Convolutional neural network architectures for predicting DNA–protein binding
Texte intégral
Figure
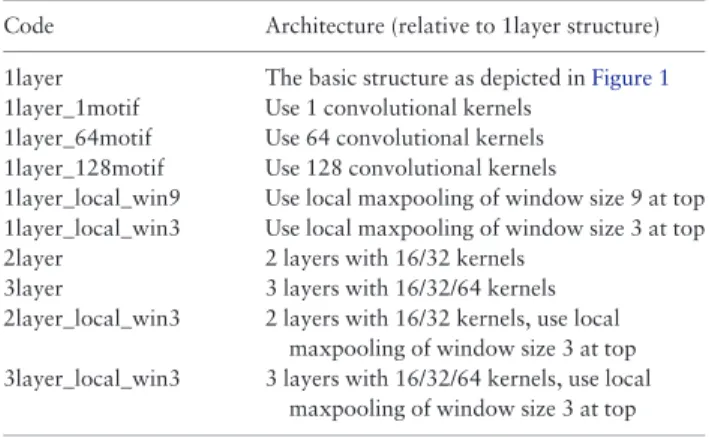


Documents relatifs
A model trained with CNN, using Word embedding as text representation, achieved a statistically significant higher training accuracy than a model trained with SVM having the
Table 1: Clinical diagnosis of DR based on various feature types with different contributions to classification. One feature in each list is required for the equivalent DR grading.
Each training example should include the following: the image obtained as a result of the Haar wavelet transform of the average cardiocycle, as well as the coordinates of the
Contact: ceslovas.venclovas@bti.vu.lt, sergei.grudinin@inria.fr protein structure | structural bioinformatics | geometric deep learning | Voronoi tessellation | convolutional
This paper proposes a novel scheme based on neural networks and deep learning, focusing on the convolutional neural network (CNN) architecture approach to enhance a copy-move
When dealing with convolutional neural networks, in order to find the best fit- ting model, we optimized 13 hyperparameters: convolutional kernel size, pooling size, stride
Some convolutional filters from the first layer of the regressor, acting on the tentative correspondence map, show preferences to spatially co-located features that
Tradition- ally, correspondences consistent with a geometric model such as epipolar geometry or planar affine transformation, are computed by detecting and matching local features