oooo
Current Genetics
9 Springer-Verlag 1991
Sequence analysis of the
ARG7 gene of
Schizosaccharomyces pombe
coding for argininosuccinate lyase
E x p r e s s i o n o f t h e g e n e in
Saccharomyces cerevisiae
Roland Loppes 1, Reiner Michels 1, Isabelle Decroupette 1, and Bernard Joris 2
1 Laboratory of Molecular Genetics, Botany Institute, B22, and z Laboratory for Protein Engineering, Chemistry Institute, B6, University of Li6ge, Sart Tilman, B-4000 Li+ge, Belgium
Received January 3, 1991
Summary.
The complete nucleotide sequence of theA R G 7 gene, coding for argininosuccinate lyase (EC 4.3.2.1), in the fission yeast (Schizosaccharomyces
pombe) has been determined. It consists of an open read- ing frame of 461 codons. The deduced protein has a molecular weight of 51 200 Da. The gene is devoid of introns which is confirmed by the fact that it is expressed in Escherichia coli after spontaneous insertion of a bacte- rial sequence probably bearing a prokaryotic promoter. A perfect "TATA" box is found at 72 and the major transcription initiation site in Saccharomyces cerevisiae is located at - 11 as shown by primer extension experiments. Comparison of the S. pombe lyase with related proteins from other organisms reveals an important degree of con- servation except in the carboxyterminal part of the polypeptide. Additionally, a deletion removing 66 amino acids of the carboxy terminus yields an enzyme exhibiting some biological activity. A unique 1 500 b transcript was found in S. cerevisiae when the intact gene was present, but the deleted version of the gene gave rise to at least three transcripts of 1 800, 2 800 and 3 900 b.
Key words:
Schizosaccharomyces pombe - Saccha-romyces cerevisiae - Argininosuccinate lyase - Sequence
Introduction
Argininosuccinate lyase (ASL, EC 4.3.2.1) catalyses the cleavage of argininosuccinate into fumarate and arginine, the last step in the biosynthesis of this amino acid. The enzyme also plays a key role in the urea cycle leading to the removal of ingested nitrogen in the mammalian liver. It is highly analogous to the 6-crystallin found exclusively in reptiles and birds (Piatigorsky et al. 1988).
Argininosuccinate lyase has been purified from beef (Lusty and Ratner 1972) and human (O'Brien and Barr 1981) liver and from microorganisms such as Saccha-
Offprint requests to: R. Loppes
romyces cerevisiae (Schweitzer 1982) and Chlamy-
domonas reinhardtii (Farrell and Overton 1987). In every case, the enzyme was shown to be a homotetramer with subunits of about 50 000 Da. In rat (Amaya et al. 1988) and man (O'Brien et al. 1986; Matuo et al. 1988) the amino acid sequence has been determined from cDNA clones. In yeast, the complete sequence (Beacham et al. 1984), and in Chlamydomonas a partial sequence (De- buchy et al. 1989), was obtained from clones obtained from genomic libraries. In Escherichia coli, only the first 151 bases of the gene are known (Parsot et al. 1988).
The A R G 4 gene ofS. cerevisiae, coding for ASL, is not under a specific control system mediated by arginine but is subject to the general control of amino acid biosynthe- sis (Delforge et al. 1975; Messenguy and Dubois 1983). A deletion analysis of the A R G 4 promoter has recently been carried out (Thiry-Blaise and Loppes 1990). This study confirmed the role of the putative UAS and revealed another interesting sequence (a stretch of 14 dA residues lying between -124 and -137 from the initiation co don) probably acting as a constitutive promoter (Struhl 1985). Nothing is known about the structure of the corre- sponding gene (ARG7) in the fission yeast Schizosaccha-
romyces pombe. This gene has been recently cloned (Remacle et al. 1988) by complementation of an arg4 mutant of S. cerevisiae. In this study, we report the com- plete sequence of the A R G 7 gene and the mapping of the transcription initiation sites in S. cerevisiae. The argini- nosuccinate lyase of S. pombe displays a high degree of similarity to the corresponding S. cerevisiae, human and rat proteins and to the chicken lens structural protein 6-crystallin.
Materials and methods
Strains and media. Saccharomyces cerevisiae UL1 (arg4 his3 leu2; Remacle et al. 1988) was used for selecting arginine prototrophs. It was grown on YNB minimal medium with 50 mg/1 of each appro- priate supplement or on YPD rich medium. Escherichia eoli C600
argH (lacking argininosuccinate lyasc, &argBH Apro-argF-lac
256 Sa 4 0 0 b p I I R Sc BH . . . 2 1 . l m I . . . i + . + + + § 2 4 7 S RP R R Sc I i I i B . . . 2pm ~ p U L G - SP2 . . . pULG- S P 3 . . . pULG-ID1
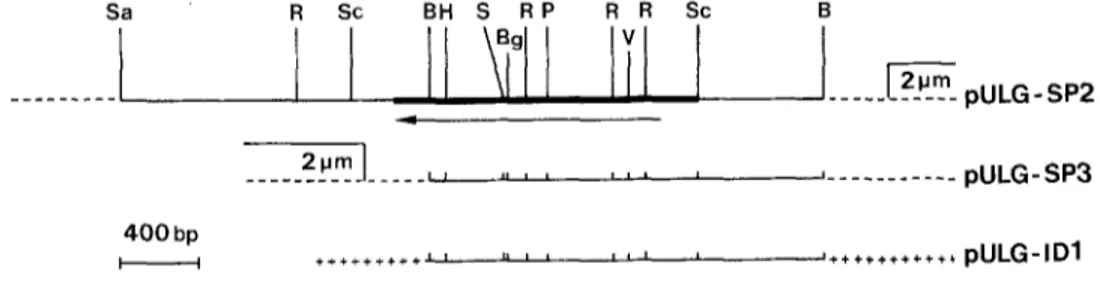
Fig. 1. Plasmids used in this study: pULG-SP2 (3,6 kb Sau3A fragment inserted at the YEp13 BamHI site; one BamHI site is re- generated), pULG-SP3 (2 kb BamHI fragment in YEp13) and pULG-ID1 (2 kb BamHI frag- ment in pUC19)_ The sequenced region is marked by black bar. The arrow shows the orientation of the gene in the three construc- tions. - - , insert; . . . , YEp13; ++++, pUC19. B, BamHI; Bg, BglII; H, HindI[I; P, PvuII; R, EcoRI; S, SacI; Sa, Sau3A; So, ScaI; V, EcoRV
argH mutation on M9 medium supplemented with proline (50 mg/ 1), thiamine (1 mg/l) and citrutline (50 mg/1). C600 pyrF (Apro- argF-lac argI thi pyrF hSrk hsdk) was used as a host for most plasmids. Both C600 strains were provided by M. Crabeel (V. U. Brussels). JM 105 (thi rpsL endA sbcB15 hsr k hsd k 9 lac proAB [F' traD36proAB laelq 9 MI5]) served as host for M13 phages and plasmid pUC19.
Plasmids. The S. pombe ARG7 gene is located on a 3.6 kb Sau3A fragment cloned at the BamHI site of YEp13 to give pULG-SP2 (Remacle et al. 1988) (see Fig. 1). Plasmids pULG-SP3 and pULG- IDI were obtained by cloning the 2 kb BamHI-BamHI fragment into YEp13 and pUC19 respectively.
DNA sequencing. DNA fragments were cloned into M13mp18 and M I 3mp19 and sequenced by the dideoxy chain termination method (Sanger et at. 1977) using the Klenow fragment of DNA polymerase I according to a procedure provided by Bethesda Research Labora- tories, Gaithersburg, USA (Focus 9, 3, 1987) and a discontinuous buffer concentration gradient during electrophoresis (Biggin et al. 1983),
RNA preparations and Northern blots. Total S. cerevisiae RNA was prepared according to a procedure adapted from Nicolet et al. (1985). About 5 • 9 cells were collected during exponential growth in YNB medium and resuspended in 4 ml of cold 200 mM Tris-HCl pH 7.5, 500 mM NaCl, 1% SDS, 10 mM EDTA in a 30 ml Corex (Du Pont, Wilmington, USA) tube. Two milliliters of glass beads (0.5 mm diameter) and 4 ml phenol-chloroform were added and the cells were disrupted by fast vortex mixing. After centrifuga- tion (15 min at 8 000 g) the aqueous phase was extracted three times with phenol-chloroform, once with ether and the nucleic acids were precipitated overnight at -20~ with ethanol, washed with 70% ethanol, dried and dissolved in 250 I~1 of water. About 0.5 mg RNA in 100 pl 50 mM Tris-HC1 pH 7.8, 5 mM MgCl 2 were treated with 40 ~tg/mt RNAse-free DNAse for 30 rain at 25 ~ After two extrac- tions with phenol-chloroform, RNA was precipitated with ethanol, washed, dried and dissolved in 100 gl 10 mM Tris-HC1 pH 8.0, 1 mM EDTA and kept at -20 ~ RNA samples (20 p.g) were denatured with glyoxal (Sambrook et al. 1989) and fractionated by electrophoresis in 1.2% agarose gels in 10 mM sodium phosphate buffer pH 7.0 at 4 ~ (3 V/cm) with continuous buffer recirculation. After treatment of the gel for 10 rain in 5 mM NaOH, RNA was transferred to Zeta-probe (Bethesda Research Laboratories) by blotting with 5 mM NaOH as described by Vrati et al. (1987). The membranes were briefly rinsed in 2 x SSC (1 x SSC= 150 mM NaC1, 15 mM trisodium citrate), 0.1% SDS and kept at -20 ~ Membranes were prehybridized at 48~ for 5 h in 50% formamide, 1.5 x SSPE ( I x S S P E = 1 8 0 m M NaC1, 10mM NaH2PO 4, l mM EDTA, pH 7,4), 1% SDS, 0.5% Blotto and 0.5 mg/ml denatured herring sperm DNA then hybridized overnight at 48 ~ in the same solution containing 10% dextran sulfate and 105 cpm/cm 2 of the probe la- belled by nick-translation (Rigby et al. 1977) and finally washed for 15 min in 2 x SSC at 25~ 15 min in 0.1 x SSC, 0.1% SDS at 25 ~ and 30 rain in 0.1 x SSC, 1% SDS at 60~
Primer extension. Primer extension reactions were carried out using total RNA. An oligonucleotide (5'-CATCAGTGGATCAG-
TAGCT-3') complementary to bases + 48 to + 66 of the ARG7 gene was labelled with T4 polynucleotide kinase and ?-[32p]ATP and purified by chromatography on Sephadex G-10 (Pharmacia, Upp- sala, Sweden) equilibrated with 50 mM ammonium bicarbonate pH 7.8 (Zoller and Smith 1982). Twenty-five txg of RNA were resus- pended in 7.5 gl of 250 mM KC1, 10 mM Tris-HC1 pH 8.3 and annealed with 2 p mole of labeled primer (1.6 x 107 cpm/p mole). The extension reaction was then allowed to proceed for 1 h at 37 ~ in a total volume of 25 gl (50 mM Tris-HC1 pH 7.5, 10 mM DTT, 3 mM MgC12, 75 mM KC1, 100 pg/ml BSA, 500 pM each of dATP, dTTP, dGTP and dCTP, 50 gg/ml actinomycin D and 500 units M-MLV reverse transcriptase). The elongated cDNA was precipi- tated by ethanol, resuspended in sequencing loading buffer and denatured for 3 min at 100~ before loading on the gel.
Computer analysis. The GCG sequence analysis system was utilized (Devereux et al. 1984). In this package, the "distances" algorithm requires aligned sequences and compares them residue by residue. The final score represents the number of matches divided by the length of the shortest sequence excluding the gaps, In the present analysis, a match was scored when the substitution had a value larger than, or equal to 1.5 in the table of Gribskov and Burgess (1986) which is a normalized form of the Dayhoff (1979) table.
Results
Location o f the A R G 7 gene within the cloned f r a g m e n t T h e A R G 7 gene o f S. p o m b e h a s p r e v i o u s l y b e e n s h o w n to be l o c a l i z e d o n a 3.6 k b S a u 3 A f r a g m e n t a n d o n a 2 k b B a m H I f r a g m e n t ( R e m a c l e et al. 1988). W h e n these frag- m e n t s were s u b c l o n e d i n t o Y E p l 3, the r e s u l t i n g p l a s m i d s ( p U L G - S P 2 a n d p U L G - S P 3 r e s p e c t i v e l y ) c o m p l e m e n t e d a n arg4 m u t a n t o f S. cerevisiae l a c k i n g a r g i n i n o s u c c i n a t e l y a s e a c t i v i t y (Fig. 1). T h e c o l o n i e s h a r b o u r i n g p U L G - SP3, h o w e v e r , g r e w m u c h m o r e s l o w l y t h a n t h o s e h a r - b o u r i n g p U L G - S P 2 , w h i c h s u g g e s t e d t h a t t h e gene was slightly a l t e r e d in p U L G - S P 3 .
T h e p o s i t i o n o f the gene in the 3.6 k b f r a g m e n t a n d the d i r e c t i o n o f t r a n s c r i p t i o n were i n d i r e c t l y i n f e r r e d f r o m t h e s t u d y o f its e x p r e s s i o n in E. coli C600 argH. T h i s s t r a i n , h a r b o u r i n g p U L G - S P 2 , w a s strictly d e p e n d e n t o n arginine. H o w e v e r , c o l o n i e s a p p e a r e d a f t e r 3 d a y s at 37 ~ o n p l a t e s l a c k i n g a r g i n i n e a t a f r e q u e n c y o f a b o u t 1 x 10 - 8 . T h e p l a s m i d ( p U L G - S P 2 - 3 ) e x t r a c t e d f r o m one o f these c l o n e s was s h o w n to t r a n s f o r m C600 a r g H to a r g i n i n e i n d e p e n d e n c e i n d i c a t i n g t h a t this p r o p e r t y w a s specified b y the p l a s m i d r a t h e r t h a n b y the h o s t g e n o m e . R e s t r i c t i o n o f p U L G - S P 2 - 3 a n d p U L G - S P 2 w i t h v a r i o u s c o m b i n a t i o n s o f e n d o n u c l e a s e s r e v e a l e d the p r e s e n c e in p U L G - S P 2 - 3 o f a 1.2 k b i n s e r t i o n in the B a m H I - B a m H I
257 - 1 8 1 A G T A C T T G C T A T C - 1 2 1 C A C G G A T G T T A T T T T G A C C C A T A T T C G T A T A G T G T T A T C T A A C T A G A G A A A A A G C T A A A A 9 " - 6 1 T G A T A A G T G C G A A T A T C T G A G G A A G A A A A A G T C A T T T C T T C A C G C T A T T A T A T A A G T A A A - 1 G G A A A A T T A G A C C A T C A T T T G T A G A C T G A A A A T A A T A T T G A A T A A A A G C A T C T T G G C A C T * 1 M e t A l a G l u L y s S e r S e r L y s L y s L e u T r p G l y G l y A r g P h e S e r G l y 1 A T G G C A G A A A A A T C A A G C A A A A A A C T A T G G G G A G G T A G A T T T T C A G G A 17 A l a T h r A s p P r o L e u M e t A l a G l u P h e A S h L y s S e t I l e T y r S e r G l y 4 9 G C T A C T G A T C C A C T G A T G G C A G A A T T C A A C A A A T C C A T C T A T A G T G G A 33 L y s G l u M e t C y s G I u G I u A s p V a l I l e G l y S e r M e t A l a T y r A l a L y s 9 7 A A G G A A A T G T G C G A A G A A G A T G T T A T T G O T T C C A T G G C G T A C G C A A A A 4 9 A l a L e u C y s G l n L y s A S h V a l I l e S e r G l u G l u G l u L e u A S h S e r I l e 1 4 5 G C C T T G T G C C A G A A A A A T G T G A T A T C T G A A G A A G A G C T G A A T A G C A T C 6 5 L e u L y s G l y L e u G l u G l n I l e G l n A r g G I U T r p A s h s e r G l y G l n P h e 1 9 3 C T A A A A G G A T T G G A A C A A A T T C A A A G A G A A T G G A A T T C G G G T C A A T T C 81 V a l L e u G I u P r o S e r A s p G I u A s p V a l H i s T h r A l a A s n G l u A r g A r g 2 4 1 G T T T T G G A A C C A T C C G A C G A A G A T G T T C A C A C A G C A A A C G A G C G C C G A 97 L e u T h r G l u I l e I l e G l y A s p V a l A l a G l y L y s L e u H i S T h r G l y A r g 2 8 9 T T A A C T G A G A T A A T C G G T G A T G T T G C T G G C A A G C T A C A T A C T G G C A G A 1 1 3 S e r A r g A s n A s p G l n V a l T h r T h r A s p L e u A r g L e u T r p L e u C y s A r g 3 3 7 A G T C G T A A T G A C C A A G T T A C C A C C G A T T T G C G T T T A T O G C T A T G C A G A 1 2 9 L y s I l e L y s G I u V a l G I u V a l T y r V a l I l e A s n L e u L e u L y s V a l P h e 3 8 5 A A A A T C A A A G A G G T T G A A G T C T A T G T C A T T A A C T T G C T T A A A G T T T T T 1 4 5 T h r A S h A r g A l a G l u M e t G I U I 1 e A s p V a l I l e M e t S e r G l y T y r T h r 4 3 3 A C C A A C A G A G C T G A G A T G G A G A T T O A T G T A A T A A T G T C A G G T T A T A C G 1 6 1 H i s L e u G l n A r g A l a G l n P r o V a l A r g T r p S e r H i s P h e L e u M e t S e r 4 8 1 C A T T T A C A A A G O G C T C A G C C T G T T C G T T G G T C C C A T T T T C T C A T G T C T 1 7 7 H i s A l a L e u P r o L e u L e u G l y A s p L e u G l y A r g L e u A r g G l n L e u T y r 5 2 9 C A C G C C T T G C C T T T A T T A G G T G A C C T T G G C A G A C T T C G T C A G C T G T A T 1 9 3 T h r A r g V a l S e r G l n L e u T h r A l a G l y A l a G l y A l a L e u A l a G l y L y s 5 7 7 A C T C O T G T A A G T C A A C T T A C C G C T G G T O C T G G T G C T T T A G C T G G C A A A 2 0 9 P r o phe A s n V a l A s p A r g G l u P h e L e u P r o L y s G I u L e u G l y P h e G l u 6 2 5 C C T T T C A A C G T C G A T C G C G A G T T C C T T C C T A A A G A G C T T G G A T T C G A A 2 2 5 G l y I l e I l e M e t A S h S e r M e t A S h A l a V a l G l y A s p A r g A s p P h e V a l 6 7 3 G G C A T T A T C A T G A A T T C C A T G A A T G C T G T T G G T G A T C O T O A T T T T G T C 2 4 1 I l e G l u P h e M e t P h e T r p A l a G l y M e t V a l M e t L e u H i s I l e s e r A r g 7 2 1 A T C G A A T T T A T G T T T T G G G C A G G C A T G G T A A T G C T T C A C A T T T C T C G C 2 5 7 P h e A l a G I u A s p L e u I l e I l e T y r S e r S e t S e t G I u P h e G l y P h e V a l 7 6 9 T T T O C T G A A O A T C T T A T C A T A T A T T C G A G C T C G G A A T T T G G A T T C G T C 2 7 3 T h r L e U S e r A s p A l a T y r S e t T h r G l y S e t S e r I l e M e t P r o G l n L y s 8 1 7 A C A C T C T C C G A T G C G T A T T C T A C G G G A A G T A G T A T T A T G C C C C A A A A A 2 8 9 L y s A S h P r o A s p S e r L e u G I u L e u L e u A r g G l y L y s S e r G l y A r g V a l 8 6 5 A A G A A C C C T G A T T C T T T A G A G C T A C T T C G G G G T A A G A G C G G T C G T G T T 3 0 5 L e u G l y A s p M e t I l e G l y L e u M e t I l e T h r V a l L y s G l y T h r P r o T h r 9 1 3 T T A G G T G A T A T G A T T G G C C T C A T G A T A A C T G T T A A A G G C A C A C C T A C A 3 2 1 T h r T y r A S h L y s A s p L e u G l n G l u A s p L y s G I u P r o L e u P h e A s p A l a 9 6 1 A C C T A T A A C A A A G A T T T G C A A G A A G A C A A G G A A C C A C T A T T T G A T G C C 3 3 7 P h e L y s T h r V a l S e r A s p S e r L e u G l n I l e L e u T h r G l y V a l V a l S e r 1 O O 9 T T T A A G A C C G T C T C T G A C T C T T T G C A A A T T T T G A C T G G C G T T G T C T C A 3 5 3 T h r L e u T h r I 1 e A s n P r o T h r L y s I l e A l a G l u S e r L e u T h r P r o A s p 1 0 5 7 A C C C T T A C C A T C A A T C C T A C A A A G A T T G C C G A A A G C T T G A C C C C C G A T 3 6 9 L e u L e u A l a S e r T h r A s p L e u A l a G I u T y r L e u V a l A r g L y s G 1 y L e u 1 1 0 5 T T A C T A G C T A G C A C T G A T T T G O C T G A G T A T C T T G T T C G T A A A G G T C T T 3 8 5 P r o P h e A r g G l n T h r H i s H i s I l e S e r ~ A 1 a V a l A r g M e t A l a 1 1 5 3 C C A T T T C G C C A A A C T C A T C A T A T T T C G ~GGA T C C I G C A G T T C G C A T G G C T 4 0 1 G l u G l u A r g A s n T h r T h r L e u A s p L y s L e u S e r V a l S e t A s p L e u G l n 1 2 0 1 G A A G A G A G A A A C A C T A C T C T C G A C A A G T T A T C A G T T T C T G A T T T G C A A 2 2 4 6 7 2 2 4 0 7 2 0 2 5 6 7 6 8 2 7 2 8 1 6 2 8 8 8 6 4 3 0 4 9 1 2 3 2 0 9 6 0 3 3 6 1 0 0 8 3 5 2 1 0 5 6 3 6 8 1 1 0 4 3 8 4 1 1 5 2 4 O O 1 2 0 0 4 1 6 1 2 4 8 4 1 7 S e r L e u H i s P r o L e u Phe A s p G I u A s p V a l S e r L y s V a l P h e A s n T y r 4 3 2 1 2 4 9 T C A T T G C A T C C T T T A T T T G A T G A A G A C G T T T C A A A A G T C T T T A A T T A C 1 2 9 6 4 3 3 G I u G I u S e r V a l G l u L y s A r g C y s S e r I l e G l y G l y T h r A l a L y s H i s 4 4 8 1 2 9 7 G A A G A A A G T G T T G A A A A A A G A T O T T C A A T T G G T G G T A C T G C T A A G C A T 1 3 4 4 4 4 9 C y s V a l G l n A s p A s n A r g A l a T y r T h r I l e S e r A S h S e r * * * 4 8 1 1 3 4 5 T G T G T T C A A G A C A A T C G A G C A T A T A C G A T C A G C A A T T C T T A A A C T G T C 1 4 9 2
Fig. 2. Nucleotide and deduced amino acid sequences of the A R G 7
gene. The nucleotide sequence is numbered from the first nucleotide of the presumed initiation codon. At the 5' end, the solid underline indicates the putative "TATA" box. Asterisks at - 11 and - 10 indicate the major transcriptional start points. At the 3' end, the
solid underline shows the deleted amino acids in pULG-SP3
fragment, more precisely between the EcoRV and ScaI sites (data not shown). This observation suggested that the expression of ARG7 in E. coli results from the inser- tion near the 5' end of the gene of a bacterial IS element bearing a prokaryotic promoter. In this case, and consid- ering that the S. cerevisiae ARG4 (1389 bp) and the
S. pombe ARG7 genes probably have similar sizes, the
origin of transcription should be localized between ScaI and EcoRV and the direction of transcription should be from ScaI to EcoRV.
Nucleotide sequence of ARG7
1 6 4 8
3 2
96 The complete nucleotide sequence of ARG7 is shown in 1,'] Fig. 2. An open reading frame of 1383 bp starts at an
64 A T G codon located 193 bp downstream from the S c a I
1 9 2
site and terminates at a TAA codon downstream from the
8 0
240 BamHI site. This sequence encodes a polypeptide of 461
96 amino acid residues. The translation initiation codon is
2 8 8
the only one (see other ATG codons in the same reading
1 1 2
9 3 6 frame at positions + 6 4 and + 1 0 3 ) for which the sur-
128 rounding region (ACTATGG) is in g o o d agreement with
3 8 4
the S. c e r e v i s i a e A / G X X A T G G consensus (Beacham
1 4 4
4 3 2 et al. 1984). This choice seems also to be correct on the
16o basis of amino acid comparisons with other eukaryotic
4 8 0
176 genes coding for argininosuccinate lyase (see Fig. 3). The 528 first stop codon (TAA) is found at position 1384. The 876182 predicted molecular weight for the polypeptide chain is 208 5 1 2 0 0 Da. Within the 193 bp of the 5' untranslated re-
6 2 4
gion, the percentage of A + T is 69% against 59% in the coding region. The high A + T content is c o m m o n to a number of S. cerevisiae (Beacham etal. 1984) and
S. pombe (Kikuchi et al. 1988; Szankosi et al. 1988; Rus-
sel and Hall 1982; Mc Leod et al. 1987) promoters. N o t e in particular a 19 bp region ( - 1 4 to - 3 2 ) containing 18 (dA + dT) residues9 A perfect TATA box (TATATAA) (Breatnach and Chambon 1981) is seen at position - 7 2 in the ARG7 sequence. The bias in codon usage is relatively low: only two (CCG, G G G ) of the 61 amino acid coding triplets are not used at all and only small differences are found in the usage patterns between the two yeast species.
Amino acid sequence comparison
The complete or partial amino acid sequences of ASL and 6-crystallins are aligned in Fig. 3. Only short gaps were introduced to obtain a consensus sequence with 168 identities (36%) and six highly analogous stretches (boxed in Fig. 3). There is a remarkable lack of similarity in the carboxy-terminal region of the protein where only 16 amino acids are shared from residues 388 to 461. This part of the protein most probably plays a minor role in catalysis. This hypothesis is confirmed by the fact that pULG-SP3, in which the ASL gene is interrupted at the
BamHI site (Fig. 2), complements a S. cerevisiae arg4
mutant.
The genetic distances between four aligned sequences have been determined (Table ~). A value approaching 1 reflects a close relationship at the evolutionary level. It can be seen that the two higher eukaryotes are very close to each other (0.8937) whereas the two yeast species are both far from each other (0.5466) and from rats and humans.
Hydrophobicity, charge plots and secondary structure predictions comparing S. cerevisiae and S. pombe en- zymes show that the two proteins are very similar over most of their sequence (data not shown).
258 1 S. p o m b e . M A E K S S K K L S. c e r e v i s i a e . M S D . G T Q K L R a t . M A S E . S G K L M a n . M A S E . S G K L C h i c k e n C d l . M A T E . G D K L C h i c k e n C d 2 . M A S E . G D K L C. r e i n h a r d t i i A A P A D N T K K L E . c o l i .MA ... L B e e f C o n s e n s u s -M . . . K L 5O S. p o m b e L C Q K N V I S E E S. c e r e v i s i a e L Q K L G L L T E T R a t L E K A G L L T K A M a n L E K A G L L T K A C h i c k e n C d l L E K A S I L T K T C h i c k e n C d 2 L E K A G I L T K T C. r e i n h a r d t i i E . c o l i L V T V G V L B e e f L E K A G L L T K C o n s e n s u s L . . . i O O S. p o m b e I I G , D V A G K L S. c e r e v i s i a e L I G R E I A G K V R a t L I G . E A A G K L M a n L I G . A T A G K L C h i c k e n C d l L I G . D I A G K L 49 W G G R F S G A T D p L M A E F N K S I Y S G K E M C E E D V I G S M A Y A K A W G G R F T G E T D P L M H L Y N A S L P Y D Y K M Y K A D L E G T K V Y T A G W G G R F A G S V D P T M D K F N S S I A Y D R H L W N V D L Q G S K A Y S R G W G G R F V G A V D P I M E K F N A S I A Y D R H L W E V D V Q G S K A Y S R G L G G R F V G S T D P I M E I L S S S I S T E Q R L T E V D I Q A S M A Y A K A W G G R F S G S T D P I M E M L N S S I A C D Q R L S E V D I Q G S M A Y A K A W G G R F T A K T D p L M E K F N E S L P F D K R L W A E D I K W G G R F T Q A A D Q R F K Q F N D S L R F D Y R L A E Q D I V G S V A W S K A G - G G R F . . . . D . . . S . . . D . . . 99 E L N S I L E G L E Q I Q R E W N S G Q F V L E P S D E D V H T A N E R R L T E E L A K I H E G L A E I K K E W D A D K F V R H P N D E D I H T A N E R R L G E E M Q Q I L Q G L D K V A E E W A Q G I F K L Y P N D E D I H T A N E R R L K E E M D Q I L H G L D K V A E E W A Q G T F K L N S N D E D I H T A N E R R L K E E L E K I L S G L E K I S E E S S K G V L V M T Q S D E D I Q T A I E R R L K E E L E K I L S G L E K I S E E W S K G V F V V K Q S D E D I H T A N E R R L K E V A E E W K A G A F V I N A G D E D I H T A N E R R L T E E - - - I - - G L . . . E . . . ~ D E D - H T A - E R R L - E l 146 H T G R S R N D Q V T T D L R L W L C R K I K E V E V Y . . V I N L L K V F T N H T G R S R N D Q V V T D L R I Y . C R D I V N D T L F P A L K G L V E V L I K H T G R S R N D Q V V T D L R L W M R Q T Y S K L S T F . . L K V L I E A M V D H T G R S R N D Q V V T D L R L W M R Q T C S T L S G . . L L W E L I R T M V D Q T G R S R N E Q V V T D L K L L L K S S I S V I S T H . . L L Q L I K T L V E C h i c k e n C d 2 L I G . D I A G K L H T G R S R N D Q V V T D L K L L L K S S I S V I S T H . . L L Q L I K T L V E C. r e i n h a r d t i i L V G . A V G G K L H T G R S R N D Q . . . C o n s e n s u s - - G - - - A G K - H T G R S R N - Q V - T D L ~ . . . L . . . 147 196 S. p o m b e R A E M E I D V I M S G Y T H L Q R A Q p V R W S H F L M S H A L P L L G D L G R L R Q L Y T R V S S. c e r e v i s i a e R A E G E I D V L M P G Y T H L Q R A Q P I R W S H W L S S Y A T Y F T E D Y K R L G Q I L H R L N R a t R A E A E C E V L F P G Y T H L Q R A Q P I R W S H W I L S H A V A L T R D L E R L K E V Q K R I N M a n R A E A E R D V L F P G Y T H L Q R A Q P I R W S H W I L S H A V A L T R D S E R L L E V R K R I N C h i c k e n C d l R A A I E I D I I M P G Y T H L Q K A L P I R W S Q F L L S H A V A L T R D S E R L G E V K K R I T C h i c k e n C d 2 R A A T E I D V I M P G Y T H L Q K A L P I R W S Q F L L S H A V A L I R D S E R L G E V K K R M S C. r e i n h a r d t i i I - - ] C . . . R A - - E . . . ~ G Y T H L Q - A - P - R W S H + - - S - A . . . D - - R L . . . R - - 1 197 246 S. p o m b e Q L T A G A G A L A G K P F N V D P ~ F L P K E L G F E G I I M N S M N A V G D R D F V I E F M F W S. c e r e v i s i a e Q S P L G A G A L A G H P Y G I D R E F L A E G L G F N S V I G N S L V A V S D R D F I V E L M F W R a t V L P L G S G A I A G N P L G V D R E F L C A E L N F G A I T L N S M D A T S E R D F V A E F L F W M a n V L P L G S G A I A G N P L G V D R E L L R A E L N F G A I T L N S M D A T S E R D F V A E F L F W C h i c k e n C d l V L P L G S G A L A G N P L E I D R E L L R S E L D M T S I T L N S I D A I S E R D F V V E L I S V C h i c k e n C d 2 V L P L G S G A L A G N P L E I D R E L L R S E L D F A S I S L N S M D A I S E R D F W E L L S V C. r e i n h a r d t i i V S D R E V V I E T V F A C o n s e n s u s . . . . G - G A - A G - P - - - D R E - L - - - L . . . N S - - A - - - R - F - - E . . . . 247 2 9 6 S. p o m b e A G M V M L H I S R F A E D L I I Y S S S E F G F V T L S D A Y S T G S S I M P Q K K N P D S L E L S. c e r e v i s i a e G T L F M N H I S R F A E D L I I Y C T A E F G F I Q L S D A Y S T G S S L M p Q K K N A D S L E L R a t A S L C M T H L S R M A E D L I L Y G T K E F N F V Q L S D A Y S T G S S L M P Q K K N P D S L E L M a n R S L C M T H L S R M A E D L I L Y C T K E F S F V Q L S D A Y S T G S S L M p Q K K N P D S L E L C h i c k e n C d l A T L L M I H L S K L A E D L I I F S T T E F G F V T L S D A Y S T G S S L L P Q K K N P D S L E L C h i c k e n C d 2 A T L L M I H L S K L A E D L I I F S T T E F G F V T L S D A Y S T G S S L L P Q K K N P D S L E L C. r e i n h a r d t i i A S L L C V H L S R W A E D L I I Y S S G P F G Y V Q C S D A Y A T G S S L M P Q K K N P D A L E L C o n s e n s u s . . . . M + H - S - - A E D L I . . . F . . . S D A Y S T G S S - - P Q K K N P D - L E L 1 297 346 S. p o m b e L R G K S G R V L G D M I G L M I T V K G T P T T Y N K D L Q E D K E P L F D A F K T V S D S L Q I S. c e r e v i s i a e L R G K S G R V F G D L T G F L M S L K G I P S T Y D K D M Q E D K E P L F D C L T T V E H S M L I R a t I R S K A R R V F G R C A G L L M T L K G L P S T Y N K D L Q E D K E A V F E V S D T M T A V L Q V M a n I R S K A G R V F G R C A G L L M T L K G L P S T Y N K D L Q E D K E A V F E V S D T M S A V L Q V C h i c k e n C d l I R S K A G R V F G R L A A I L M V L K G I P S T F S K D L Q E D K E A V L D V V D T L T A V L Q V C h i c k e n C d 2 I R S K A G R V F G R L A A V L M V L K G L P S T Y N K D L Q E D K E A V F D V V D T L T A V L Q V C. r e i n h a r d t i i I C . . . - R - K - - R V - G ] . . . I K G - P . . . . K D - Q E D K E } T 347 3 9 6 S. p o m b e L T G V V S T L T I N P T K I A E S L T P D L L A S T D L A E Y L V R K G L P F R Q T H H I S G S A S. c e r e v i s i a e A T G V I S T L T V N K E K M E A A L T M D M L A . T D L A D Y L V R K G V P F R E T H H I S G E C R a t A T G V I S T L Q I H R E N M A Q A L S P D M L A . T D L A Y Y L V R K G M P F R Q A H E A S G K A M a n A T G V I S T L Q I H Q E N M G Q A L S P D M L A . T D L A u R Q A H E A S G K A C h i c k e n C d l A T G V I S T L Q V N K E N M E K A L T P E L L S . T D L A L Y L V R K G M P I R Q A Q T A S G K A C h i c k e n C d 2 A T G V I S T L Q V N K E N M E K A L T P E L L S . T D L A L Y L V R K G M P F R Q A H V A S G K A C. r e i n h a r d t i i L S A D M L A . T D L A E Y L V R K G V P F R E T H H H C . . . ~ . . . L- - - ~ L - - T D L A - Y L V R K G - P - R - - H ~ - S G - - 397 445 S. p o m b e V R M A E E R N . T T L D K L S V S D L Q S L H P L F D E D V S K V F N Y E E S V E K R C S I G G T S. c e r e v i s i a e V A T A E R L G L S G I D K L T L E Q Y Q K I D S R F G Q D L F E T F N F E Q S V E R R D A T G G T R a t V V V A E M K G V . A L N Q L S L Q E L Q T V S P L F S S D V N L V W D Y S H S V E Q Y T A L G G T M a n V F M A E T K G V . A L N Q L S L Q E L Q T I S P L F S G D V I C V W D Y G H S V E Q Y G A L G G T C h i c k e n Cdl V H L A E T K G I . T I N N L T L E D L K S I S P L F A S D V S Q V F S W N S V E Q Y T A V G G T C h i c k e n C d 2 V H L A E T K G I . A I N K L T L E D L K S I S P L F A S D V S Q V F N I V N S V E Q Y T A V G G T C. r e i n h ~ r d t i i V W D F N R S A E M R D T E G G T C o n s e n s u s V - - A E . . . L . . . F - - D 446 461 S. p o m b e A K H C V Q D N R A Y T I S N S . . . S. c e r e v i s i a e A K S A V L K Q L D N L K S Q L N . . . R a t A Q S S V E W Q ~ S Q V R A L L Q M . . . M a n A R S S V D W Q I R Q V R A L L Q A Q Q A.. C h i c k e n C d l A K S S V T A Q I E Q L R E L L K K Q K E Q A C h i c k e n C d 2 A K S S V T A Q I E Q L R E L L K K Q K E Q A C. r e i n h a r d t i i S K R S V L E Q V Q K M R T Y L A A E G QH. C o n s e n s u s A - - - V . . . . . . S -E . . . G C T
Fig. 3. Alignments of the amino acid sequences of argininosucci- nate lyases and of chicken 6-1 and 6-2 crystallins. The human sequence is from Matuo et al. (1988). The numbering corresponds to the S. pombe sequence without gaps
Table 1. Comparison matrix of argininosuccinate lyases. The values were calculated on the basis of the "distances" algorithm and repre- sent the fractions of identical residues (see Materials and methods)
Species S. pombe S. cerevisiae Rat Man
S. pombe 1.0000 0.5466 0.5141 0.5184
S. cerevisiae 1.0000 0.5401 0.5335
Rat 1.0000 0.8937
Man 1.0000
pH rate profiles and chemical modifications of beef ASL indicate that a histidine and a carboxylate group are essential for catalytic activity (Garrard et al. 1985). On the other hand, experiments with site-directed agents on the same enzyme suggest that a lysine residue (lysine 52 in Fig. 3) plays a key role in the binding of argininosucci- nate (Lusty and Rather 1987). The position of this amino acid is not strictly conserved in S. pombe. In addition, no lysine residue is present in that region of the E. coli se- quence, and we thus propose that it is not essential for the binding of the substrate.
The introduction of chicken 6-crystallins in the com- parison does not greatly affect the consensus sequence. Delta-1 (CD 1) and delta-2 (CD2) crystallins are very sim- ilar proteins (91% identity) but only CD2 has retained ASL activity. The analysis of substitutions of the CD1 sequence versus the consensus sequence provides inter- esting information. Acidic residues are well conserved or else display conservative substitutions (asp~glu, g l u ~ a s p ) except in the case of glutamine 149 which is substituted by an alanine in both crystallins. Six histidine residues are conserved in the four ASL enzymes at posi- tions 90, 109, 161, 172, 253 and 390. Four of them are replaced by glutamine (at positions 90, 109, 172 and 390) in CDI but only histidine 390 is replaced by glutamine in fully active CD2. Accordingly, an essential difference be- tween the inactive CD1 and the fully active ASLs is the absence of the histidine residues 90, 109 and 390 in CDI, which confirms the role of histidine residues in catalytic activity.
Northern blot and primer extension analyses
The expression of the ARG7 gene at the transcriptional level was examined by Northern analysis. R N A s were extracted from S. cerevisiae UL1, harbouring plasmid pULG-SP2 (full gene) or pULG-SP3 (gene lacking the C-terminal part). Total R N A was electrophoresed, blot- ted and probed with 32p-labeled pULG-ID1. N o signal was visualized in the control (host strain UL1 without plasmid). A single hybridization band was detected in pULG-SP2 at a position (1 500 b) in agreement with the length of the gene. At least three transcripts of about 1 800, 2800 and 3900 b were observed in pULG-SP3 (Fig. 4). As the orientiation of the truncated gene in pULG-SP3 is opposite to the orientation of the gene in pULG-SP2 (Fig. 1), the transcripts must find their 3' ends in the 2 gm part of the plasmid.
The transcription initiation sites have been determined by primer extension using as a primer a 19 b oligonucle-
259 transcription initiation site is 61 bases, a value which is very close to that found in the S. cerevisiae A R G 4 gene (TATA box at -119, initiation site at -57) and which is thus in agreement with the location of the initiation "win- dow" in budding yeast (Furter-Graves and Hall 1990).
Fig. 4. Northern blots of RNA prepared from S. cerevisiae strain
UL1 without plasmid (lane 1) and with plasmids pULG-SP2
(lane 2) or pULG-SP3 (lane 3). The probe was [3zP]-labeled (nick- translation) pULG-ID1 (1.14 x 108 cpm/gg DNA). Autoradiogra- phy was performed at -70~ with intensifying screen for 24 h
Fig. 5. Initiation site determination of the ARG7 mRNAs by prim-
er extension and DNA sequence analysis; lane i, pULG-SP3; lane 2, pULG-SP2. In parallel, DNA sequencing of the SacI-EcoRI frag- ment (see Fig. 1) was performed by the method of Sanger using the same oligonucleotide as that used for primer extension
otide complementary to the region + 48 to + 66 from the first translation codon (Fig. 5). Extended products were run in parallel with the products of the dideoxy sequenc- ing of the non-transcribed strand (in the region ScaI-
EcoRI, Fig. 1) using the primer extension oligonucle- otide. In pULG-SP2, as well as in pULG-SP3, the main 5' terminus was found at position -11. Two other initia- tion sites were present at 10 and -6. Accordingly, the distance between the TATA box (-72) and the major
Discussion
9 The A R G 7 gene of Schizosaccharomyces pombe is natu- rally expressed in Saccharomyces cerevisiae (Remacle et al. 1988). In Escherichia coli, the gene is inactive but can be turned on by a short D N A sequence inserted close to its 5' end. The identity of this element has not been determined but could correspond to IS2 which is able to reactivate yeast genes originally silent in E. coli (Walz et al. 1978; Harashima et al. 1981) owing to the presence in this element of a bacterial promoter (Charlier et al. 1982). The fact that A R G 7 complements an E. coli mu- tant lacking argininosuccinate lyase indicates that this gene is devoid of introns. The analysis of the A R G 7 cod- ing sequence confirms this assumption.
In the 5' non-coding region of A R G 7 we have detected a perfect "TATA" box (Breatnach and Chambon 1981) located 61 bp upstream of the major transcription initia- tion site, a distance which is favourable to the expression of the gene in S. cerevisiae (Furter-Graves and Hall 1990). It should be stressed, however, that the initiation sites were determined in S. cerevisiae and that they do not necessarily correspond to those operating in S. pombe (Russel 1983). How A R G 7 is regulated in S. pombe is unknown and the upstream activator sequence 5'- TGACTC-3' involved in the general control of S. cere-
visiae A R G 4 gene has not been found in the short (193 bp) upstream sequence analyzed. Anyway, these ac- tivator sequences are dispensable and the deletions re- moving the A R G 4 UAS do not completely abolish the expression of this gene in S. cerevisiae (Thiry-Blaise and Loppes 1990).
The A R G 7 protein displays good similarity with other related proteins except in their carboxy-terminal part. In the course of this work, a deletion removing the 66 C-ter- minal amino acids was examined and shown to retain some enzymatic activity, which indicates that this part of the protein is not directly involved in catalysis. Such a situation has been described, for example, in the S. pombe
mei3 gene (Mc Leod et al. 1987) where a deletion remov- ing 20 C-terminal amino acids of the protein (148 amino acids) retains full mei3 activity. In the A R G 7 truncated gene present in pULG-SP3 (Fig. 1), the protein is synthe- sized up to serine 393. Translation should go on briefly through pBR322 in the direction BamHI to HindIII and stop (TGA) after incorporation of seven amino acids.
The signal for termination of the wild-type A R G 7 gene is unknown but it should not be very far from the stop codon at position + 1387 since the m R N A transcript is about 1500 bases long. The truncated gene in pULG-SP3 gives rise to at least three transcripts of higher molecular weight which should terminate at various points of the 2 ~tm fragment. It will probably be difficult to identify the corresponding signals since no clear consensus for termi-
260
n a t i o n o f t r a n s c r i p t i o n in y e a s t has b e e n e s t a b l i s h e d so far ( Z a r e t a n d S h e r m a n 1982; H e n i k o f f et al. 1983).
F i n a l l y , it is i n t e r e s t i n g to n o t e t h a t , o n the basis o f the genes c o d i n g for a r g i n i n o s u c c i n a t e lyase,
S. pombe
d o e s n o t a p p e a r to be m o r e closely r e l a t e d toS. cerevisiae
t h a n to m a m m a l s a n d t h a t , in c o n t r a s t , fission a n d b u d d i n g y e a s t s are e q u a l l y d i s t a n t f r o m these even if s e v e r a l fea- tures d e s c r i b e d in the l i t e r a t u r e ( K / i u f e r et al. 1985; T o d a et al. 1984) s u p p o r t the h y p o t h e s i s t h a tS. pombe
is m o r e closely r e l a t e d to h i g h e r e u k a r y o t e s t h a n isS. cerevisiae.
Acknowledgements.
This work was supported by a grant from the Belgian ER.F.C. (2.4521.89). The authors are grateful to Dr. J.-M. Fr~re for revising the manuscript and to F. Hayet for expert typing. R.L. and B.J. are Research Associates of the National Foundation for Scientific Research (Belgium).References
Amaya Y, Matsubasa T, Takiguchi M, Kobayashi K, Saheki T, Kawamoto S, Mori M (1988) J Biochem 103:177-181 Beacham IR, Schweitzer BW, Warrick HM, Carbon J (1984) Gene
29:271-279
Biggin MD, Gibson TJ, Hong GF (1983) Proc Natl Acad Sci USA 80:3963 3965
Breathnach R, Chambon P (1981) Annu Rev Biochem 50:349-383 Charlier D, Piette J, Glansdorff N (1982) Nucleic Acids Res
10:5935 5948
Dayhoff MO (1979) Atlas of protein sequence and structure. Vol 5, suppl 3, National Biochemical Research Foundation, Silver Spring, Md., USA
Debuchy R, Purton S, Rochaix JD (1989) EMBO J 8:2803-2809 Delforge J, Messenguy IV, Wiame JM (1975) Eur J Biochem 57:231 -
239
Devereux J, Haeberli P, Smithies O (1984) Nucleic Acids Res 12:387-395
Farrell K, Overton S (1987) Biochem J 242:261-266
Furter-Graves E, Hall BD (1990) Mol Gen Genet 223:407-416 Garrard LJ, Bui QTN, Nygaard R, Raushel FM (1985) J Biol Chem
260:5548-5553
Gribskov M, Burgess RR (1986) Nucleic Acids Res 14:6745-6763 Harashima S, Sidhu RS, Toh-e A, Oshima Y (1981) Gene 16: 335-
341
Henikoff S, Kelly JD, Cohen EH (1983) Cell 33:607-614 K/iufer NF, Simanis V, Nurse P (1985) Nature 318:78 80 Kikuchi Y, Katazawa Y, Shimatake H, Yamamoto M (1988) Curr
Genet 14:375-379
Lusty CJ, Ratner S (1972) J Biol Chem 247:7010-7022
Lusty C J, Rather S (1987) Proc Natl Acad Sci USA 84:3176-3180 Matuo S, Tatsuno M, Kobayashi K, Saheki T, Miyata T, Iwanaga
S, Amaya Y, Mori M (1988) FEBS Lett 234:395-399 Mc Leod M, Stein M, Beach D (1987) EMBO J 6:729-736 Messenguy F, Dubois E (1983) Mol Gen Genet 189:148-156 Nicolet CM, Chenevert JM, Friedberg EC (1985) Gene 36:225-234 O'Brien WE, Barr R (1981) Biochemistry 20:2056-2060
O'Brien WE, Mc Innes R, Kalumuck K, Adcock M (1986) Proc Natl Acad Sci USA 83:7211-7215
Parsot C, Boyen A, Cohen GN, GlansdorffN (1988) Gene 68: 275- 283
Piatigorsky J, O'Brien WE, Norman BL, Kalumuck K, Wistow G J, Borras T, Nickerson JM, Wawrousek EF (1988) Proc Natl Acad Sci USA 85:3479-3483
Remacle JE, Breyer D, Loppes R (1988) Curr Genet 14:381 385 Rigby PWJ, Dieckmann M, Rhodes C, Berg P (1977) J Mol Biol
113:237-251
Russell PR (1983) Nature 301:167-169
Russell PR, Hall BD (1982) Molec Cell Biol 2:106-116
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Sanger F, Nicklen S, Coulson AR (1977) Proc Natl Acad Sci USA 74:5463 5467
Schweitzer BW (1982) Ph.D Thesis, University of California, Santa Barbara, Calif., USA
Struhl K (1985) Proc Natl Acad Sci USA 82:8419-8423
Szankosi P, Heyer WD, Schuchert P, Kohli J (1988) J Mol Biol 204:917-925
Thiry-Blaise LM, Loppes R (1990) Mol Gen Genet 223:474-480 Toda T, Adachi Y, Hiraoka Y, Yanagida M (1984) Cell 37:233-242 Vrati S, Mann DA, Reed KC (1987) Mol Biol Reports (Bio-Rad
Laboratories) 1, 3 : 1 - 4
Walz A, Ratzkin B, Carbon J (1978) Proc Natl Acad Sci USA 75:6172 6176
Zaret K, Sherman F (1982) Cell 28:563 573
Zoller MJ, Smith M (1982) Nucleic Acids Res 10:6487-6500