HAL Id: hal-03105993
https://hal.sorbonne-universite.fr/hal-03105993
Submitted on 11 Jan 2021
HAL is a multi-disciplinary open access
archive for the deposit and dissemination of
sci-entific research documents, whether they are
pub-lished or not. The documents may come from
teaching and research institutions in France or
abroad, or from public or private research centers.
L’archive ouverte pluridisciplinaire HAL, est
destinée au dépôt et à la diffusion de documents
scientifiques de niveau recherche, publiés ou non,
émanant des établissements d’enseignement et de
recherche français ou étrangers, des laboratoires
publics ou privés.
Co-infection of SARS-CoV-2 with other respiratory
viruses and performance of lower respiratory tract
samples for the diagnosis of COVID-19
Sonia Burrel, Pierre Hausfater, Martin Dres, Valérie Pourcher,
Charles-Edouard Luyt, Elisa Teyssou, Cathia Soulié, Vincent Calvez,
Anne-Geneviève Marcelin, David Boutolleau
To cite this version:
Sonia Burrel, Pierre Hausfater, Martin Dres, Valérie Pourcher, Charles-Edouard Luyt, et al..
Co-infection of SARS-CoV-2 with other respiratory viruses and performance of lower respiratory tract
samples for the diagnosis of COVID-19. International Journal of Infectious Diseases, Elsevier, 2021,
102, pp.10 - 13. �10.1016/j.ijid.2020.10.040�. �hal-03105993�
Short
Communication
Co-infection
of
SARS-CoV-2
with
other
respiratory
viruses
and
performance
of
lower
respiratory
tract
samples
for
the
diagnosis
of
COVID-19
Sonia
Burrel
a,b,
Pierre
Hausfater
c,d,
Martin
Dres
e,f,
Valérie
Pourcher
b,g,
Charles-Edouard
Luyt
h,i,
Elisa
Teyssou
a,b,
Cathia
Soulié
a,b,
Vincent
Calvez
a,b,
Anne-Geneviève
Marcelin
a,b,
David
Boutolleau
a,b,*
a
AP-HP,SorbonneUniversité,HôpitalPitié-Salpêtrière,ServicedeVirologie,Paris,France
b
SorbonneUniversité,INSERMU1136,InstitutPierreLouisd’EpidémiologieetdeSantéPublique(IPLESP),Paris,France
c
AP-HP,SorbonneUniversité,HôpitalPitié-Salpêtrière,Serviced’AccueildesUrgences,Paris,France
d
SorbonneUniversitésGRC-14BIOSFASTetINSERMUMR-S1166,Paris,France
e
SorbonneUniversité,INSERM,UMRS1158Neurophysiologierespiratoireexpérimentaleetclinique,Paris,France
fAP-HP,SorbonneUniversité,HôpitalPitié-Salpêtrière,ServicedePneumologie,Médecineintensive–Réanimation(Département‘R3S’),Paris,France g
AP-HP,SorbonneUniversité,HôpitalPitié-Salpêtrière,ServicedeMaladiesInfectieusesetTropicales,Paris,France
h
AP-HP,SorbonneUniversité,HôpitalPitié-Salpêtrière,ServicedeMédecineIntensiveRéanimation,InstitutdeCardiologie,Paris,France
i
SorbonneUniversité,INSERM,UMRS_1166-ICANInstitutdeCardiométabolismeetNutrition,Paris,France
ARTICLE INFO Articlehistory: Received10July2020
Receivedinrevisedform19October2020 Accepted22October2020
Keywords: SARS-CoV-2
Otherrespiratoryviruses Co-infection
Lowerrespiratorytract
ABSTRACT
Objectives:This studywasperformedduringtheearlyoutbreakperiodofcoronavirusdisease2019 (COVID-19)andtheseasonalepidemicsofotherrespiratoryviralinfections,inordertodescribethe extentofco-infectionsofsevereacuterespiratorysyndromecoronavirus2(SARS-CoV-2)withother respiratoryviruses.Italsocomparedthediagnosticperformancesofupperrespiratorytract(URT)and lowerrespiratorytract(LRT)samplesforSARS-CoV-2infection.
Methods:From25Januaryto29March2020,allURTandLRTsamplescollectedfrompatientswith suspectedCOVID-19receivedinthevirologylaboratoryofPitié-SalpêtrièreUniversityHospital(Paris, France)weresimultaneouslytestedforSARS-CoV-2andotherrespiratoryviruses.
Results:Atotalof1423consecutivepatientsweretested:677(47.6%)males,746(52.4%)females,median age50(range,1–103)years.Twenty-one(1.5%)patientswerepositiveforbothSARS-CoV-2andother respiratoryviruses.ThedetectionrateofSARS-CoV-2wassignificantlyhigherinLRTthaninURT(53.6% vs.13.4%;p<0.0001).Theanalysisofpairedsamplesfrom117(8.2%)patientsshowedthatSARS-CoV-2 loadwaslowerinURTthaninLRTsamplesin65%ofcases.
Conclusion:Thedetectionofotherrespiratoryvirusesinpatientsduringthisepidemicperiodcouldnot ruleout SARS-CoV-2co-infection.Furthermore,LRTsamples increasedtheaccuracyofdiagnosisof COVID-19.
©2020PublishedbyElsevierLtdonbehalfofInternationalSocietyforInfectiousDiseases.Thisisanopen accessarticleundertheCCBY-NC-NDlicense(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
The severeacuterespiratorysyndromecoronavirus 2 (SARS-CoV-2) responsible for coronavirus disease 2019 (COVID-19) emergedinWuhan,China,inDecember2019(Zhuetal.,2020). A few studies have reported proportions of SARS-CoV-2 co-infections with other respiratory viruses (range, 0–20%)
(Chen et al.,2020; Kimet al., 2020;Leuzingeret al.,2020; Lin et al.,2020a;Wee et al.,2020).Furthermore,lowerrespiratory tract(LRT)sampleshavebeenfoundtosignificantlyimprovethe efficiency of diagnosis compared with upper respiratory tract (URT)samplesfornon-SARS-CoV-2respiratoryinfections(Branche etal.,2014;Falseyetal.,2012).
This study, performed during the early outbreak period of COVID-19and theseasonalepidemics ofotherrespiratory viral infections,aimedtodescribetheextentofco-infectionsof SARS-CoV-2withotherrespiratoryviruses.Asecondobjectiveofthis studywastocomparethediagnosticperformancesofURTandLRT samplesforSARS-CoV-2infection.From25Januaryto29March
*Corresponding authorat: ServicedeVirologie,BâtimentCERVI (5eétage), HôpitalPitié-Salpêtrière,83Boulevarddel’hôpital,75652Pariscedex13,France.
E-mailaddress:david.boutolleau@aphp.fr(D.Boutolleau).
https://doi.org/10.1016/j.ijid.2020.10.040
1201-9712/©2020PublishedbyElsevierLtdonbehalfofInternationalSocietyforInfectiousDiseases.ThisisanopenaccessarticleundertheCCBY-NC-NDlicense(http:// creativecommons.org/licenses/by-nc-nd/4.0/).
ContentslistsavailableatScienceDirect
International
Journal
of
Infectious
Diseases
2020, all URT and LRT samples collected from patients with suspectedCOVID-19receivedinthevirologylaboratoryof Pitié-Salpêtrière University Hospital (Paris, France) were tested for SARS-CoV-2andotherrespiratoryviruses.Thestudypopulation consisted of hospitalised symptomatic adults. All patients had respiratorysymptoms,asmentionedinthedefinitionofpossible COVID-19 cases at that time in France. SARS-CoV-2 RNA was detectedbyreal-timeRT-PCR(Cormanetal.,2020).SARS-CoV-2 loads were estimated with cycle threshold (Ct) values. Other respiratory viruses were tested with the Filmarray1 RP2plus (BIOMERIEUX):influenzavirus,respiratorysyncytialvirus, meta-pneumovirus, adenovirus, parainfluenza virus, non-SARS-CoV-2 coronavirus, and rhinovirus/enterovirus. For statistical analyses, continuous variables were expressed as median (range) and categoricalvariablesasnumbers(percentages).Comparisonswere performedusingMann-WhitneyUtest(continuousvariables)and Chi-squaredtest(categoricalvariables).P<0.05wasconsideredto bestatisticallysignificant.
Duringthestudyperiod,1423consecutivehospitalisedpatients (677 males, 746females, median age50 years) wereincluded. Patients originated from Pitié-Salpêtrière hospital (n=1167) or fromotherclosehospitals(n=256).Mostofpatientsfrom Pitié-Salpêtrière hospital werehospitalised within 10 daysafter the onset of symptoms and weretested for SARS-CoV-2 and other respiratoryviruseswithin24hofadmission.Amongpatientsfrom Pitié-Salpêtrière hospital,233/1167 (20.0%)werehospitalised in theintensivecareunit(ICU).Detectionratesofrespiratoryviruses amongpatientswere:724(50.9%)negativeforrespiratoryviruses, 398 (28.0%) positive for other respiratory viruses, 280 (19.7%) positiveforSARS-CoV-2,and21(1.5%)positiveforSARS-CoV-2and otherrespiratoryviruses(Table 1).Amongpatientsnegativefor SARS-CoV-2 (n=1122), those who were positive for other respiratory viruses were statistically younger than those who werenegative(median,39vs.52 years,p<0.0001).Conversely, among patientswho werepositivefor SARS-CoV-2(n=301),no significantagedifferencewasobservedbetweenthosewhowere positiveforotherrespiratoryvirusesandthosewhowerenegative (median,56vs.54years,notsignificant).Amongthe21patients co-infectedwithSARS-CoV-2,otherrespiratoryvirusesthatwere detectedwerenon-SARS-CoV-2coronavirus(n=6),influenzavirus (n=5), adenovirus (n=3), rhinovirus/enterovirus (n=3), para-influenza virus (n=3), and adenovirus+rhinovirus/enterovirus (n=1).Fourof21(19.0%)co-infectedpatientswerehospitalisedin ICU, which was significantly lower than the 113/280 (46.4%) patientsinfectedwithSARS-CoV-2alone(p=0.020)(Table1).
Atotalof1160URTand379LRTsampleswerecollected.The detectionrateofSARS-CoV-2wassignificantlyhigherinLRT(203, 53.6%)thaninURT(1160,13.4%;p<0.0001).Pairedsamplesfrom
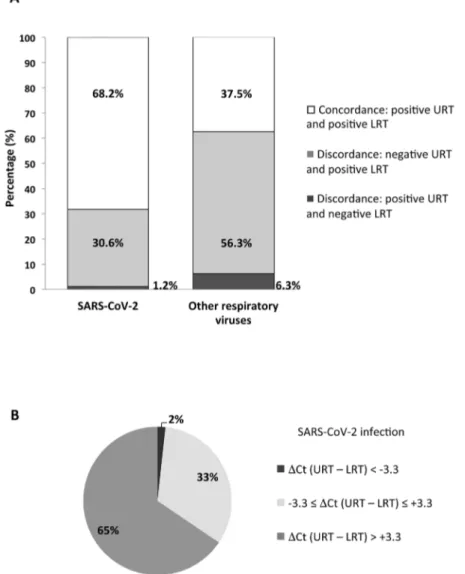
URTandLRTwereobtainedfrom117(8.2%)patients.Amongthe85 patientswhowerepositive forSARS-CoV-2,52 (68.2%)showed concordantpositiveresultsinURTandLRTsamples(Figure1A),but SARS-CoV-2 loadwas at least1 loghigher in LRTthan in URT samplesfor38patients(65%)(Figure1B).Moreover,26(30.6%) patients showed discordant SARS-CoV-2 results, with negative URTandpositiveLRTsamples(Figure1A).Asimilarprofilewas observed for the 32 patients infected with other respiratory viruses:concordantpositiveresultsinURTandLRTfor12(37.5%), while18(56.3%)showeddiscordantresults,withnegativeURTand positive LRT samples (Figure 1A). The proportion of patients positiveforSARS-CoV-2withdiscordantresultswassignificantly lowerthantheoneofpatientspositiveforotherrespiratoryviruses withdiscordantresults:30.6%vs.56.3%(p=0.012).
In the present study, 7% (21/301) of SARS-CoV-2-positive patientswereco-infectedwithotherrespiratoryviruses.This co-infectionrateissimilartosomepreviouslyreportedrates(range, 0–6.5%)(Chenetal.,2020;Leuzingeretal.,2020;Linetal.,2020a), butlowerthanothers(upto20%)(Kimetal.,2020).Thismightbe due todifferent study populations or potential spatiotemporal variationsinviralepidemiology.Inparticular,itisverylikelythat thelockdowninFrance,whichstartedon17Marchandlasteduntil 11May2020,hadno(orverylittle)influenceonthecirculationof respiratoryvirusesduringthepresentstudyperformedfrom25 Januaryto29March2020.Inlinewithpreviousstudies,different types of otherrespiratory viruses weredetected togetherwith SARS-CoV-2amongco-infectedpatients,including non-SARS-CoV-2coronavirus,influenzavirus,adenovirus,rhinovirus/enterovirus, andparainfluenzavirus(Kimetal.,2020;Leuzingeretal.,2020;Lin etal.,2020a;Weeetal.,2020).Patientsco-infectedwith SARS-CoV-2andotherrespiratoryvirusesdidnotsignificantlydifferin ageand genderfromthose infectedwithSARS-CoV-2 alone, as previouslydescribed(Kimetal.,2020).AmongICUpatients,the proportionofco-infectedpatientswassignificantlylowerthanthe oneinfectedwithSARS-CoV-2alone,possiblyindicatingthat co-infection with other respiratory viruses might not worsen the severityofSARS-CoV-2-associatedrespiratorydisease,whichwas inaccordancewithapreviousstudy(Weeetal.,2020).Thecurrent studyfoundahigherefficiencyofLRTthanURTsamplesfor COVID-19diagnosis,withasignificantlyhigherrateofdetectionof SARS-CoV-2 and a 1 loghigher SARS-CoV-2 loadfor the majorityof infectedpatients.Thisdiscrepancycouldbepartlyexplainedbythe variabilityinthedelaybetweentheonsetofsymptomsandthe samplingamongpatients.However,thishigherdiagnostic perfor-manceofLRTsamplesforrespiratoryinfections,including COVID-19,hasbeenpreviouslyreported(Brancheetal.,2014;Falseyetal., 2012;Linetal.,2020b,Wangetal.,2020).Inthecurrentstudy,the proportionof patientspositive for SARS-CoV-2 withdiscordant
Table1
CharacteristicsofsuspectedCOVID-19patients,accordingtorespiratoryvirologicalfindings. SARS-CoV-2( )
Otherrespiratoryviruses( )
SARS-CoV-2( )
Otherrespiratoryviruses(+)
SARS-CoV-2(+)
Otherrespiratoryviruses( )
SARS-CoV-2(+)
Otherrespiratoryviruses(+)
No.(%)ofpatients 724(50.9%) 398(28.0%) 280(19.7%) 21(1.5%)
Demographiccharacteristics
Male 328(45.3%) 178(44.7%) 160(57.1%) 11(52.4%)
Female 396(54.7%) 220(55.3%) 120(42.9%) 10(47.6%)
Medianage(y)(range) 52(0–103)a
39(0–101)a 56(12–94) 54(30–94) HospitalisationinICUb Yes 89(15.3%) 27(8.4%) 113(46.7%)c 4(19.0%)c No 493(84.7%) 295(91.6%) 129(53.3%) 17(81.0%)
Abbreviations:( ),negative;(+),positive;ICU,intensivecareunit;y,years.
a
p<0.0001(Mann–WhitneyUtest).
b
RatesofhospitalisationinICUonlyforthepatientsfromPitié-SalpêtrièreUniversityHospital(n=1167).
c
p=0.020(Chi-squaredtest).
S.Burrel,P.Hausfater,M.Dresetal. InternationalJournalofInfectiousDiseases102(2021)10–13
resultswassignificantlylowerthantheoneofpatientswhowere positiveforotherrespiratoryviruseswithdiscordantresults.Those differentprofilesofcompartmentalisationofPCRpositivitywithin therespiratorytractmaysuggestsomedifferencesin pathophysi-ologyofSARS-CoV-2infectioncomparedwithinfectionsbyother respiratoryviruses.
In conclusion, the detection of other respiratory viruses in patients during this epidemic period could not rule out SARS-CoV-2 co-infection, and LRT samples increased the accuracyofdiagnosisofviralrespiratoryinfections,including COVID-19.
Conflictofinterest
DBandPHreceivedlecturehonorariumandfeesforcongress transportation and registration from Biomerieux. MD received honorarium for fees for travel expenses from Lungpacer. CEL receivedlecturehonorariumfromBiomerieux.Otherauthorshave noconflictofinteresttodeclare.
Funding
Thisresearchdidnotreceiveanyspecificgrantfromfunding agenciesinthepublic,commercialornot-for-profitsectors. Ethicalapproval
Duetotheobservationalandretrospectivenatureofthestudy, theneedforspecificinformedconsentfromindividualpatients wasnotrequired.Alldataweregeneratedfromroutinestandard clinicalmanagementofthepatients.Aninformedconsenttothe treatmentofpersonaldatawasacquireduponhospitaladmission fromthepatientsortheirlegalrepresentatives.
Acknowledgements
Wethankallhealthcareworkersinvolvedinthediagnosisand treatment of COVID-19 patients in Pitié-Salpêtrière University Hospital(Paris,France).
Figure1.Comparisonofupperrespiratorytract(URT)andlowerrespiratorytract(LRT)fordiagnosinginfectionswithSARS-CoV-2andotherrespiratoryviruses,usingpaired samplesfromCOVID-19suspectedpatients.(A)DistributionofpairedrespiratorysamplesaccordingtotheconcordanceofdetectionofSARS-CoV-2andotherrespiratory virusesinURTandLRT.(B)DistributionofpairedrespiratorysamplesaccordingtothedifferenceinSARS-CoV-2loadbetweenURTandLRT.SARS-CoV-2loadswereestimated withcyclethreshold(Ct)valuesandthedifferenceofCtvaluesobtainedbetweenURTandLRT(DCtURT–LRT)wascalculated.Pairedsampleswerethendistributedaccording totheDCtvalueof3.3,giventhatthisdifferenceapproximatelycorrespondswithaSARS-CoV-2loadof1log.
References
BrancheAR,WalshEE,FormicaMA,FalseyAR.Detectionofrespiratoryvirusesin sputum fromadults byuseofautomatedmultiplexPCR.J ClinMicrobiol 2014;52:3590–6.
ChenN,ZhouM,DongX,QuJ,GongF,HanY,etal.Epidemiologicalandclinical characteristicsof99casesof2019novelcoronaviruspneumoniainWuhan, China:adescriptivestudy.Lancet2020;395:507–13.
Corman VM,Landt O, Kaiser M, Molenkamp R,Meijer A, ChuDKW, et al. Detection of 2019 novelcoronavirus(2019-nCoV)byreal-timeRT-PCR.EuroSurveill2020;25:1–8.
FalseyAR,FormicaMA,WalshEE.Yieldofsputumforviraldetectionbyreverse transcriptasePCRinadultshospitalizedwithrespiratoryillness.JClinMicrobiol 2012;50:21–4.
KimD,QuinnJ,PinskyB,ShahNH,BrownI.Ratesofco-infectionbetween SARS-CoV-2andotherrespiratorypathogens.JAMA2020;323:2085–6.
LeuzingerK,RoloffT,GosertR,SogaardK,NaegeleK,RentschK,etal.Epidemiology of Severe Acute Respiratory Syndrome Coronavirus 2 emergence amidst community-acquiredrespiratoryviruses.JInfectDis2020;222:1270–9.
LinD,LiuL,ZhangM,HuY,YangQ,GuoJ,etal.Co-infectionsofSARS-CoV-2with multiplecommonrespiratorypathogensininfectedpatients.SciChinaLifeSci 2020a;63:606–9.
LinC,XiangJ,YanM,LiH,HuangS,ShenC.Comparisonofthroatswabsandsputum specimensforviralnucleicaciddetectionin52casesofnovelcoronavirus (SARS-Cov-2)-infected pneumonia (COVID-19). Clin Chem Lab Med 2020b;58:1089–94.
WangW,XuY,GaoR,LuR,HanK,WuG,etal.DetectionofSARS-CoV-2indifferent typesofclinicalspecimens.JAMA2020;323:1843–4.
WeeLE,KoKKK,HoWQ,KwekGTC,TanTT,WijayaL.Community-acquiredviral respiratoryinfections amongst hospitalized inpatients during a COVID-19 outbreak in Singapore: co-infection and clinical outcomes. J Clin Virol 2020;128:104436.
ZhuN,ZhangD,WangW,LiX,YangB,SongJ,etal.Anovelcoronavirusfrompatients withpneumoniainChina,2019.NEnglJMed2020;382:727–33.
S.Burrel,P.Hausfater,M.Dresetal. InternationalJournalofInfectiousDiseases102(2021)10–13