigne 1992), and the recent introduction of the North African wood mouse (Apodemus sylvaticus) from western Europe (Libois et al. 2001), it is possible that human activities led to the trans-location of C. russula from Morocco to Spain. Our molecular dating analysis suggested that this species was transferred from Morocco to Europe across the Strait of Gibraltar about 60 000 (50 000–80 000) bp. This estimate coincides with pos-sible human dispersal across the Strait of Gibraltar in the middle and upper Pleistocene. This question has been debated periodically for decades and still remains a hot issue for current archaeologists (Straus 2001). The records of human contacts between North Africa and Iberia are patchy and ambiguous, but there are data indicating that humans have migrated between these two areas since the middle Pleistocene (Alimen 1975). According to Straus (2001), the Strait of Gibraltar was a very effective natural barrier to human movements throughout the Pleistocene (between c. 30 000 and 40 000 bp). The first solid evidence of humans crossing the Strait of Gibraltar (similarity between lithic assemblages, marine fishing, and probable navigation) appears in the terminal Palaeolithic, about 25 000 bp (Bouzouggar et al. 2002; Collina-Girard 2003).
The discrepancy between our molecular timing analysis and the oldest documented trans-Strait of Gibraltar human movements may result from an overestimation of population divergence time from our molecular data, or just from lack of archaeological data. It is worth noting that haplotype divergence analyses systematically overestimate the amount of time that has passed before populations separated because haplotypes can diverge within the ancestral popu-lation well before the popupopu-lations separate (Taberlet et al. 1998). A number of factors linked to demography, genetic drift, and genetic structure within the source population may have strong effects, making it hard to estimate the amplitude of the discrepancy between haplotype and popu-lation splits (Nichols 2001).
The phylogeographical structure within the Maghreb
The genetic differentiation between the western and eastern clades is particularly high (mean = 8.5%) in com-parison with the general range of variation observed between mammalian taxa at the intraspecific level (Avise et al. 1998; Johns & Avise 1998; Bradley & Baker 2001). This genetic differentiation is far greater than that observed within each of these clades (mean = 0.4% for western clade, and mean = 0.6% for eastern clade). This explains also the high Nei’s genetic distances observed in allozyme studies between continental European populations (western clade)
P H Y L O G E O G R A P H Y I N T H E W E S T E R N M E D I T E R R A N E A N 1159 and insular populations (eastern clade) of Sardinia (D =
0.084, Catzeflis 1984), Ibiza (D = 0.102, Catalan et al. 1988), and Pantelleria (D = 0.15, Vogel et al. 2004). Based on our molecular data, the split occurred around 2.2 Ma, a time of great palaeoclimatic variability in the area, which was char-acterized by rapid alternations of dry and humid periods (Kowalski 1991). These climatic fluctuations may have played a central role in local adaptation and gene flow disruption between ancient C. russula populations in Algeria.
Because of the lack of sampling in Algeria, we do not know exactly where the two observed lineages meet in the field. The investigation by Sarà & Vogel (1996), based on multivariate analysis of mandibles and skulls collected in several sites from Tunisia to Morocco, showed a stepped cline with a transition zone situated in eastern Algeria. The fact that one outlier specimen from a Tunisian locality (Lac Melleguè) grouped with the Moroccan cluster (Sarà & Zanca 1992) may be an indication of a patchy pattern of distribution of morphotypes within the area. Further investigations along the Tunisian–Algerian border are needed to determine the characteristics of the contact zone of both clades and morphotypes in the field.
The colonization of the Atlantic and Mediterranean
islands
According to our data, the Atlantic islands were recently colonized by source populations from the western clade (Europe–Morocco). C. russula probably reached some of the continental islands along the west coast of France during the regression of the Holocene sea, given its presence on all islands separated from the continent by a shallow stretch of sea (Cosson et al. 1996). Some more remote islands, subjected to regular boat traffic with the continent, were colonized later on with the accidental help of humans. This was probably the case for Gran Canaria following the Spanish colonization (Vogel et al. 2003) and for the tiny Sein Island in Brittany (Cosson et al. 1996).
Greater white-toothed shrews found on the Mediter-ranean islands (Ibiza, Sardinia, and Pantelleria) were all derived from the eastern clade (Tunisia). The most recently discovered taxon was Crocidura cossyrensis Contoli, which was found on Pantelleria (an island situated between Sicily and Tunisia) in 1989. There is some debate about whether this is indeed a real species (Contoli et al. 1989; Contoli 1990, 1992; Vogel et al. 1992; Hutterer 1993; Vogel et al. 2004). Our study shows that C. cossyrensis Contoli is closely related with C. russula populations from Tunisia.
No fossils of C. russula from before the Holocene, and more precisely from before the development of regular and intensive boat traffic between the islands and the con-tinents, have been found on the Mediterranean islands. Whether introduction was passive or deliberate is still a matter of debate, but there is good evidence and a general
consensus that C. russula, like other mammal species, was introduced by humans (Alcover 1980; Cheylan 1984; Vigne & Alcover 1985). Small mammals like shrews probably reached islands during the trade of agricultural goods, as has previously been suggested for the introduction of Crocidura suaveolens in Crete during the Minoan period (Vogel et al. 1986). According to Alcover & Vesmanis (1985), C. russula arrived in Sardinia around 8000 bp. Its arrival in Ibiza is not documented (Alcover 1980).
Insights into the Quaternary legacy within the Maghreb
Hewitt (2000) used the term ‘Quaternary legacy’ to encom-pass evolutionary events related to climatic fluctuations during the Quaternary. In contrast to other continents and even sub-Saharan Africa, very little is known about the footprints left by the Quaternary climatic fluctuations in North Africa. The Maghreb region provides an interesting example in this regard. The region is limited to the west by the Atlantic Ocean and to the north by the Mediterranean Sea. The geographically close Iberian Peninsula is sep-arated by the Strait of Gibraltar to the north. East and south, the Maghreb is bordered by an arid zone that extends for several thousand kilometres across the Sahara Desert. Palaeontological records indicate that numerous major ecological changes have occurred within the region in the last million years (Jamet 1991). Recent climatic models and studies of Holocene plant fossils suggest that the western Sahara desert was considerably smaller during the time between 2 and 3 Ma. A long-lasting wet phase that ended around 1.6 Ma was immediately followed by a hyperarid period around 1.5 Ma. The size and aridity of the Sahara Desert then continued to fluctuate consider-ably, with a frequency of about 41 000 years over between 0.9 and 1.5 Ma, then slowly (100 000 years) from 0.1 to 0.9 Ma, and more rapidly again thereafter. The western Sahara was probably particularly small in the early and mid-Holocene (6000–9000 bp) with the expansion of grass-land into the Sahara (Jolly et al. 1998; Claussen et al. 2003). Throughout the Quaternary, North Africa and the Sahara were characterized by an Artemisia steppe intermingled with very big lakes (Suc et al. 1995; Faure 1987). This en-vironment expanded and retracted according to climatic oscillations, until the last glacial Würmian age, when two refuge areas in northwestern and northeastern Africa existed (Brown & Gibson 1983; Jolly et al. 1998).
It is possible that Maghreb biota regularly expanded to the south during mesic periods when most of the western part of the Sahara was vegetated by xerophytic woods/ shrub and warm grass (Claussen & Gayler 1997). During arid periods, the isolation of populations in separate Maghreb refuges (Brown & Gibson 1983) may have led to allopatric differentiation and strong phylogeographical structures. This phenomenon is currently poorly understood,
1160 J . - F . C O S S O N E T A L .
but is thought to explain morphological variations in the lizard Acanthodactylus erythrurus (Bons & Geniez 1995) and the land snail Helix aspersa (Madec et al. 1996), as well as genetic differentiation in the agamid lizards Agama impalearis (Brown et al. 2002) and H. aspersa (Guiller et al. 2001). Our study of the C. russula group provides further evidence of morphological and genetic variations related to the Quatern-ary period within the Maghreb.
Acknowledgements
PV acknowledges the former collaboration of F. Catzeflis and T. Maddalena. We are grateful to Lori Handley for critical remarks and to Sylvain Piry for help in preparing figures. Funding was provided by the Institut National de la Recherche Agronomique. The work of RH in Sardinia was funded by the Alexander-Koenig-Stiftung.
References
Alcover JA (1980) Note on the origin of the present mammalian fauna from the Balearic and Pityusic islands. Miscellánia
Zooló-gica, 6, 141–149.
Alcover JA, Vesmanis I (1985) Sobre les restes subfossils de la musaranya de dents blanques Crocidura russula (Hermann, 1780) de la Grotta su Guanu, illa de Sardenya (Mammalia, Insecti-vora). Endins, 10, 63–70.
Alimen H (1975) Les «Isthmes» hispano-marocain et sicilo-tunisien aux temps acheuléens. L’Anthropologie, 79, 399–436.
Avise JC, Walker D, Johns GC (1998) Speciation and Pleistocene effects on the vertebrate phylogeography. Proceedings of the
Royal Society of London. Series B, Biological Sciences, 265, 1707–1712.
Beerli P, Hotz H, Uzzell H (1996) Geologically dated sea barriers calibrate a protein clock for the Aegean water frogs. Evolution, 50, 1676–1687.
Blondel J, Aronson J (1999) Biology and Wildlife of the Mediterranean
Region. Oxford University Press.
Bons J, Geniez P (1995) Contribution to the systematics of the lizard Acanthodactylus erythrurus (Sauria, Lacertidae) in Morocco.
Herpetological Journal, 5, 271–280.
Bouzouggar A, Kozlowski JK, Otte M (2002) Study of the Aterian lithic assemblages from El Aliya cave in Tangier (Morocco).
L’Anthropologie, 106, 207–248.
Bradley RD, Baker RJ (2001) A test of the genetic species concept: cytochrome-b sequences and mammals. Journal of Mammalogy, 82, 960–973.
Brown JH, Gibson AC (1983) Biogeography. Mosby Co, St. Louis. Brown RP, Suárez NM, Pestano J (2002) The Atlas mountains as a
biogeographical divide in North-West Africa: evidence from mtDNA evolution in the agamid lizard Agama impalearis.
Mole-cular Phylogenetics and Evolution, 24, 324–332.
Castella V, Ruedi M, Excoffier L, Ibanez C, Arlettaz R, Hausser J (2000) Is the Gibraltar Strait a barrier to gene flow for the bat
Myotis myotis (Chiroptera: Vespertillionidae)? Molecular Ecology,
9, 1761–1772.
Catalan J, Poitevin F, Fons R, Guerasimov S, Croset H (1988) Biologie évolutive des populations ouest-européennes de crocidures (Mammalia, Insectivora). III. Structures génétiques des popula-tions continentales et insulaires de Crocidura russula (Hermann, 1780) et de Crocidura suaveolens (Pallas, 1811). Mammalia, 52, 387–400.
Catzeflis F (1984) Systématique biochimique, taxonomie et phylogénie
des musaraignes d’Europe (Soricidae, Mammalia). PhD Thesis,
Uni-versity of Lausanne, Switzerland.
Cheylan G (1984) Les mammifères des îles de Provence et de Méditerranée occidentale: un exemple de peuplement insulaire non équilibré? Revue d’Ecologie-La Terre et la Vie, 39, 37–54. Cheylan G (1990) Patterns of Pleistocene turnover, current
dis-tribution and speciation among Mediterranean mammals. In:
Biogeography of Mediterranean Invasions (eds Groves RH, Di Castri
F), pp. 227–262. Cambridge University Press, Cambridge. Claussen M, Brovkin V, Ganopolski A, Kubatzki C, Petoukhov V
(2003) Climate change in northern Africa: the past is not the future. Climatic Change, 57, 99–118.
Claussen M, Gayler V (1997) The greening of the Sahara during the mid-Holocene: results of an interactive atmosphere-biome model. Global Ecology and Biogeography Letters, 6, 369–377. Clément M, Posada D, Crandall KA (2000) tcs: a computer program
to estimate gene genealogies. Molecular Ecology, 9, 1657–1659. Collina-Girard J (2001) Atlantis off the Gibraltar Strait? Myth and
geology. Comptes Rendus de l’Académie Des Sciences Serie, II. 333, 233–240.
Contoli L (1990) Ulteriori dati su Crocidura cossyrensis Contoli, 1989, con particolare riguardo a soecie congeneri dell’arena mediterranea (Mammalia, Soricidae). Hystrix, 2, 53–58. Contoli L (1992) De Cossyrae crocidura (Crocidura cossyrensis
Con-toli, 1989). Atti Della Società Italiana Scienze Naturali e del Museo
Civico di Storia Naturale di Milano, 132, 61–68.
Contoli L, Aloise G (2001) On the taxonomy and distribution of
Crocidura cossyrensis and Crocidura russula (Insectivora, Soricidae)
in Maghreb. Hystrix, 12, 11–18.
Contoli L, Benincasa-Stagni B, Marenzi AR (1989) Morfologia di
Crocidura Wagler, 1832 (Mammalia, Soriciae) in Italia, Sardegna
e Sicilia, con il metodo dei descrittori di Fourier: primi dati.
Hystrix, 1, 113–129.
Corbet GB (1978) The Mammals of the Palaearctic Region: a Taxonomic
Review. British Museum (Natural History), London.
Cosson JF, Pascal M, Bioret F (1996) Origine et répartition des musaraignes du genre Crocidura dans les îles bretonnes. Vie et
Milieu–Life and Environment, 48, 233–244.
De Jong H (1998) In search of historical biogeographic patterns in the western Mediterranean terrestrial fauna. Biological Journal of
the Linnean Society, 65, 99–164.
Dobson M (1998) Mammal distributions in the western Mediterr-anean: the role of human intervention. Mammal Review, 28, 77–88. Dobson M, Wright A (2000) Faunal relationships and zoo-geographical affinities of mammals in north-west Africa. Journal
of Biogeography, 27, 417–424.
Edwards SV (1997) Relevance of microevolutionary processes to higher level molecular systematics. In: Avian Molecular Evolution
and Systematics (ed. Mindell DP), pp. 251–278. Academic Press,
New York.
Edwards SV, Beerli P (2000) Perspectives: gene divergence, population divergence, and the variance in coalescence times in phylogeographic studies. Evolution, 54, 1839–1854.
Ellerman JR, Morrison-Scott TCS (1966) Checklist of Palearctic and
Indian Mammals, 1758–1946. 2d Trustees British Museum
(Natural History). London.
Faure H (1987) Il quadro cronologico delle fasi pluviali e glaciali dell’Africa. In: Storia Generale dell’Africa. Metodologia e Preistoria
dell’Africa (ed. Ki-Zerbo J), pp. 400–409. Jaca Book Milano.
Flemming N, Bailey G, Courtillot V et al. (2003) Coastal and marine palaeo-environments and human dispersal points across the
P H Y L O G E O G R A P H Y I N T H E W E S T E R N M E D I T E R R A N E A N 1161 Africa–Eurasia boundary. In: Maritime Heritage (eds Brebbia
CA, Gambin T), pp. 13. Wessex Institute of Technology, South-ampton, UK and University of Malta, Malta.
Fumagalli L, Taberlet P, Stewart DT, Gielli L, Hausser J, Vogel P (1999) Molecular phylogeny and evolution of Sorex shrews (Soricidae: Insectivora) inferred from mitochondrial DNA sequence data. Molecular Phylogenetics and Evolution, 11, 222–235. Gantenbein B, Largiadèr CR (2003) The phylogeographic
import-ance of the Strait of Gibraltar as a gene flow barrier in terrestrial arthropods: a case study with the scorpion Buthus occitanus as a model organism. Molecular Phylogenetics and Evolution, 28, 119– 130.
Genoud M, Hutterer R (1990) Crocidura russula (Hermann, 1780) – Hausspitzmaus. In: Handbuch der Säugetiere Europas (eds Niethammer J, Krapp F), pp. 429–452. Aula-Verlag, Wiesbaden. Geraads D (1995) Rongeurs et Insectivores (Mammalia) du Pliocène final de Ahl Oughlam (Casablanca, Maroc). Geobios, 28, 99–115.
Guiller A, Coutellec-Vreto MA, Madec L, Deunff J (2001) Evolu-tionary history of the land snail Helix aspersa in the western Mediterranean: preliminary results inferred from mitochondrial DNA sequences. Molecular Ecology, 10, 81–87.
Harris DJ, Sá-Sousa P (2002) Molecular phylogenetics of the Iberian wall lizard (Podarcis): is Podarcis hispanica a species com-plex? Molecular Phylogenetics and Evolution, 23, 75–81.
Heaney LR (1986) Biogeography of mammals in SE Asia: estimates of rates of colonisation, extinction and speciation. Biological Journal
of the Linnean Society, 28, 127–165.
Hewitt GM (1996) Some genetic consequencies of ice ages and their role in divergence and speciation. Biological Journal of the
Linnean Society, 58, 247–276.
Hewitt GM (1999) Post-glacial recolonisation of European biota.
Biological Journal of the Linnean Society, 68, 87–112.
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages.
Nature, 405, 907–913.
Hewitt GM (2001) Speciation, hybrid zones and phylogeography — or seen genes in space and time. Molecular Ecology, 10, 537– 549.
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society of
London. Series B, Biology Sciences, 359, 183–195.
Hsü KJ, Montader L, Bernouilli D et al. (1977) History of the Medi-terranean salinity crisis. Nature, 267, 399–403.
Hutterer R (1993) Order Insectivora. In: Mammal Species of the
World, A Taxonomic and Geographic Reference (eds Wilson DE,
Reeder DM), pp. 69–130. Smithsonian Institution Press, Washing-ton, D.C.
Jamet P (1991) Flore et faune du Sahara depuis 18 000 bp. Actes
Congres National Des Societés Savantes, 116, 55–69.
Johns GC, Avise C (1998) A comparative summary of genetic dis-tances in the vertebrates from the mitochondrial cytochrome b gene. Molecular Biology and Evolution, 15, 1481–1490.
Jolly D, Harrisson SP, Damnati B, Bonnefille R (1998) Simulated climate and biomes of Africa during the late Quaternary: com-parison with pollen and lake status data. Quaternary Science
Reviews, 17, 629–657.
Kowalski K (1991) L’histoire de la faune de rongeurs de la zone aride de l’ancien monde pendant le Quaternaire. In: Le Rongeur
et l’Espace (eds Le Berre M, Le Guelte L), pp. 167–175. Chabaud,
Paris.
Krijgsman W (2002) The Mediterranean: Mare Nostrum of Earth sciences. Earth and Planetary Science Letters, 205, 1–12.
Libois RM, Michaux JR, Ramalhinho MG, Maurois C, Sarà M (2001) On the origin and systematics of the North African wood mouse (Apodemus sylvaticus) populations. A comparative study of mtDNA restriction patterns. Canadian Journal of Zoology, 79, 1503–1511.
Lo Brutto S, Arculeo M, Sarà M (2004) Mitochondrial simple sequence repeats (SSR) and 12S-rRNA gene reveal two distinct lineages of Crocidura russula (Mammalia, Soricidae). Heredity, 92, 527–533.
Maddison DR (1991) The discovery and importance of multiple islands of most-parsimonious trees. Systematic Zoology, 40, 315– 328.
Madec L, Bellido A, Guiller A (1996) Statistical and biogeograph-ical significances of patterns of morphologbiogeograph-ical and biochembiogeograph-ical variation in the land snail Helix aspersa. Comptes Rendus de
l’Academie Des Sciences Paris Série, III. 319, 225–229.
Michaux JR, Magnanou E, Paradis E, Nieberding C, Libois R (2001) Mitochondrial phylogeography of the woodmouse (Apodemus sylvaticus) in the western Palearctic region. Molecular
Ecology, 12, 685–697.
Molina O, Brown RP, Suarez NM, Pestano JJ (2003) The origin of the osorian shrew (Crocidura osorio) from Gran Canaria resolved using mtDNA. Italian Journal of Zoology, 70, 179–181.
Nehren R (1992) Zur Prähistorie der Maghrebländer (Marokko —
Algerien — Tunesien). 1. Teil. Mainz, Philipp von Zabern.
Nichols R (2001) Gene trees and species trees are not the same.
Trends in Ecology and Evolution, 16, 358–364.
Palmer M, Cambefort Y (2000) Evidence for reticulate paleogeo-graphy: beetle diversity linked to connection–disjunction cycles of the Gibraltar Strait. Journal of Biogeography, 27, 403–416. Poitevin F, Catalan J, Fons R, Croset H (1986) Biologie évolutive
des populations ouest-européennes de crocidures. 1. Critères d’identification et répartition biogéographique de Crocura
russula (Hermann, 1780) et Crocidura suaveolens (Pallas, 1811). Revue d’Ecologie-la Terre et la Vie, 41, 299–314.
Posada D, Crandall KA (1998) modeltest: testing the model of DNA substitution. Bioinformatics, 14, 817–819.
Prüser F, Mossakowski D (1998) Low substitution rates in mitochon-drial DNA in Mediterranean carabid species. Insect Molecular
Biology, 7, 121–128.
Quezel P, Barbero M (1993) Variations climatiques au Sahara et en Afrique sèche depuis le Pliocene: enseignements de la flore et de la végétation actuelles. Revue d’Ecologie-la Terre et la Vie, 24, 191– 202.
Reumer JWF, Meylan A (1986) New developments in vertebrate cytotaxonomy IX. Chromosome numbers in the order Insectivora (Mammalia). Genetica, 70, 119–151.
Robinson M, Gouy M, Gautier C, Mouchiroud D (1998) Sensitivity of the relative-rate test to taxonomic sampling. Molecular Biology
and Evolution, 15, 1091–1098.
Rozas J, Rozas R (1999) dnasp version 3: an integrated program for molecular population genetics and molcular evolution analysis.
Bioinformatics, 15, 174–175.
Rzebik-Kowalska B (1988) Studies on the genus Crocidura (Insec-tivora, Mammalia) in Algeria. Acta Zoologica Cracoviensia, 31, 176–192.
Sanmartín I (2003) Dispersal vs. vicariance in the Mediterranean: historical biogeography of the Palearctic Pachydeminae (Cole-optera, Scarabaeoidea). Journal of Biogeography, 30, 1883–1897. Sarà M, Vogel P (1996) Geographic variation of the greater
white-toothed shrew (Crocidura russula Hermann, 1780 Mammalia, Soricidae). Biological Journal of the Linnean Society, 116, 377–392.
1162 J . - F . C O S S O N E T A L .
Sarà M, Zanca L (1992) Metric discrimination and distribution of the species of Crocidura occurring in Tunisia. Acta Theriologica, 37, 103–116.
Straus LG (2001) Africa and Iberia in the Pleistocene. Quaternary
International, 75, 91–102.
Street A, Gasse F (1981) Recent developments in research into the Quaternary climatic history of the Sahara. In: Sahara: Ecological
Change and Early Economic History (ed. Allen JA), pp. 7–28.
MENAS Press, London.
Suc JP, Bertini A, Combourieu-Nebout N et al. (1995) Structure of the west Mediterranean vegetation and climate since 5.3 Ma.
Acta Zoologica Cracoviensia, 38, 3–16.
Swofford DL (1998) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b1. Sinauer Associates,
Sunder-land, Massachusetts.
Taberlet P, Fumagalli L (1996) Owl pellets as a source of DNA for genetic studies of small mammals. Molecular Ecology, 5, 301–305. Taberlet P, Fumagalli L, Wust-Saucy AG, Cosson JF (1998) Com-parative phylogeography and postglacial colonization routes in Europe. Molecular Ecology, 7, 453–464.
Tchernov E (1996) Rodent faunas, chronostratigraphy and paleo-biogeography of the southern Levant during the Quaternary.
Acta Zoologica Cracoviensia, 39, 513–530.
Vigne JD (1992) Zooarchaeology and the biogeographical history of the mammals of Corsica and Sardinia, since the last ice age.
Mammal Review, 22, 87–96.
Vigne JD, Alcover JA (1985) Incidence des relations historiques entre l’homme et l’animal dans la composition actuelle du
peuplement amphibien, reptilien et mammalien des îles de Méditerranée occidentale. Actes 110ème Congrès national des
Sociétés savantes, Montpellier, Sciences, fasc. 28, 79–91.
Vogel P, Cosson JF, Lopez Jurado LF (2003) Taxonomic status and origin of the shrews (Soricidae) from the Canary Islands inferred from mtDNA comparison with the European Crocidura species. Molecular Phylogenetics and Evolution, 27, 271–282. Vogel P, Maddalena T (1987) Note sur la répartition altitudinale et
la fréquence de la musaraigne musette (Crocidura russula
yebalensis) au Maroc. Mammalia, 51, 465–467.
Vogel P, Maddalena T, Catzeflis F (1986) A contribution to the taxonomy and ecology of shrews from Crete and Turkey (Crocidura
zimmermanni and C. suaveolens). Acta Theriologica, 31, 537–545.
Vogel P, Maddalena T, Sarà M (1992) The taxonomic status of
Crocidura cossyrensis Contoli, 1989 and its relationship to African
and European Crocidura russula (Mammalia, Insectivora). Israel
Journal of Zoology, 38, 424.
Vogel P, Maddalena T, Sarà M (2004) Crocidura cossyrensis Contoli, 1989 (Mammalia, Soricidae) karyotype, biochemical genetics and hybridization experiments. Revue Suisse de Zoologie, 111, 925–934.
Vogel P, Maddalena T, Schembri PJ (1990) Cytotaxonomy of shrews of the genus Crocidura from Mediterranean islands. Vie
et Milieu–Life and Environment, 40, 124–129.
Zardoya R, Doadrio I (1998) Phylogenetics relationships of Iberian cyprinids: systematics and biogeographical implications.
Pro-ceedings of the Royal Society of London. Series B, Biological Sciences,