Development and validation of the Axiom (R) Apple480K SNP genotyping array
Texte intégral
Figure


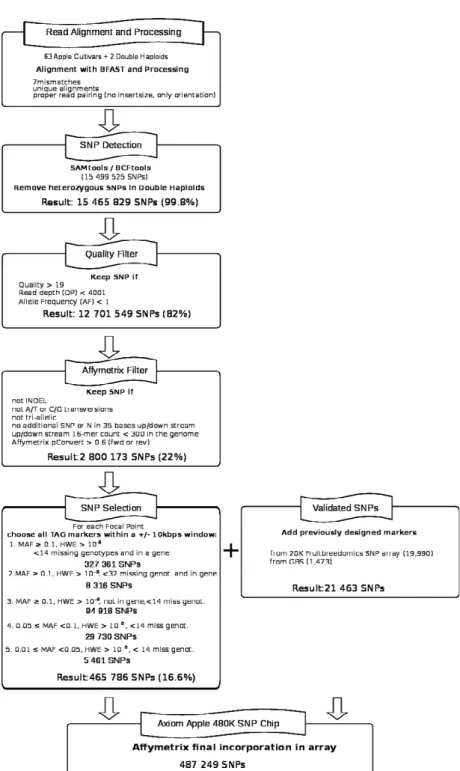

Documents relatifs
Using the focal point approach detailed in the methods, 16,330 SNPs were identified from the re-sequencing of 14 genotypes of the discovery panel, as well as 3,670 validated
The objectives of the work presented here are (1) to use SNPs previously identified in maize to develop a first reliable and standardized large scale SNP genotyping array; (2)
Sandra and Van Abbenyen (2009) assume a full-form representation of inflected word forms in Dutch as well as two memory systems that might be causally involved in errors of
« Je déclare que quiconque veut exceller un jour en quoi que ce soit, doit s’appliquer à cet objet dès l’enfance en trouvant à la fois son amusement et son occupation dans
(14) This method corresponds to positing a linear model where, for a hypothetical quantitative trait, the genetic value of an individual is the sum of independent marker
Total number of haplotypes, haplotype diversity, nucleotide diversity and haplotype-based polymorphism information content of validated cacao genes.. Disease
• GWAS conducted with 16480 SNP markers (GBS) • on 570 cocoa trees from Brazil, Cameroun, Ecuador, Trinidad, evaluated for self incompatibility. Genome Wide Association
The results show SNP markers presents/or near in putative transcription factor binding sites (TFBS) genotiped with success in two populations of mapping. RESULTS AND DISCUSSION