ContentslistsavailableatScienceDirect
Journal
of
Infection
and
Public
Health
j o u r n al ho me p ag e :h t t p : / / w w w . e l s e v i e r . c o m / l oc a t e / j i p h
Review
Article
The
outbreak
of
the
novel
severe
acute
respiratory
syndrome
coronavirus
2
(SARS-CoV-2):
A
review
of
the
current
global
status
Mbarka
Bchetnia
a,
Catherine
Girard
a,
Caroline
Duchaine
b,c,
Catherine
Laprise
a,∗aUniversitéduQuébecàChicoutimi(UQAC),Départementdessciencesfondamentales,Centreintersectorielensantédurable,Saguenay,Canada bCentrederecherche,InstitutuniversitairedecardiologieetdepneumologiedeQuébec,UniversitéLaval(IUCPQ-UL),Québec,Canada cDépartementdebiochimie,demicrobiologieetdebioinformatique,UniversitéLaval,Québec,Canada
a
r
t
i
c
l
e
i
n
f
o
Articlehistory:Received10May2020
Receivedinrevisedform13July2020 Accepted21July2020 Keywords: SARS-CoV-2 COVID-19 Emergence Transmission Prevention Treatment
a
b
s
t
r
a
c
t
Thereiscurrentlyanongoingworldwidepandemicofanovelvirusbelongingtothefamilyof
Coro-naviruses(CoVs)whicharelarge,enveloped,plus-strandedRNAviruses.Coronavirusesbelongtothe
orderofNidovirales,familyofCoronavirinaeandaredividedintofourgenera:alphacoronavirus,
beta-coronavirus,gammacoronavirusanddeltacoronavirus.CoVscausediseasesinawidevarietyofbirdsand
mammalsandhavebeenfoundinhumanssince1960.Todate,sevenhumanCoVswereidentified
includ-ingthealpha-CoVsHCoVs-NL63andHCoVs-229Eandthebeta-CoVsHCoVs-OC43,HCoVs-HKU1,the
severeacuterespiratorysyndrome-CoV(SARS-CoV),theMiddleEastrespiratorysyndrome-CoV
(MERS-CoV)andthenovelvirusthatfirstappearedinDecember2019inWuhan,China,andrapidlyspreadto
213countriesasofthewritingthispaper.Itwasofficiallynamedsevereacuterespiratorysyndrome
coronavirus2(SARS-CoV-2)bytheinternationalcommitteeontaxonomyofviruses(ICTV)andthe
dis-ease’snameisCOVID-19forcoronavirusdisease2019.SARS-CoV-2isverycontagiousandiscapableof
spreadingfromhumantohuman.Infectionroutesincludedropletandcontact,andaerosoltransmission
iscurrentlyunderinvestigation.Itisassociatedwitharespiratoryillnessthatmaycausesevere
pneumo-niaandacuterespiratorydistresssyndrome(ARDS).SARS-CoV-2becameanemergencyofinternational
concern.AsofJuly12,2020,thevirushasbeenresponsiblefor12,698,995confirmedcasesand564,924
deathsworldwideandthenumberisstillincreasing.Upuntilnow,nospecifictreatmenthasyetbeen
proveneffectiveagainstSARS-CoV-2.Sincethebeginningofthisoutbreak,severalinterestingpaperson
SARS-CoV-2andCOVID-19havebeenpublishedtoreportonthephylogeneticevolution,epidemiology,
pathogenesis,transmissionaswellasclinicalcharacteristicsofCOVID-19andpossibletreatmentsagents.
ThispaperisasystematicreviewoftheavailableliteratureonSARS-CoV-2.Itwasperformedin
accor-dancewithPRISMA(PreferredReportingItemsforSystematicReviewsandMeta-Analyses)andaims
tohelpreadersaccessthelatestknowledgesurroundingthisnewinfectiousdiseaseandtoprovidea
referenceforfuturestudies.
ᄅ2020TheAuthor(s).PublishedbyElsevierLtdonbehalfofKingSaudBinAbdulazizUniversityfor
HealthSciences.ThisisanopenaccessarticleundertheCCBY-NC-NDlicense(http://creativecommons.
org/licenses/by-nc-nd/4.0/).
Contents
Introduction...1602
Methods...1602
Searchstrategy...1602
Inclusionandexclusioncriteria...1602
Dataextractionandsynthesis...1602
Results...1602
SARS-CoV-2emergence...1602
Clinicalfeaturesandpathogenesis...1605
∗ Correspondingauthor.
E-mailaddress:catherine.laprise@uqac.ca(C.Laprise). https://doi.org/10.1016/j.jiph.2020.07.011
1876-0341/ᄅ2020TheAuthor(s).PublishedbyElsevierLtdonbehalfofKingSaudBinAbdulazizUniversityforHealthSciences.Thisisanopenaccessarticleunderthe CCBY-NC-NDlicense(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Convalescentplasmatransfusion(CP)...1607
GlobalSARS-CoV-2preventionmeasures ... 1607
Conclusion ... 1608 Funding...1608 Competinginterests ... 1608 Ethicalapproval...1608 Authorcontributions...1608 Acknowledgements...1608 References ... 1608 Introduction
Coronaviruses(CoV)arethelargestknownRNAviruses.Their sizevariesfrom65to125nmindiameterandtheirnucleicacid genomeissingle-strandedRNA,sizerangingfrom26to32kbin length[1].Since1960,sixcoronaviruseshavebeenfoundtocause diseasesinhumans;SARS-CoV-2istheseventhone,afterSARS-CoV andMERS-CoV[2].WhileHKU1,NL63,OC43and229Eare associ-atedwithmildsymptomsinhumans,SARS-CoV,MERS-CoV,and SARS-CoV-2,belongingtothebetacoronavirusgenus,causesevere todeadlypneumoniainhumans[3].Fever,drycough,difficulty breathingandfatigueusuallyaccompanythispneumonia[4,5].The fatalityrates ofSARS-CoV,MERS-CoVand SARS-CoV-2are9.5%, 34.4%,and 2.3%respectively[6].COVID-19showssome particu-larpathogenic,epidemiologicalandclinicalfeatures whichhave arenotcompletelyunderstoodtodateaswellasitswideandhigh transmissioninthecommunityversusnosocomialspreadofSARS andMERSanditsmilderinfectionandlowmortalitycomparedto theseverephenotypeandhighermortalitycausedbythetwo oth-ersviruses[7].Todate,notherapeuticorvaccineswereapproved againstanyoftheknownhumancoronavirusesandonly protec-tivemeasureswereputinplace.Basedonthecurrentpublished literature,wesummarizeinthispapertheoriginofthisnovelvirus anditslifecycle,theclinicalcharacteristicsofthedisease,the possi-bletransmissionroutes,thepathogenesis,thepreventionmeasures and the undergoing treatments of this emerging infectious disease.
Methods
Searchstrategy
The present study was conducted following the PRISMA guidelines[8]. Weperformeda systematicsearchfor accessible peer-reviewedandfullarticlespublishedfromDecember2019to May2020.TheliteraturesearchwasupdatedinJuly2020while reviewingthepaperpriortoitsresubmission.Articlesforreview wereselectedfromthefollowingdatabasesMEDLINE(PubMed), WebofScienceandGoogleScholar.Thesearchtermsincluded com-binationsof“COVID-19,SARS-CoV-2,newcoronavirus,emergence, symptoms,multiplicationcycle,transmission,diagnosistests, pre-vention,andtreatment”.Full-textversionsoftheincludedpapers wereretrieved. Thereferencelistsofrelevant studieswerealso assessed.
Inclusionandexclusioncriteria
Inclusion and exclusion criteria were recorded following PRISMAguidelinespresentedintheformofa PRISMAflow dia-gram(Fig.1).Briefly, theretrievedliteraturewasimportedinto Endnotesoftware(v.× 9.0)and screenedforexclusion criteria. First,duplicateliteraturewasremovedbyEndnote,andthen obvi-ouslyinappropriateoneswereeliminatedbasedonthetitleand abstract.Finally,remaininginappropriateentrieswereeliminated byreadingthefullarticles.Theexclusioncriteriaforarticleswere non-Englishstudies,entrieswithonlyanabstract,thosewithno relevanttopic,andstudiescontainingnousefulorduplicateddata frompreviouslypublishedstudies.Theinclusioncriteriaare arti-clesreportingconfirmedSARS-CoV-2positivepatients,andstudies presentingoriginaldataaswellasclearandpreciseend-point out-comes.
Dataextractionandsynthesis
The authors independently extracted important information fromeacharticlefulfillingtheinclusioncriteriaandreviewedeach papertoverifytheaccuracyandcoherenceofcollecteddata. Argu-mentsordisagreementswereresolvedfollowingdiscussions.The consensusextracteddataweresynthesizedinthepresentpaper. Ethicalapprovalwasnotrequiredforthisreviewofexisting peer-reviewedpapers.
Results
We identified 170papers through PubMed, Web of science and GoogleScholar databasesand 20 papers throughreference cross-checkand internetresearchof conferenceabstracts. After duplicateremoval,atotalof130paperswerescreenedfor rele-vance.Abstractsandtitlesscreeningidentified32studiesthatmet theinclusioncriteria.Afterthefull-textanalyses,12ofthese stud-ieswereexcluded.Hence,twentystudieswereeligibleaccording toourcriteriaandwereincludedinthisreview(Fig.1).
SARS-CoV-2emergence
Coronaviruseshavebeendescribedascausingseveralsystemic infections in theirselected animal host [9]. However, some of themcanadaptdramaticallyandjumpthespeciesbarrierby nat-uralrecombinationcausingepidemicsorpandemics.Infectionin humanoftenleadstosevereclinicalsymptomsandhigh mortal-ity(https://www.who.int/emergencies/mers-cov/en/).Wepresent
Fig.1.PRISMAflowchartofliteraturesearchstrategy.
Fig.2. Emergenceofhumancoronaviruses:AsofJuly12,2020,sevenCoVsareknowntobehumanpathogensincludingthealpha-CoVsHCoVs-NL63(1200-1500)and HCoVs-229E(1700-1800)andthebeta-CoVsHCoVs-OC43(1890),HCoVs-HKU1(1950),severeacuterespiratorysyndrome-CoV(SARS-CoV)(2002),MiddleEastrespiratory syndrome-CoV(MERS-CoV)(2012)andthenovelSARS-CoV-2(2019).
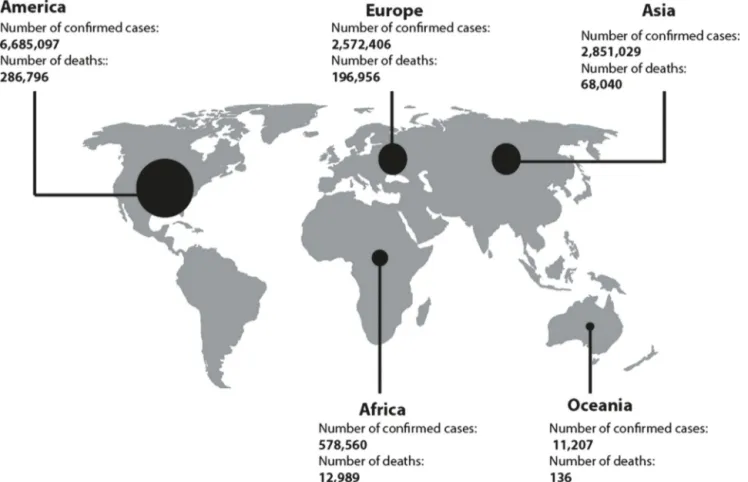
Fig.3.SARS-CoV-2situationupdateworldwide:AsofJuly12,2020,12,698,995casesofCOVID-19havebeenreportedintheworldincluding564,924deaths.Themost affectedcontinentistheAmericawith6,685,097confirmedcasesand286,796deathsonthisday(https://www.ecdc.europa.eu/).
Table1
ComparativeanalysisofSARS-CoV,MERS-CoV,andSARS-CoV-2.
Coronavirus SARS-CoV MERS-CoV SARS-CoV-2
Emergencyyear 2002 2013 2019
Emergencyarea Guangdongprovince,China Arabianpeninsula Wuhan,China
Numberofinfectedcountries 29 27 213
Animalreservoir Bat Bat Bat
Intermediatehost Palmcivets Camels Unknown
incubationtimeinhumans(days) 2–7 2–14 2–14
Causeddisease Severeacuterespiratorysyndrome
(SARS)
MiddleEastrespiratorysyndrome (MERS)
Coronavirusdisease2019 (COVID-19)
Clinicalsymptoms Malaise,diarrhea,cough,feverand
shortnessofbreath
Pneumonia,acuterespiratory distresssyndrome,renalfailure
Cough,feverandshortnessof breath
Numberofinfectedpatients 8098 2494 3,646,304
Numberofdeaths 776 858 252,425
Entryreceptorinhumancells Angiotensin-convertingenzyme2
(ACE2)
Dipeptidylpeptidase4(DPP4) Angiotensin-convertingenzyme2
(ACE2)
Usedtherapy Supportivecare Supportivecare Supportivecare
inFig.2theappearanceofthesevenhumancoronavirusesover thetime(Fig.2).In 2002,inGuangdong(China),theSARS-CoV virusemerged and spreadtothefivecontinentsinfecting8098 peopleand causing 774deaths(9.5%of cases).In 2012, MERS-CoVemerged intheArabianPeninsulainfecting2494peoplein 27countriesandcausing858deaths(34.4%ofcases)[10].Inlate December2019,SARS-CoV-2emergedinaseafoodmarket(where severalwildlifespeciesaresoldincludingbats,rabbits,snakes,birds andfrogs)inWuhanCity,Hubeiprovince,China[11].AsofJuly 12,2020,thisvirushasbeenresponsiblefor12,698,995confirmed casesand564,924deathsworldwide(2.3%).SARS-CoV-2spread rapidlyto213countries(atthetimeofrevision), andthemost affectedcontinentsaretheAmerica andEuropewith6,685,097 and 2,572,406confirmed cases respectively since 31 December
2019andasofJuly12,2020.InAmerica,286,796personswere diedand196,096onesinEuropeuntilthisday(https://www.ecdc. europa.eu/)(Fig.3).OnJanuary30,2020,theWorldHealth Orga-nization (WHO) declared theSARS-CoV-2 epidemic asa public healthemergencyofinternationalconcern[12].OnMarch11,2020, theWHOissued anannouncementof thechangeinCOVID-19’s statusfroman epidemictopandemicdisease.It wassuggested thatMERS-CoV,SARS-CoVandSARS-CoV-2originatedfrombats [13].Phylogenetic analysesshowedthat SARS-CoV-2,SARS-CoV andSARS-likecoronavirusesisolatedinbatsbelongtoadifferent cladethanMERS-CoV,withacompletegenomenucleotide iden-titybetween SARS-CoV-2 and SARS-CoVof 79.5% and between SARS-CoV-2andbatSARScoronavirus(SARSr-CoV-RaTG13)of96% [13,14].Palmcivetsandracoondogswereidentifiedasthelikely
reservoirhostfuellingspillovertohumansforSARS-CoV[15,16], whileMERS-CoV’sintermediatehostisunequivocallydromedary camels[17,18].ForSARS-CoV-2,pangolinsandsnakesarethought tobepotentialintermediatehostsbutthisrequiresfurther con-firmation[19].Moreevidenceisneededtoconfirmtheoriginof thisnovelvirusanditstransmissiontohumans,tounderstandthe bestwaytopreventandslowdownitstransmissionandto bet-tercontroloffuturezoonoticevents.InTable1,wesummarizethe principalcharacteristicsofSARS-CoV,MERS-CoVandSARS-CoV-2. Briefly,batsseemtobethecommonnaturaloriginofSARS-CoV-2, SARS-CoVandMERS-CoV.Theclinicalfeaturesofthethreeviruses arequitesimilar.However,unlikeSARS-CoVandMERS-CoV, SARS-CoV-2ismorecontagiousandspreadsrapidly,currentlyaffecting morethan213countries(https://www.who.int).SARS-CoV-2uses theSARS-CoVreceptorangiotensin-convertingenzyme2(ACE2) onhosttargetcells;however,MERS-CoVbindstothedipeptidyl peptidase4receptor(DDP4)[20].Accordingtothelateststudies, SARS-CoV-2hasthehighestnumberofcasualtiesbutits mortal-ityrateis lower(2.3%) comparedtoSARS-CoV(9.5%)andMERS (34.4%)[6].Therefore,SARS-CoV-2resemblesbothSARS-CoVand MERS-CoV,butappearsuniqueinitshightransmissionrates.
Clinicalfeaturesandpathogenesis
COVID-19isahighlycontagiousdisease.Itsclinical manifesta-tionsrangefrommildtoseverebutmostinfectedcasespresent amildformofthediseaseandthereforehavenosevereclinical features[21].Basedoncurrentdata,81%ofthecasesexhibitmild symptomsand1.2%areasymptomatic(http://weekly.chinacdc.cn). SARS-CoV-2 canspread rapidly in thecommunity contrarilyto SARSCoVandMERS-CoVthathaveahighermortalityratebuta strongernosocomialthancommunitytransmissibility.Thisislikely duetothefactthattheycauseamoresevereclinicalphenotype thanCOVID-19[6].ItwasreportedthatCOVID-19average incuba-tionperiodis5.2days(95%confidenceinterval(CI),4.1–7.0)with the95thpercentileat12.5days[22].Anotherstudyestimatedit at6.4 days(95%CI,5.6–7.7) [23].The medianageofCOVID-19 casesrangesfrom49to57 years[6] andmediantimefromthe firstsymptomtodeathis14days[24].Whileadetailedclinical landscapecontinuestobeestablished,themostcommonclinical symptomsofSARS-CoV-2observedinpatientswerefever(87.9%), cough(67.7%) and fatigue(38.1%), whereasdiarrhea (3.7%) and vomiting(5.0%)wereoccasional [24,25]. Allpatientshad pneu-moniaandabouthalfhaddyspnea[26].SomeCOVID-19patients showedarrhythmia,acuteheart injury,impaired renalfunction andabnormalliverfunction(50.7%)atadmission[27,28].In addi-tion,thereisevidenceofocularsurfaceinfectioninpatientswith COVID-19asSARS-CoV-2RNAwasdetectedineyesecretionsof patients[29,30]. Aretrospectivecase seriesstudyconductedon 214infectedpatientsfromWuhanshowedthat78(36.4%)patients displayedneurologicmanifestations[31].Furthermore,diminished ability tosmellor tasteobserved insomepatients[32,33]was foundtoresultfromaneurotropicorneurovirulentviralinfection oftheolfactorysystem[34].Theolderpopulationandindividuals withunderlyinghealthcomplicationsascardiovasculardiseases anddiabeteswerereportedtopresenttheseverediseasesymptoms [35].Childrenwerefoundlessvulnerablethantheelderpopulation [36,37].PregnantwomenmaybemorevulnerabletoSARS-CoV-2 asthis virusmayalter theimmuneresponses atthe maternal-fetalinterface,andaffectthewell-beingofmothersand infants [38].Aretrospectivestudybasedonninepregnantwomeninfected byCOVID-19showednoevidenceofintrauterinevertical trans-missionbetweenmothersandinfantsinthelatepregnancy[39]. ToavoidSARS-CoV-2newbornsinfectionsafterbirth,immediate preventioninstructionsshouldbeimplementedforthesewomen and theirneonates, including a 14-day isolation for newborns
and avoiding breast feedingduringthis period [40].Laboratory findingsshowedtypicalCTresultsincludingbilateralpulmonary parenchymalgroundglassandconsolidatedpulmonaryopacities sometimeswitharoundedmorphologyandperipherallung distri-bution[41].Ground-glass-likelungimagesareprobablyduetothe severeinflammationoflungcellsbecomingunabletoexchange car-bondioxideandoxygenafterSARS-CoV-2infection[42].Arecent studyshowedthatthatSARS-CoV-2couldinfectTcellsexplaining thelymphocytopeniacommonlyfoundinCOVID-19patients[43]. Itwasalsoobservedthatmostcriticallyillpatientsinfectedwith SARS-CoV-2hadelevatedlevelsof inflammatorycytokines(IL-6 andIL-10)[44],indicatingpotentialbacterialco-infectioncaused by dysregulated immunesystem [45]. Moreover, Nguyenteam, byusinginsilicoanalysis,showedthatgeneticvariabilityacross thethreemajorhistocompatibilitycomplex(MHC)classIgenes (humanleukocyteantigen[HLA]A,B,andC)mayaffect suscepti-bilitytoandseverityofCOVID-19whichneedfurtherexperimental investigation[46].
SARS-CoV-2transmission
UnderstandingtransmissionpathwaysofSARS-CoV-2has sig-nificant implications for intervention and prevention. It was initiallysuggestedthatChinesepatientsinfectedwith SARS-CoV-2 may havevisited theseafood market in WuhanCity or may haveconsumedinfectedanimals.However,furtherinvestigation revealed that some individuals contracted the COVID-19 with-outvisitingthemarket.Indeed,anepidemiologicalstudyinearly casesinthiscityshowedthatonly22%ofpatientsweredirectly exposed tothemarketplace,32% ofcaseswereinclosecontact withthesuspectedcasesand51%hadnocontactwitheithersource [47].Thissuggestsahuman-to-humantransmissionofthevirus and anabilitytopropagate,resultingin diseaseclustersfroma singleindexpatient [48,49].The WHOestimatedthe reproduc-tive number (R0) of SARS-CoV-2 to range between 2 and 2.5, which is higher than SARS (1.7–1.9) and MERS (<1). This sug-geststhat SARS-CoV-2hasa higherpandemicpotential[50,51]. Three transmission ways of SARS-CoV-2 in humans were pro-posedwithincubationtimesof2–14days:1)contactwithliquid dropletsproducedbyinfectedpatientsand/or2)closecontactwith infectedindividualsand3)contactwithsurfacesandmaterial con-taminated with SARS-CoV-2 (https://www.cdc.gov/coronavirus/ 2019ncov/about/transmission).Inexperimentalsetups,infectious virusescouldbedetectedupto24honcardboard,upto2–3days onplastic and stainlesssteel and upto3 hpostaerosolization (vanDoremalenetal.2020).Certainscientistsrecentlyhighlighted another possible transmission route, the airbornetransmission through dropletnuclei (oraerosols), meaningthe possibilityof thediseasespreadinginmuchsmallerparticlesfromexhaledair, knownasaerosols.Theyaresuggestingthataerosolsarealsomore likelythandropletstobeproducedbytalkingandbreathingand mightposeahigherprobabilityoftransmissionthancoughingand sneezing[52].Inlabexperiments,infectiousSARS-CoV-2particles weredetectedinaerosolsfor3h[53].LiuandcolleaguesatWuhan Universitycollectedsamplesofaerosolsinandaroundhospitals treatingCOVID-19patientsandfoundviralRNAfrom SARS-CoV-2 onprotective apparel andfloor surface and theirsubsequent resuspension.Inthisstudy,viralRNAconcentrationinaerosol sam-pleswaslow(0–42genomes/cubicmetreofair)[54].AnAmerican teamstudiedthepresenceofSARS-CoV-2inairsamplesand sur-facesfrom11isolationroomsofCOVID-19patientsandshowed thatmany(63%)ofairsampleshadevidenceofviral contamina-tion,withhigherairbornevirusconcentration(2860 copiesper cubicmetreofair)[55].Itisnoteworthytomentionthatinfectious viruseshavenotbeenrecoveredfromaerosolsinanystudy.Ina recentlypublishedpaper,tenairsamplesofpatientroomswith
spaces[57].
SARS-CoV-2structureandcellsinfection
SARS-CoV-2RNAgenomeis29.9kb[58].Itcontains14open readingframes (ORFs),encoding 27proteins. Atthe5’-terminal regionofthegenome,theORF1andORF2encode15non-structural proteinsimportantforvirusmultiplication.The3’-terminalregion ofthegenomeencodesfunctionalstructuralproteins,namelyspike (S),envelopeprotein(E),membraneprotein(M)andnucleocapsid (N),plus8accessoryproteins[58,59]. Phylogeneticand compu-tational genomic analyses suggest that to enter in host’s cells, SARS-CoV-2sharesthesamehumancellreceptorwithSARS-CoV (ACE2), while MERS-CoVuses another(DPP4) [20]. ACE2 is an ectoenzyme anchored tothe plasma membrane of thecells of severaltissues,particularlyinthelowerrespiratorytract,heart, kidneysand gastrointestinaltract[60]. Astructure model anal-ysis shows that SARS-CoV-2 binds ACE2 with above 10 folds greateraffinitythanSARS-CoV,andmuchhigherthanthethreshold requiredforviralinfection[61].TheSpike(S)protein(ofabout150 kDa)isthemajorantigenpresentedonthesurfaceofSARS-CoV-2. TheSproteinformsatransmembranehomotrimerprotrudingfrom theviralsurfacetoattachtothehostcellularreceptorACE2.S com-prisestwofunctionalsubunits:subunitS1responsibleforbinding tothecellsurfacereceptorACE2andsubunitS2responsibleviral fusiontothecellmembrane[62].
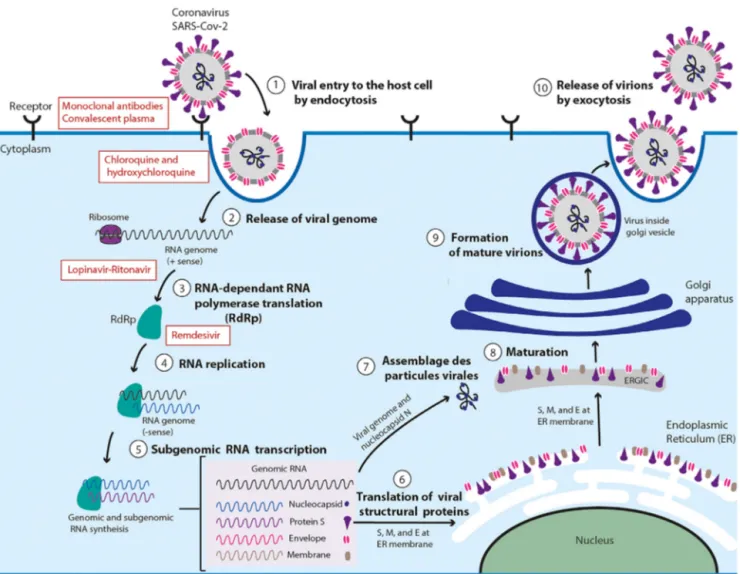
SARS-CoV-2hijackshostcells(suchaslungcells)byendocytosis [63,64].First,SproteinbindstothecellularreceptorACE2[65].This attachmentisfollowedbyactivationoftheSprotein,which initi-atesfusionoftheviralmembranewiththemembraneofthehost cell[10].Thisfusionallowsthevirustoenterthecells[66]. SARS-CoV-2releasesitsgeneticmaterialintothecellcytoplasmwhere itistranslatedintotheviralreplicasepolyproteinspp1aand1ab. Pp1aandp1abarethencleavedbyviralproteinasestoform func-tionalnon-structuralproteins(NSPs)suchasahelicase(Hel)and theRNA-dependentRNApolymerase(RdRp)whichisresponsible forreplicationofstructuralproteinRNA[67].Theplus-stranded RNAgenomeofSARS-CoV-2willservetosynthetizesubgenomic negative-strandtemplatesthatserveastemplatesformRNA syn-thesis.StructuralproteinsS1,S2,E,andMarethentranslatedby ribosomesthatareboundtotheendoplasmicreticulum(ER)[68]. Viralnucleocapsids(N)areassembledfromgenomicRNA,followed bybuddingintothelumenoftheendoplasmicreticulum(ER)–Golgi intermediatecompartment(ERGIC)[69].Thenucleocapsidsfuse withthevirionprecursor.Formedvirionswillthenbetransported fromtheERthroughtheGolgiapparatustothecellsurfaceviasmall vesiclesandreleasedfromthecellthroughexocytosis[70](Fig.4).
SARS-CoV-2diagnosistests
Diagnostictestsweredevelopedrapidlyafterthestartofthe SARS-CoV-2 outbreak allowing earlyrecognition and detection of this novel virus. Nasopharyngeal swabs are the recom-mended specimen for molecular analysis. As of 19 March 2020, the CDC made oropharyngeal, mid-turbinate, and nasal swabs acceptable specimen types if nasopharyngeal swabs are not available (https://www.cdc.gov/coronavirus/2019-nCoV/lab/ guidelines-clinical-specimens.html). Samples are collectedfrom theupperrespiratory tract(oropharyngealandnasopharyngeal)
(ELISA) kits for detection of IgM and IgG antibodies against N and other SARS-CoV-2 proteins have also recently been made available.Severalotherdiagnostictestsaredeveloped todetect otherregionsoftheSARS-CoV-2genomeortargetingRdRp,Hel, S,EandN genes[72]. Anothereasy-to-implementand accurate CRISPR–Cas12-basedlateralflowassayfordetectionofSARS-CoV-2 fromrespiratoryswabRNAextractsinjust30minisunder develop-ment[73].Currentlythereare628SARS-CoV-2testscommercially availableorindevelopmentforthediagnosisofCOVID-19https:// www.finddx.org/covid-19/pipeline/.
SARS-CoV-2treatment
Todate,novaccinesortherapieshavebeenapprovedtotreat anyoftheknownhumancoronaviruses.Therapidglobalspreadof COVID-19hasemphasizedtheneedforthedevelopmentofnew coronavirusvaccines andtherapeuticsforthisfamilyof viruses. Treatmentwillreducetheeconomicimpactontheworldas SARS-CoVhadahistoryoftaxingtheglobaleconomy30US$to100US $billion[74].SincethebeginningoftheCOVID-19outbreak,the WHOhasencouragedresearchersallovertheworldtodevelopa cureforthisdisease.Herewepresentsomeoftheseinitiativesthat arestillinearlystagesofdevelopment.
Antiviralagents
Randomizedcontrolledtrialswereinitiatedandarebeing con-ductedformanyantiviralagents.Lopinavir(LPV)wasshownto inhibittheproteaseactivityofcoronavirusinvitroandinanimal modelsandalreadyusedforSARSandMERSincombinationwith ritonavir,anotherantiviraldrug[75,76].However,arecent trial showedlopinavir-ritonavirhasnotreatmentbenefitforseverely infectedpatientsbySARS-CoV-2[77].Ribavirinisaguanosine ana-logue,usedtotreatseveralviralinfectionsincludingthosecaused bythehepatitisCandrespiratorysyncytialvirusesbytargettingthe RdRpcomplex[78].MessengerRNA(mRNA)vaccinetechnologyis alsounderdevelopment(inphase1clinicaltrialbytheUSNational InstituteofAllergyandInfectiousDiseases)[79].
Remdesivir, a nucleotide analogantiviral inhibitor that may competeforRdRp,wasdesignedfortheEbolavirusandwaswith efficientagainstMERSandSARS[80].Remdesivirhasbeenreported toinhibitinvitroSARS-CoV-2proliferationandthereforehas clin-icaltherapeuticpotential[81].Recently,Remdesivirwasusedina clinicaltrialincluding53COVID-19patients.Resultsshowed clini-calimprovementin36ofthe53patients(68%).However,thisdrug needstobeusedespeciallyforpatientsnotreceivinginvasive ven-tilationasthemortalityratewas18%whenreceivingventilation, comparedto5%whennotreceiving[82].
Chloroquineandhydroxychloroquine
Chloroquineis antimalarialandautoimmunediseasedrug.It blocksviralinfectionbyincreasingendosomalpHlimitingvirusto cellfusionaswellasinterferingwiththeglycosylationofcellular receptorACE2[83]. Hydroxychloroquineisananalogof chloro-quine.Bothdrugshaveimmunomodulatoryeffectandcansupress theimmuneresponseofIL-6andIL-10thathavebeenreported tobeincreasedinresponsetoSARS-CoV-2[84].Clinicalcontrolled trialshaveshownthatchloroquinewasprovedtobeeffectivein
Fig.4.SARS-CoV-2lifecycleininfectedcellsandinhibitiontargets:SARS-CoV-2beginsitslifecyclebybindingoftheSproteinpresentedonthesurfaceofthevirusto thecellularreceptorACE2onthetargetcell.Afterreceptorbinding,theSproteinchangesconformation,facilitatingviralenvelopefusionwiththeinfectedcellmembrane throughendocytosis.SARS-CoV-2thenreleasesitsgeneticmaterialintothehostcell.GenomicRNAistranslatedintoviralreplicasepolyproteinspp1aand1ab,whichare thencleavedintosmallproductsbyviralproteinases.Bydiscontinuoustranscription,thepolymeraseproducesaseriesofsubgenomicmRNAsthataretranslatedintoviral proteins.Thepositive-sensegenomicRNAisthenpackagedintoaribonucleocapsidandisassembledintoviralparticlesintheERandGolgiapparatuswheretheyundergo maturation.Virionsarefinallytransportedviasmallvesiclesandreleasedoutofthecellthroughexocytosis.Inhibitiontargetsarepresentedinred.
thetreatmentofCOVID-19byreducingpneumoniaexacerbation andwasincludedintherecommendationsforthepreventionand treatmentsofSARS-CoV-2[85].
Corticosteroids
Corticosteroidscouldsupresslunginflammationbuttheiruse for thetreatmentof COVID-19lung injury is not supportedby clinical evidence as the clearance of viral infection is delayed andalsoduetotheoccurrenceofsidecomplications[86,87].The WHOadvisesagainsttheuseofcorticosteroidsunlessindicatedfor anotherreason[72].
Antibodies
ThespikeproteinSis theprincipal targetofantibodies.The SARS-CoVmonoclonalantibodyCR3022,aneutralizingantibody previouslyisolatedfromaconvalescentSARSpatient,was identi-fiedtobindpotentlywiththisprotein[88].Thisantibodymaybea potentialtherapeuticcandidate.
Convalescentplasmatransfusion(CP)
Convalescentplasmatransfusion(CP)therapywassuccessfully usedinthetreatmentofSARS,MERSandduringthe2009H1N1
pandemicwithacceptableefficacyandsafety[89–91].Itconsistsof collectingconvalescentplasmafrompatients2weeksafter recov-ery,toensureneutralisationandahighantibodiestiterfollowed byitsadministrationtoinfectedpatients.Duanandhiscolleagues (2020) performed a pilotstudy in three participating hospitals in Chinatoexplore thefeasibilityof CPtreatmentin 10severe COVID-19patients.Theyshowedthatclinicalsymptoms signifi-cantlyimprovedwiththeincreaseofoxyhemoglobin saturation within 3 days, accompanied by rapid neutralizationof viremia [92].Anotherstudyperformedonanuncontrolledcaseseriesof fivecriticallyinfectedpatients,showedimprovementintheir clin-icalsymptoms[93].DespiteCPbeinganeffectivewaytoimprove survivalrateofseverelyinfectedpatients,itdoesnotpermitthe patienttoacquireaSARS-CoV-2immuneprotectionandthesafety ofplasmaglobulinproductsspecifictoSARS-CoV-2deserves fur-therconsideration[94].
GlobalSARS-CoV-2preventionmeasures
Given the lack of available effective vaccine or treatments, it is primordial to control the source of infection and cut off the transmission routeof SARS-CoV-2 by implementing robust preventativemeasuresagainstthisvirus.Weknowthatclose
con-ofrespiratoryillnessinordertodecreasetheriskofspreadingby breakingthetransmissionchain.Thus,manycountriessuspended alltypes(cultural,social,religious,scientific,sporting,andpolitical) ofmassgatheringsandoptedforvideoconferences,and telecom-muting.TheWHOalsorecommendedtomaintainpersonalhygiene especially regular hand washing with soap and water or hand sanitizercontainingatleast60% alcohol,a healthylifestyle and adequatenutritionalintake[95].Outside,peopleneedtorespect minimum2msocialdistancinganditispreferredtowear protec-tivemasks.Tolimitaerosoltransmission,itisimportanttokeep regularroomventilationandeffectivesanitization[24].
In the face of this pandemic, some countries showed good COVID-19curvecontrolbecausetheyrapidlydeployedintensecase findingmeasurestostopvirustransmission.Forexample,South Koreadramaticallyslowedtheepidemicbyperformingmorethan 300000diagnostictests(5,828.6testspermillion)inthe9weeks afterthefirst case wasdescribed. Individualswho tested posi-tivewereidentifiedandisolated[96].Singaporeusedabroadcase definition,aggressivecontacttracing,andisolationbytestingall patientswithpneumoniaandinfluenza-likeillnessesinprimary caresettingsandhospitals,severelysickpatientsinintensivecare, anddeathswithapossibleinfectiousdisease[97].TaiwanandHong Kongusedsimilarstrategies[98].
Conclusion
TheinternationalalertabouttheCOVID-19infectionhashelped inthecontainmentofSARS-CoV-2.Atthedateofwriting, COVID-19showedpromisingsignsofending.Manycountriesseemtobe efficientlycontrollingthisSARS-CoV-2pandemicwaveandhave considerablylimitedthemortalityratethankstoknowledge gar-neredinthepastfromSARSandMERSepidemics,allowingforthe rapidinstitutionofmoreefficientpreventivemeasures.However, SARS-CoV-2isfarfrombeingeradicatedandmanyresearchers pre-dictnovelwavesin thefuture. Thatiswhy researcheffortson SARS-CoV-2andCOVID-19needtoberedoubledtodiscover effi-cienttreatmentsassoonaspossible.Severalpromisingcompetitive therapeuticoptionsarecurrentlyunderdevelopmentalloverthe worldbut requiretime tofor validationandcommercialization. ThereisstillmuchtolearnaboutCOVID-19anditcriticalthat sci-entistaroundtheworldcollaborateandshareinformationinorder tofacethis newglobalthreatand todevelop asuitablecureto benefitallofhumanity.
Funding Nofundingsources. Competinginterests Nonedeclared. Ethicalapproval Notrequired. tion. Acknowledgements
CatherineLaprise(C.L)isthedirectoroftheCentre intersecto-rielensantédurabledel’UQACandthechairholderoftheCanada ResearchChairtier1(CRC1)intheEnvironmentandGeneticsof RespiratoryDisordersandAllergies(www.chairs.gc.ca).Catherine LapriseisoneoftheprincipalresearchersoftheBiobanque Québé-coisedelaCOVID-19(bqc19.ca).CarolineDuchaineisholderofthe Tier-1CanadaResearchChaironBioaerosols.MbarkaBchetniais professorundergrantintheLapriselaboratorywiththesupportof CRC1.
References
[1]ShereenMA, KhanS,KazmiA,BashirN,SiddiqueR. COVID-19infection: origin,transmission,andcharacteristicsofhumancoronaviruses.JAdvRes 2020;24:91–8.
[2]ZhuN,ZhangD,WangW,LiX,YangB,SongJ,etal.Anovelcoronavirusfrom patientswithpneumoniainChina,2019.NEnglJMed2020;382:727–33. [3]CormanVM,LienauJ,WitzenrathM.Coronavirusesasthecauseofrespiratory
infections.Internist(Berl)2019;60:1136–45.
[4]ChangTH,WuJL,ChangLY.Clinicalcharacteristicsanddiagnosticchallenges ofpediatricCOVID-19:asystematicreviewandmeta-analysis.JFormosMed Assoc2020;119(5):982–9.
[5]HuangY,TuM,WangS,ChenS,ZhouW,ChenD,etal.Clinicalcharacteristicsof laboratoryconfirmedpositivecasesofSARS-CoV-2infectioninWuhan,China: aretrospectivesinglecenteranalysis.TravelMedInfectDis2020:101606. [6]PetrosilloN,ViceconteG,ErgonulO,IppolitoG,PetersenE.COVID-19,SARSand
MERS:aretheycloselyrelated?ClinMicrobiolInfect2020;26(6):729–34. [7]MunsterVJ,KoopmansM,vanDoremalenN,vanRielD,deWitE.Anovel
coronavirusemerginginChina-keyquestionsforimpactassessment.NEnglJ Med2020;382:692–4.
[8]MoherD,LiberatiA,TetzlaffJ,AltmanDG.Preferredreportingitemsfor sys-tematicreviewsandmeta-analyses:thePRISMAstatement.AnnInternMed 2009;151(264–9):W64.
[9]SuS,WongG,ShiW,LiuJ,LaiACK,ZhouJ,etal.Epidemiology,genetic recombi-nation,andpathogenesisofcoronaviruses.TrendsMicrobiol2016;24:490–502. [10]Walls AC, Park YJ,Tortorici MA, Wall A,McGuire AT, VeeslerD. Struc-ture,function,andAntigenicityoftheSARS-CoV-2spikeglycoprotein.Cell 2020;181(281–292):e6.
[11]WangH,LiX,LiT,ZhangS,WangL,WuX,etal.Thegeneticsequence,origin, anddiagnosisofSARS-CoV-2.EurJClinMicrobiolInfectDis2020:1–7. [12]LiC,YangY,RenL.Geneticevolutionanalysisof2019novelcoronavirusand
coronavirusfromotherspecies.InfectGenetEvol2020;82:104285. [13]ZhouP,YangXL,WangXG,HuB,ZhangL,ZhangW,etal.Apneumonia
outbreakassociatedwithanewcoronavirusofprobablebatorigin.Nature 2020;579:270–3.
[14]LuR,ZhaoX,LiJ,NiuP,YangB,WuH,etal.Genomiccharacterisationand epidemiologyof2019novelcoronavirus:implicationsforvirusoriginsand receptorbinding.Lancet2020;395:565–74.
[15]KanB,WangM,JingH,XuH,JiangX,YanM,etal.Molecularevolution analysisandgeographicinvestigationofsevereacuterespiratorysyndrome coronavirus-likevirusinpalmcivetsatananimalmarketandonfarms.JVirol 2005;79:11892–900.
[16]WangLF,ShiZ,ZhangS,FieldH,DaszakP,EatonBT.ReviewofbatsandSARS. EmergInfectDis2006;12:1834–40.
[17]MemishZA,MishraN,OlivalKJ,FagboSF,KapoorV,EpsteinJH,etal.Middle Eastrespiratorysyndromecoronavirusinbats,SaudiArabia.EmergInfectDis 2013;19:1819–23.
[18]RajVS,OsterhausAD,FouchierRA,HaagmansBL.MERS:emergenceofanovel humancoronavirus.CurrOpinVirol2014;5:58–62.
[19]LamTT,ShumMH,ZhuHC,TongYG,NiXB,LiaoYS,etal.IdentifyingSARS-CoV-2 relatedcoronavirusesinMalayanpangolins.Nature2020;583(7815):282–5. [20]WanY,ShangJ,GrahamR,BaricRS,LiF.Receptorrecognitionbythenovel
coronavirusfromWuhan:ananalysisbasedondecade-longstructuralstudies ofSARScoronavirus.JVirol2020:94.
[21]WangY,WangY,ChenY,QinQ.Uniqueepidemiologicalandclinicalfeaturesof theemerging2019novelcoronaviruspneumonia(COVID-19)implicatespecial controlmeasures.JMedVirol2020;92(6):568–76.
[22]LiP,FuJB,LiKF,ChenY,WangHL,LiuLJ,etal.TransmissionofCOVID-19 intheterminalstageofincubationperiod:afamilialcluster.IntJInfectDis 2020;96:452–3.
[23]BackerJA,KlinkenbergD,WallingaJ.Incubationperiodof2019novel coro-navirus(2019-nCoV)infectionsamongtravellersfromWuhan,China,20-28 January2020.EuroSurveill2020:25.
[24]GuanWJ,ZhongNS,ReplyClinicalcharacteristicsofCovid-19inChina.NEngl JMed2020;382(19):1861–2.
[25]YangW,CaoQ,QinL,WangX,ChengZ,PanA,etal.Clinicalcharacteristicsand imagingmanifestationsofthe2019novelcoronavirusdisease(COVID-19):a multi-centerstudyinWenzhoucity,Zhejiang,China.JInfect2020;80:388–93. [26]WuD,WuT,LiuQ,YangZ.TheSARS-CoV-2outbreak:whatweknow.IntJ
InfectDis2020;94:44–8.
[27]WangD,HuB,HuC,ZhuF,LiuX,ZhangJ,etal.Clinicalcharacteristicsof138 hos-pitalizedpatientswith2019novelcoronavirus–InfectedpneumoniainWuhan, China.JAMA2020;323:1061–9.
[28]LiZ,WuM,YaoJ,GuoJ,LiaoX,SongS,etal.Cautiononkidney dysfunc-tionsofCOVID-19patients.medRxiv2020,http://dx.doi.org/10.1101/2020.02. 08.20021212.
[29]XieHT,JiangSY,XuKK,LiuX,XuB,WangL,etal.SARS-CoV-2intheocular surfaceofCOVID-19patients.EyeVis(Lond)2020;7:23.
[30]ZhangX,ChenX,ChenL,DengC,ZouX,LiuW,etal.TheevidenceofSARS-CoV-2 infectiononocularsurface.OculSurf2020;18(3):360–2.
[31]MaoL,JinH,WangM,HuY,ChenS,HeQ,etal.Neurologicmanifestationsof hospitalizedpatientswithcoronavirusdisease2019inWuhan,China.JAMA Neurol2020;77(6):1–9.
[32]WolfelR,CormanVM,GuggemosW,SeilmaierM,ZangeS,MullerMA,etal. Virological assessment of hospitalizedpatients with COVID-2019. Nature 2020;581(7809):465–9.
[33]SpinatoG,FabbrisC,PoleselJ,CazzadorD,BorsettoD,HopkinsC,etal. Alter-ationsinsmellortasteinmildlysymptomaticoutpatientswithSARS-CoV-2 infection.JAMA2020;323(20):2089–90.
[34]Xydakis MS,Dehgani-Mobaraki P, Holbrook EH, Geisthoff UW,Bauer C, Hautefort C, et al, S1473-3099(20)30293 Smelland taste dysfunction in patientswithCOVID-19.LancetInfectDis2020,http://dx.doi.org/10.1016/ S1473-3099(20)30293-0.
[35]NiuS,TianS,LouJ,KangX,ZhangL,LianH,etal.Clinicalcharacteristicsofolder patientsinfectedwithCOVID-19:adescriptivestudy.ArchGerontolGeriatr 2020;89:104058.
[36]LuX,ZhangL,DuH,ZhangJ,LiYY,QuJ,etal.SARS-CoV-2infectioninchildren. NEnglJMed2020;382:1663–5.
[37]DongY,WangL,BurgnerDP,MillerJE,SongY,RenX,etal.Infectiousdiseasesin childrenandadolescentsinChina:analysisofnationalsurveillancedatafrom 2008to2017.BMJ2020;369:m1043.
[38]LiuH,WangLL,ZhaoSJ,Kwak-KimJ,MorG,LiaoAH.Whyarepregnantwomen susceptibletoCOVID-19?Animmunologicalviewpoint.JReprodImmunol 2020;139:103122.
[39]ChenH,GuoJ,WangC,LuoF,YuX,ZhangW,etal.Clinical characteris-ticsandintrauterineverticaltransmissionpotentialofCOVID-19infection inninepregnantwomen:aretrospectivereviewofmedicalrecords.Lancet 2020;395:809–15.
[40]XuL,YangQ,ShiH,LeiS,LiuX,ZhuY,etal.Clinicalpresentationsandoutcomes ofSARS-CoV-2infectedpneumoniainpregnantwomenandhealthstatusof theirneonates.SciBull(Beijing)2020;65(18):1537–42.
[41]ChungM,BernheimA,MeiX,ZhangN,HuangM,ZengX,etal.CTimaging featuresof2019novelcoronavirus(2019-nCoV).Radiology2020;295:202–7. [42]LiuW,LiH.COVID-19:attacksthe1-betachainofhemoglobinandcaptures
theporphyrintoinhibithumanhememetabolism.ChemRxiv;2020. [43]WangX,XuW,HuG,XiaS,SunZ,LiuZ,etal.SARS-CoV-2infectsT
lympho-cytesthroughitsspikeprotein-mediatedmembranefusion.CellMolImmunol 2020;17(8):894.
[44]MooreBJB,JuneCH.CytokinereleasesyndromeinsevereCOVID-19.Science 2020;368(490):473–4.
[45]Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immuneresponseinpatientswithCOVID-19inWuhan,China.ClinInfectDis 2020;71(15):762–8.
[46]Nguyen A, David JK, Maden SK, Wood MA, Weeder BR, Nellore A, etal.HumanleukocyteantigensusceptibilitymapforSARS-CoV-2.JVirol 2020;94(13):e00510–20.
[47]LiR,TianJ,YangF,LvL,YuJ,SunG,etal.Clinicalcharacteristicsof225 patientswithCOVID-19inatertiaryHospitalnearWuhan,China.JClinVirol 2020;127:104363.
[48]PhanT.GeneticdiversityandevolutionofSARS-CoV-2.InfectGenet Evol 2020;81:104260.
[49]RiouJ,AlthausCL.Patternofearlyhuman-to-humantransmissionofWuhan 2019novelcoronavirus(2019-nCoV),December2019toJanuary2020.Euro Surveill2020:25.
[50]ChenTM,RuiJ,WangQP,ZhaoZY,CuiJA,YinL.Amathematicalmodelfor simulatingthephase-basedtransmissibilityofanovelcoronavirus.InfectDis Poverty2020;9:24.
[51]LiuY,GayleAA,Wilder-SmithA,RocklovJ.Thereproductivenumberof COVID-19ishighercomparedtoSARScoronavirus.JTravelMed2020:27.
[52]LewisD.Isthecoronavirusairborne?Expertscan’tagree.Nature2020;580:175. [53]van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, WilliamsonBN,etal.AerosolandsurfacestabilityofSARS-CoV-2ascompared withSARS-CoV-1.NEnglJMed2020;382:1564–7.
[54]LiuY,NingZ,ChenY,GuoM,LiuY,GaliNK,etal.Aerodynamicanalysisof SARS-CoV-2intwoWuhanhospitals.Nature2020;582(7813):557–60. [55]SantarpiaJL,RiveraDN,HerreraV,MorwitzerMJ,CreagerH,SantarpiaGW,
etal.TransmissionpotentialofSARS-CoV-2inviralsheddingobservedatthe UniversityofNebraskaMedicalCenter.medRxiv2020,http://dx.doi.org/10. 1101/2020.03.23.20039446.
[56]FaridiS,NiaziS,SadeghiK,NaddafiK,YavarianJ,ShamsipourM,etal.Afield indoorairmeasurementofSARS-CoV-2inthepatientroomsofthelargest hospitalinIran.SciTotalEnviron2020;725:138401.
[57]MorawskaL,CaoJ.AirbornetransmissionofSARS-CoV-2:theworldshouldface thereality.EnvironInt2020;139:105730.
[58]WuF,ZhaoS,YuB,ChenYM,WangW,SongZG,etal.Anewcoronavirus associatedwithhumanrespiratorydiseaseinChina.Nature2020;579:265–9. [59]MalikYS,SircarS,BhatS,SharunK,DhamaK,DadarM,etal.Emergingnovel coronavirus(2019-nCoV)-currentscenario,evolutionaryperspectivebasedon genomeanalysisandrecentdevelopments.VetQ2020;40:68–76.
[60]Imai Y, KubaK, Ohto-NakanishiT, PenningerJM.Angiotensin-converting enzyme2(ACE2)indiseasepathogenesis.CircJ2010;74:405–10.
[61]WrappD,WangN,CorbettKS,GoldsmithJA,HsiehCL,AbionaO,etal. Cryo-EMstructureofthe2019-nCoVspikeintheprefusionconformation.Science 2020;367:1260–3.
[62]LetkoM,MarziA,MunsterV.Functionalassessmentofcellentryandreceptor usageforSARS-CoV-2andotherlineageBbetacoronaviruses.NatMicrobiol 2020;5:562–9.
[63]QinfenZ,JinmingC,XiaojunH,HuanyingZ,JichengH,LingF,etal.Thelifecycle ofSARScoronavirusinVeroE6cells.JMedVirol2004;73:332–7.
[64]Weiss SR, Navas-Martin S. Coronavirus pathogenesis and the emerging pathogensevereacuterespiratorysyndromecoronavirus.MicrobiolMolBiol Rev2005;69:635–64.
[65]RabiFA,AlZoubiMS,KasasbehGA,SalamehDM,Al-NasserAD.SARS-CoV-2 andcoronavirusdisease2019:whatweknowsofar.Pathogens2020:9. [66]SimmonsG,ZmoraP,GiererS,HeurichA,PohlmannS.Proteolyticactivation
oftheSARS-coronavirusspikeprotein:cuttingenzymesatthecuttingedgeof antiviralresearch.AntiviralRes2013;100:605–14.
[67]ZhavoronkovA.Geroprotectiveandsenoremediativestrategiestoreducethe comorbidity,infectionrates,severity,andlethalityingerophilicandgerolavic infections.Aging(AlbanyNY)2020:12.
[68]ShinJS,JungE,KimM,BaricRS,GoYY.Saracatinibinhibitsmiddleeast respi-ratorysyndrome-coronavirusreplicationinvitro.Viruses2018:10.
[69]StertzS,ReicheltM,SpiegelM,KuriT,Martinez-SobridoL,Garcia-SastreA,etal. TheintracellularsitesofearlyreplicationandbuddingofSARS-coronavirus. Virology2007;361:304–15.
[70]Masters PS. The molecular biology of coronaviruses. Adv Virus Res 2006;66:193–292.
[71]CormanVM,LandtO,KaiserM,MolenkampR,MeijerA,ChuDK,etal. Detec-tionof2019novelcoronavirus(2019-nCoV)byreal-timeRT-PCR.EuroSurveill 2020:25.
[72]ZhaiP,DingY,WuX,LongJ,ZhongY,LiY.Theepidemiology,diagnosisand treatmentofCOVID-19.IntJAntimicrobAgents2020:105955.
[73]BroughtonJP,DengX,YuG,FaschingCL,ServellitaV,SinghJ,etal. CRISPR-Cas12-baseddetectionofSARS-CoV-2.NatBiotechnol2020;38(7):870–4. [74]SmithRD.Respondingtoglobalinfectiousdiseaseoutbreaks:lessonsfromSARS
ontheroleofriskperception,communicationandmanagement.SocSciMed 2006;63:3113–23.
[75]YaoTT,QianJD,ZhuWY,WangY,WangGQ.Asystematicreviewoflopinavir therapyforSARScoronavirusandMERScoronavirus-Apossiblereferencefor coronavirusdisease-19treatmentoption.JMedVirol2020;92(6):556–63. [76]ZhuS,GuoX,GearyK,ZhangD.EmergingtherapeuticstrategiesforCOVID-19
patients.Discoveries(Craiova)2020;8:e105.
[77]CaoB,WangY,WenD,LiuW,WangJ,FanG,etal.AtrialofLopinavir-Ritonavirin adultshospitalizedwithsevereCovid-19.NEnglJMed2020;382(19):1787–99. [78]ElfikyAA.Ribavirin,remdesivir,Sofosbuvir,Galidesivir,andTenofoviragainst SARS-CoV-2RNAdependentRNApolymerase(RdRp):amoleculardocking study.LifeSci2020;253:117592.
[79]AhmedSF,QuadeerAA,McKayMR.Preliminaryidentificationofpotential vac-cinetargetsfortheCOVID-19coronavirus(SARS-CoV-2)basedonSARS-CoV immunologicalstudies.Viruses2020:12.
[80]GordonCJ,TchesnokovEP,WoolnerE,PerryJK,FengJY,PorterDP,etal. Remde-sivirisadirect-actingantiviralthatinhibitsRNA-dependentRNApolymerase fromsevereacuterespiratorysyndromecoronavirus2withhighpotency.JBiol Chem2020;295(20):6785–97.
[81]WangM,CaoR,ZhangL,YangX,LiuJ,XuM,etal.Remdesivirand chloro-quineeffectivelyinhibittherecentlyemergednovelcoronavirus(2019-nCoV) invitro.CellRes2020;30:269–71.
[82]GreinJ,OhmagariN,ShinD,DiazG,AspergesE,CastagnaA,etal. Compas-sionateuseofremdesivirforpatientswithsevereCovid-19.NEnglJMed 2020;382(24):2327–36.
[83]VincentMJ,BergeronE,BenjannetS,EricksonBR,RollinPE,KsiazekTG,etal. ChloroquineisapotentinhibitorofSARScoronavirusinfectionandspread. VirolJ2005;2:69.
[84]SavarinoA,BoelaertJR,CassoneA,MajoriG,CaudaR.Effectsofchloroquine onviralinfections:anolddrugagainsttoday’sdiseases?LancetInfectDis 2003;3:722–7.
[85]GaoJ,HuS.Updateonuseofchloroquine/hydroxychloroquinetotreat coro-navirusdisease2019(COVID-19).BiosciTrends2020;14(2):156–8.
2005;24:44–6.
[90]ZhouJ,LiuJH,JinY,OuyangXL,YangLG.[ProtectiveeffectsofDMSOon func-tionoflyophilizedhumanplatelets].ZhongguoShiYanXueYeXueZaZhi 2007;15:1284–8.
[91]Hung KT, Berisha SZ, Ritchey BM, Santore J, Smith JD.Red blood cells playaroleinreversecholesteroltransport.ArteriosclerThrombVascBiol 2012;32:1460–5.
[92]DuanK,LiuB,LiC,ZhangH,YuT,QuJ,etal.Effectivenessof convales-centplasmatherapyinsevereCOVID-19patients.ProcNatlAcadSciUSA 2020;117(17):9490–6.
[93]Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 critically ill patients with COVID-19with convalescent plasma. JAMA 2020;323(16):1582–9.
Therapy,KoreanSocietyforHealthcare-associatedInfectionControland Pre-vention, KoreaCentersforDiseaseControlandPrevention.Reportonthe epidemiologicalfeaturesofcoronavirusdisease2019(COVID-19)outbreakin theRepublicofKoreafromJanuary19toMarch2,2020.JKoreanMedSci 2020;35:e112.
[97]LeeVJ,ChiewCJ,KhongWX.InterruptingtransmissionofCOVID-19:lessons fromcontainmenteffortsinSingapore.JTravelMed2020;27(3):taaa039. [98]WangCJ,NgCY,BrookRH.ResponsetoCOVID-19inTaiwan:bigdataanalytics,
newtechnology,andproactivetesting.JAMA2020,http://dx.doi.org/10.1001/ jama.2020.3151.