Representation of functional information in the SWISS-PROT Data Bank
Texte intégral
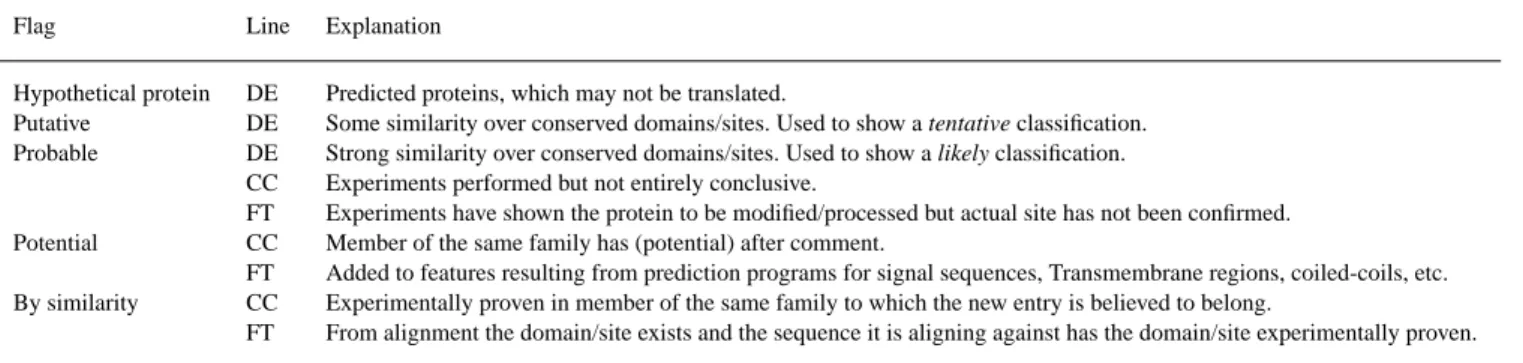
(2) Representation of functional information in the SWISS-PROT Data Bank. Table 1. Flags describing the evidence level of SWISS-PROT annotation (representation of functional information in the SWISS-PROT Data Bank). Flag. Line. Explanation. Hypothetical protein Putative Probable. DE DE DE CC FT CC FT CC FT. Predicted proteins, which may not be translated. Some similarity over conserved domains/sites. Used to show a tentative classification. Strong similarity over conserved domains/sites. Used to show a likely classification. Experiments performed but not entirely conclusive. Experiments have shown the protein to be modified/processed but actual site has not been confirmed. Member of the same family has (potential) after comment. Added to features resulting from prediction programs for signal sequences, Transmembrane regions, coiled-coils, etc. Experimentally proven in member of the same family to which the new entry is believed to belong. From alignment the domain/site exists and the sequence it is aligning against has the domain/site experimentally proven.. Potential By similarity. alignment is statistically highly significant, the active site is conserved, and so we classify it as a ‘PROBABLE MITOCHONDRIAL NUCLEASE’. All predicted protein sequences lacking any significant sequence similarity to characterized proteins are labeled as ‘hypothetical proteins’. The majority of these cases come from the genome sequencing projects. Example:. DE HYPOTHETICAL 33.8 KD PROTEIN C5H10.01 DE IN CHROMOSOME I. Most of the SWISS-PROT annotation can be found in the CC and FT lines. The CC (Comment) lines and the Feature Table (FT lines) give an indication about the level of characterization of a protein. All non-experimentally verified information is flagged. Let us have again a look at Q10480, where you can find the following CC lines: CC -!- FUNCTION: THIS ENZYME HAS BOTH CC RNASE AND DNASE ACTIVITY(BY SIMILARITY). CC -!- COFACTOR: REQUIRES MANGANESE OR CC MAGNESIUM (BY SIMILARITY). CC -!- SUBUNIT: HOMODIMER (BY SIMILARITY). CC -!- SUBCELLULAR LOCATION: MITOCHONDRIAL CC INNER MEMBRANE (POTENTIAL). CC -!- SIMILARITY: BELONGS TO THE DNA/RNA CC NON-SPECIFIC ENDONUCLEASES FAMILY. The function, cofactor and subunit comments are all labeled ‘by similarity’. This indicates that these have been assigned due to similarity to an existing characterized entry, in this case the mitochondrial nuclease from Saccharomyces cerevisiae. The label ‘potential’ is also used to indicate the assignment by comparative analysis. In general this label is used if there is no experimental proof for the information given in a CC topic for a protein, but similarity searches or other prediction methods allow potential comments (as with the subcellular location comment in Q10480). If comparative analysis reveals highly likely comments, then the label ‘probable’ is used: CC -!- SUBUNIT: HOMOTRIMER (PROBABLE).. All features in the FT lines, which have been derived from sequence similarity searches and prediction programs, but are lacking final experimental verification, are flagged. If a feature is highly likely, i.e. the experiments performed are not entirely conclusive, then the label ‘probable’ is used. The label ‘potential’ is used to indicate the assignment by comparative analysis and/or prediction programs for signal sequences, transmembrane regions, coiled-coils etc. Another label used to indicate that a feature has not been experimentally proven but only inferred through sequence analysis is ‘by similarity’:. FT ACT SITE 142 142 BY SIMILARITY. This example comes again from Q10480, the ‘PROBABLE MITOCHONDRIAL NUCLEASE’ of Schizosaccharomyces pombe. The label ‘by similarity’ indicates that this feature has been assigned due to similarity to an existing characterized entry, in this case the mitochondrial nuclease from Saccharomyces cerevisiae. Table 1 outlines the rules used to flag annotation added to entries where no, or only limited, experimental evidence is available. The addition of functional information to SWISS-PROT entries is a labor-intensive process where efforts are taken to add all relevant, available data. Great care is taken to ensure that the information added can be traced either to the relevant data source or back to the entry (or entries) reporting the experimentally determined characteristics. For a more detailed description of these assignments please visit our documentation files at www.expasy.ch/ sprot/sp-docu.html. Finally we also want to point out the importance of using the original SWISS-PROT data produced by SIB and EBI. The SWISS-PROT database is widely redistributed; it is integrated into other public data sets, and is also distributed by many vendors in conjunction with software packages. Users should be aware that the database supplied may not be the latest data and may not include all of the information available in the original.. 1067.
(3)
Figure
Documents relatifs
This process relies on three periplasmic proteins: (i) SurA, a chaperone that also has two peptidyl-prolyl isomerase domains and that is the main OMP-targeting factor in the
On the other hand, the dephosphorylation of MAPK3/4/6 by SpvC is clearly coupled to the attenuation of the plant defense responses. When present in Arabidopsis protoplasts, SpvC
When forming their value proposition, which is needed for acquiring new customers, the company may focus one of the three main assets that are needed to run process instances:
HAL is a multi-disciplinary open access archive for the deposit and dissemination of sci- entific research documents, whether they are pub- lished or not. The documents may come
(a) The easiest thing to do is to apply the algorithm from the lecture: take the matrix (A | I) and bring it to the reduced row echelon form; for an invertible matrix A, the result
A cross-reference table between the Protein Data Bank of macromolecular structures and the National Biomedical Research Foundation-Protein Identification Resource amino acid
SWISS-PROT is an annotated protein sequence database established in 1986 and maintained collaboratively, since 1988, by the Department of Medical Biochemistry of the University
It is important to provide the users of biomolecular databases with a degree of integration between the three types of sequence- related databases (nucleic acid sequences,